CRISPR/Cas9 Gene Editing of NtAITRs, a Family of Transcription Repressor Genes, Leads to Enhanced Drought Tolerance in Tobacco
Abstract
1. Introduction
2. Results
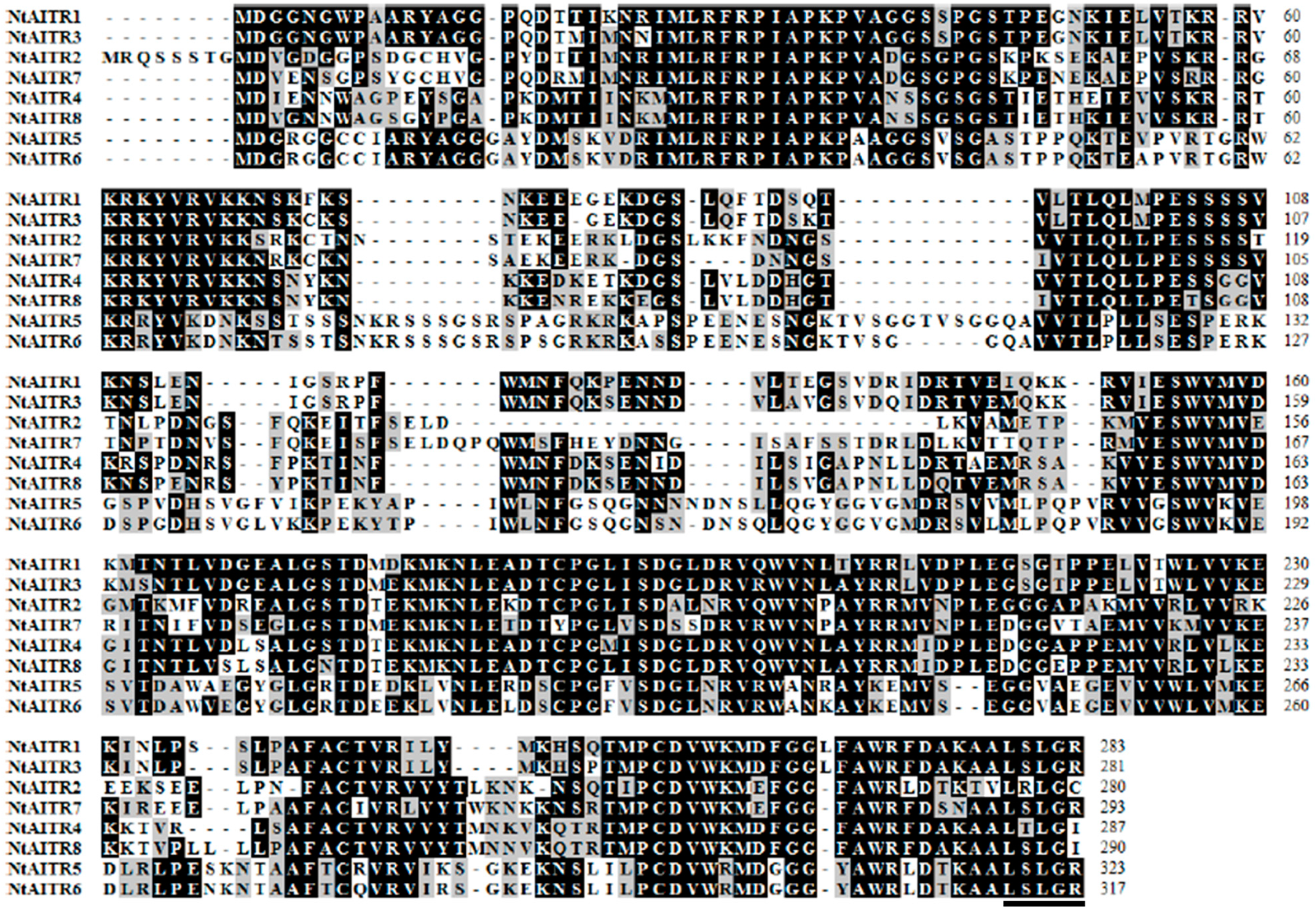
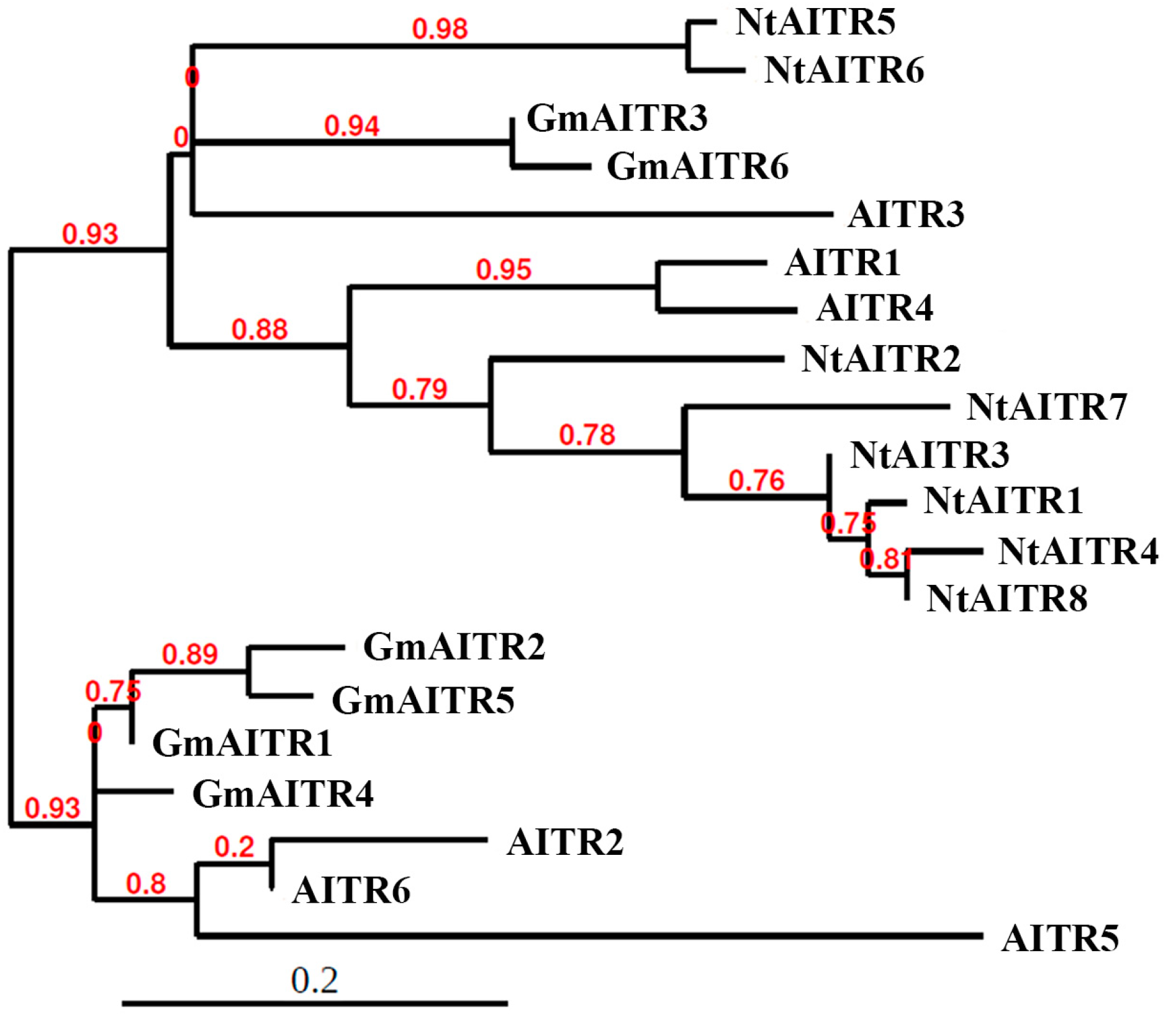
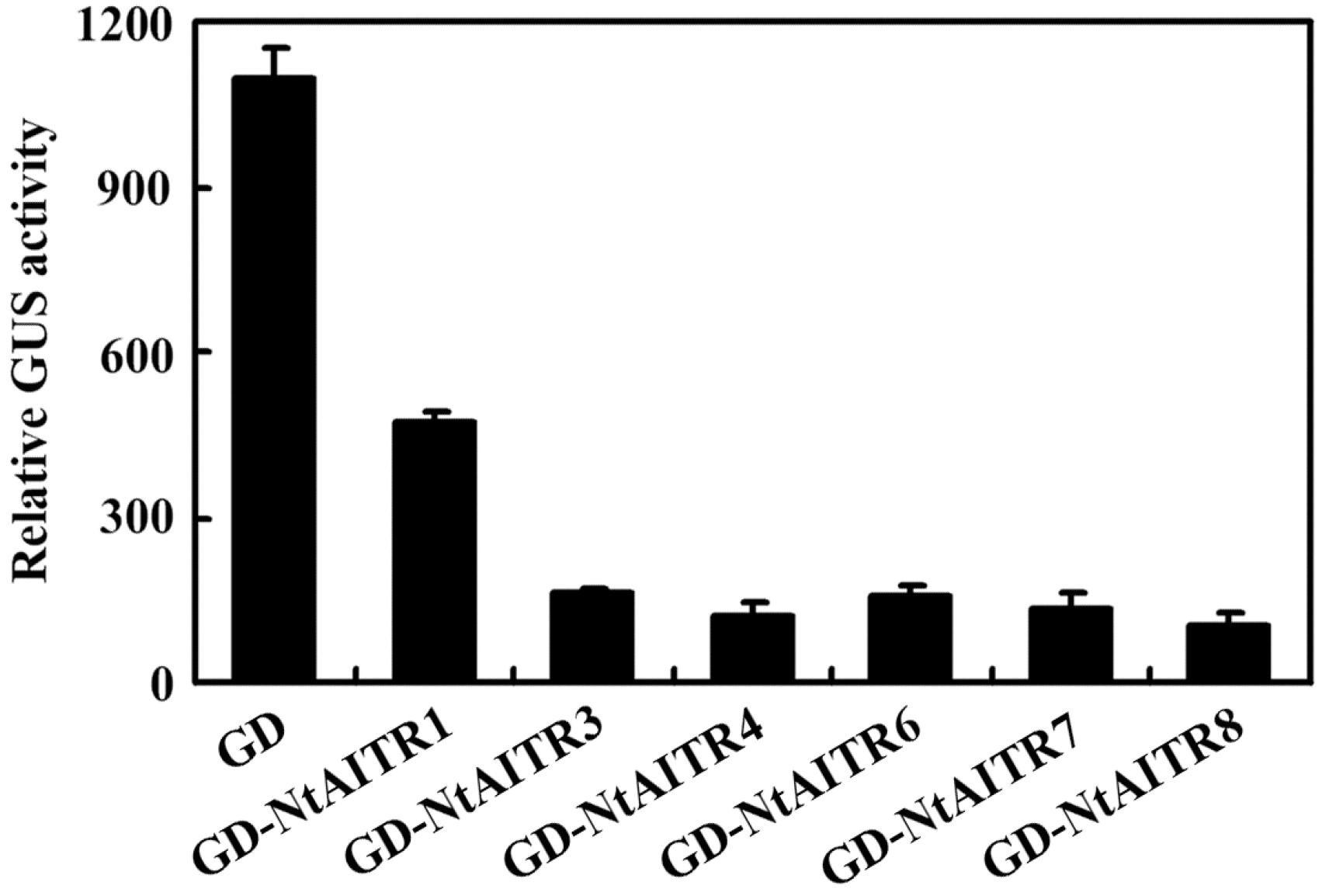
2.1. Identification of AITRs in Tobacco
2.2. Expression Levels of NtAITRs Are Increased in Response to ABA and Drought Treatments
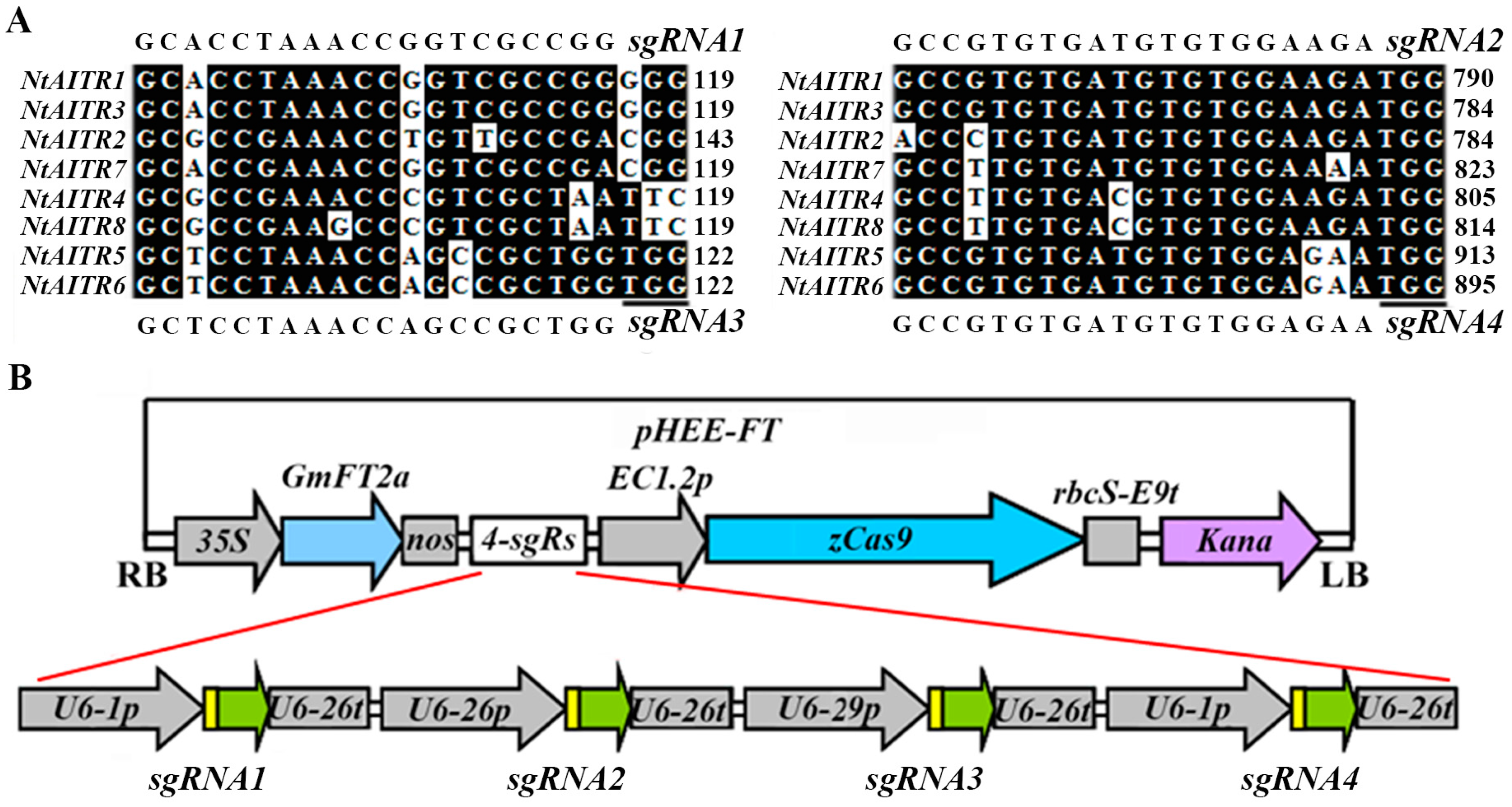
2.3. Generation of Transgene-Free Ntaitr Mutants
2.4. The ntaitr12356 Mutant Plants Show Enhanced Tolerance to Drought
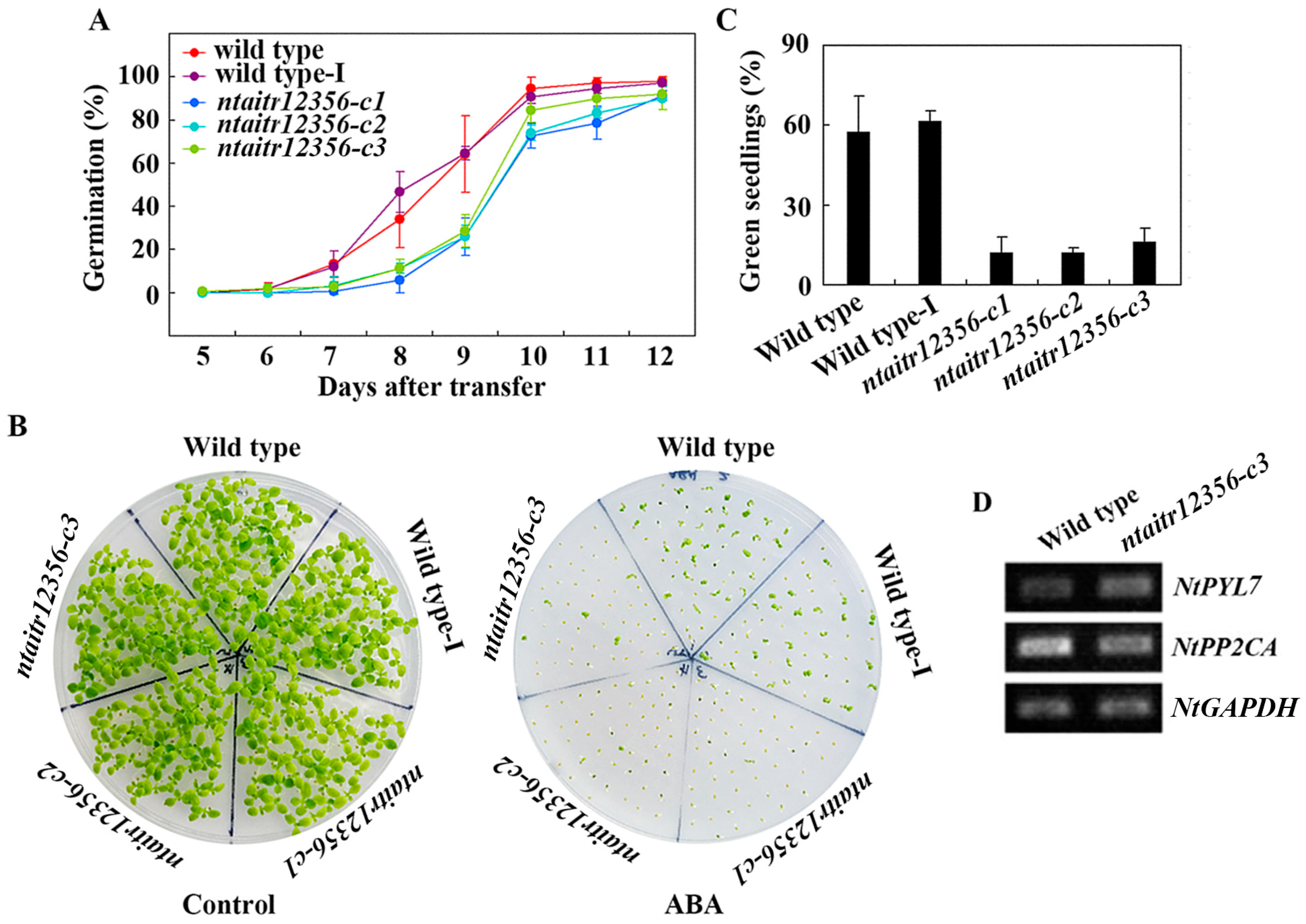
2.5. The ntatir12356 Mutants Are Hypersensitive to ABA
3. Discussion
4. Materials and Methods
4.1. Identification of AITRs in Tobacco
4.2. Plant Materials and Growth Conditions
4.3. ABA and Drought Treatment, RNA Isolation and RT-PCR
4.4. Constructs
4.5. Plasmid Isolation, Protoplast Isolation and Transient Transfection
4.6. Generation of Transgene-Free ntaitr Mutants
4.7. Seed Germination and Seedling Greening Assays
4.8. Drought Tolerance Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Xu, J. Abiotic stress responses in plant roots: A proteomics perspective. Front. Plant Sci. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Rodriguez, P.L.; Leube, M.P.; Grill, E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol. Biol. 1998, 38, 879–883. [Google Scholar] [CrossRef]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.; Vartanian, N.; Giraudat, J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999, 11, 1897–1910. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- Guo, J.; Yang, X.; Weston, D.J.; Chen, J.G. Abscisic acid receptors: Past, present and future. J. Integr. Plant Biol. 2011, 53, 469–479. [Google Scholar] [CrossRef]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 2017, 40, 2958–2971. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Boil. 2021, 21, 137. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Peterson, F.C.; Mosquna, A.; Yao, J.; Volkman, B.F.; Cutler, S.R. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 2015, 520, 545–548. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signaling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Muñoz-Bertomeu, J.; Ibañez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014, 80, 1057–1071. [Google Scholar] [CrossRef]
- Seo, K.I.; Lee, J.H.; Nezames, C.D.; Zhong, S.; Song, E.; Byun, M.O.; Deng, X.W. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 2014, 26, 695–711. [Google Scholar] [CrossRef]
- Yu, F.; Lou, L.; Tian, M.; Li, Q.; Ding, Y.; Cao, X.; Wu, Y.; Belda-Palazon, B.; Rodriguez, P.L.; Yang, S.; et al. ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol. Plant. 2016, 9, 1570–1582. [Google Scholar] [CrossRef]
- Belda-Palazon, B.; Rodriguez, L.; Fernandez, M.A.; Castillo, M.C.; Anderson, E.A.; Gao, C.; Gonzalez-Guzman, M.; Peirats-Llobet, M.; Zhao, Q.; De Winne, N.; et al. FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 2016, 28, 2291–2311. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Liu, H.X.; Stone, S.; Callis, J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013, 75, 965–976. [Google Scholar] [CrossRef]
- Liu, H.; Stone, S.L. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 2010, 22, 2630–2641. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Stone, S.L. Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 2013, 288, 20267–20279. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Liu, S.; Hu, Q.; Luo, S.; Li, Q.; Yang, X.; Wang, X.; Wang, S. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 2015, 6, 388. [Google Scholar] [CrossRef]
- Huang, Y.; Feng, C.Z.; Ye, Q.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genetics 2016, 12, e1005833. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, W.; Wang, Y.; Zhou, G.; Liu, S.; Li, D.; Adnan; Hussain, S.; Ahmed, S.; Zhang, C.; et al. AIW1 and AIW2, two ABA-induced WD40 repeat-containing transcription repressors function redundantly to regulate ABA and salt responses in Arabidopsis. J. Plant Interact. 2020, 15, 196–206. [Google Scholar] [CrossRef]
- Cheng, Y.; Ma, Y.; Zhang, N.; Lin, R.; Yuan, Y.; Tian, H.; Hussain, S.; Chen, S.; Yang, W.; Li, Y.; et al. The R2R3 MYB transcription factor MYB71 regulates abscisic acid response in Arabidopsis. Plants 2022, 11, 1369. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Dong, Q.; Wang, W.; Chen, S.; Cheng, Y.; Tian, H.; Li, X.; Hussain, S.; Wang, L.; Gong, L.; et al. Evolution of AITR family genes in cotton and their functions in abiotic stress tolerance. Plant Biol. 2021, 23 (Suppl. 1), 58–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xun, H.; Wang, W.; Ding, X.; Tian, H.; Hussain, S.; Dong, Q.; Li, Y.; Cheng, Y.; Wang, C.; et al. Mutation of GmAITR genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front. Plant Sci. 2021, 12, 779598. [Google Scholar] [CrossRef] [PubMed]
- Andrianov, V.; Borisjuk, N.; Pogrebnyak, N.; Brinker, A.; Dixon, J.; Spitsin, S.; Flynn, J.; Matyszczuk, P.; Andryszak, K.; Laurelli, M.; et al. Tobacco as a production platform for biofuel: Overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 2010, 8, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Barla, F.G.; Kumar, S. Tobacco biomass as a source of advanced biofuels. Biofuels 2016, 10, 335–346. [Google Scholar] [CrossRef]
- de las Mercedes Dana, M.; Pintor-Toro, J.A.; Cubero, B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006, 142, 722–730. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, Y.; Han, L.; Guan, Z.; Chai, T. A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep. 2008, 27, 795–803. [Google Scholar] [CrossRef]
- Yang, C.; Wang, R.; Gou, L.; Si, Y.; Guan, Q. Overexpression of Populus trichocarpa mitogen-activated protein kinase kinase4 enhances salt tolerance in tobacco. Int. J. Mol. Sci. 2017, 18, 2090. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, J.; Duan, W.; Ma, X.; Qu, L.; Xu, Z.; Yang, Y.; Xu, J. NtRAV4 negatively regulates drought tolerance in Nicotiana tabacum by enhancing antioxidant capacity and defence system. Plant Cell Rep. 2022, 41, 1775–1788. [Google Scholar] [CrossRef]
- Sun, H.; Sun, X.; Wang, H.; Ma, X. Advances in salt tolerance molecular mechanism in tobacco plants. Hereditas 2020, 157, 5. [Google Scholar] [CrossRef]
- Xie, H.; Bai, G.; Lu, P.; Li, H.; Fei, M.; Xiao, B.; Chen, X.; Tong, Z.; Wang, Z.; Yang, D. Exogenous citric acid enhances drought tolerance in tobacco (Nicotiana tabacum). Plant Biol. 2022, 24, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, N.; Hussain, S.; Ahmed, S.; Wang, S. Integration of a FT expression cassette into CRISPR/Cas9 construct enables fast generation and easy identification of transgene-free mutants in Arabidopsis. PLoS ONE 2019, 14, e0218583. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Xie, H.; Yao, H.; Li, F.; Chen, X.; Zhang, Y.; Xiao, B.; Yang, J.; Li, Y.; Yang, D.H. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genomics 2019, 20, 575. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, S.; Wu, M.; Li, Z.; Liu, P.; Li, F.; Chen, Q.; Yang, A.; Yang, J. Evolutionary and functional analyses of the 2-oxoglutarate-dependent dioxygenase genes involved in the flavonoid biosynthesis pathway in tobacco. Planta 2019, 249, 543–561. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef]
- Wang, S.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Auxin response factor 7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 2005, 17, 1979–1993. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Hu, Q.; Dai, X.; Tian, H.; Zheng, K.; Wang, X.; Mao, T.; Chen, J.G.; Wang, S. Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 2015, 83, 300–311. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, L.; Zhang, W.; Cai, L.; Guo, H.; Tian, H.; Schiefelbein, J.; Wang, S. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2016, 39, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, J.; Yuan, C.; Guo, Y.; Yu, H.; Li, Y.; Huang, C. Cas9-PF, an early flowering and visual selection marker system, enhances the frequency of editing event occurrence and expedites the isolation of genome-edited and transgene-free plants. Plant Biotechnol. J. 2019, 17, 1191–1193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Han, Y.; Zhang, M.; Zhou, S.; Kong, X.; Wang, W.; Min, Y.Z. Overexpression of the wheat expansin gene TaEXPA2 improved seed production and drought tolerance in transgenic tobacco plants. PLoS ONE 2016, 11, e0153494. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Xu, Z.; Wei, Y.; Wang, M. Overexpression of the maize sulfite oxidase increases sulfate and GSH Levels and enhances drought tolerance in transgenic tobacco. Front. Plant Sci. 2018, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Q.; Guo, C.; Sun, J.; Li, Z.; Wang, Y.; Yang, A.; Pu, W.; Guo, Y.; Gao, J.; et al. NtNAC053, a novel NAC transcription factor, confers drought and salt tolerances in tobacco. Front. Plant Sci. 2022, 13, 817106. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Ma, Y.; Wang, X.; Cheng, N.; Meng, D.; Chen, S.; Wang, W.; Wang, X.; Hu, X.; Yan, L.; et al. CRISPR/Cas9 Gene Editing of NtAITRs, a Family of Transcription Repressor Genes, Leads to Enhanced Drought Tolerance in Tobacco. Int. J. Mol. Sci. 2022, 23, 15268. https://doi.org/10.3390/ijms232315268
Li G, Ma Y, Wang X, Cheng N, Meng D, Chen S, Wang W, Wang X, Hu X, Yan L, et al. CRISPR/Cas9 Gene Editing of NtAITRs, a Family of Transcription Repressor Genes, Leads to Enhanced Drought Tolerance in Tobacco. International Journal of Molecular Sciences. 2022; 23(23):15268. https://doi.org/10.3390/ijms232315268
Chicago/Turabian StyleLi, Guimin, Yanxing Ma, Xiaoping Wang, Nini Cheng, Deyu Meng, Siyu Chen, Wei Wang, Xutong Wang, Xiaojun Hu, Li Yan, and et al. 2022. "CRISPR/Cas9 Gene Editing of NtAITRs, a Family of Transcription Repressor Genes, Leads to Enhanced Drought Tolerance in Tobacco" International Journal of Molecular Sciences 23, no. 23: 15268. https://doi.org/10.3390/ijms232315268
APA StyleLi, G., Ma, Y., Wang, X., Cheng, N., Meng, D., Chen, S., Wang, W., Wang, X., Hu, X., Yan, L., & Wang, S. (2022). CRISPR/Cas9 Gene Editing of NtAITRs, a Family of Transcription Repressor Genes, Leads to Enhanced Drought Tolerance in Tobacco. International Journal of Molecular Sciences, 23(23), 15268. https://doi.org/10.3390/ijms232315268






