Detection of SARS-CoV-2 Virus by Triplex Enhanced Nucleic Acid Detection Assay (TENADA)
Abstract
1. Introduction
2. Results and Discussion
2.1. Design and Synthesis of PPRH and Reporter Probes
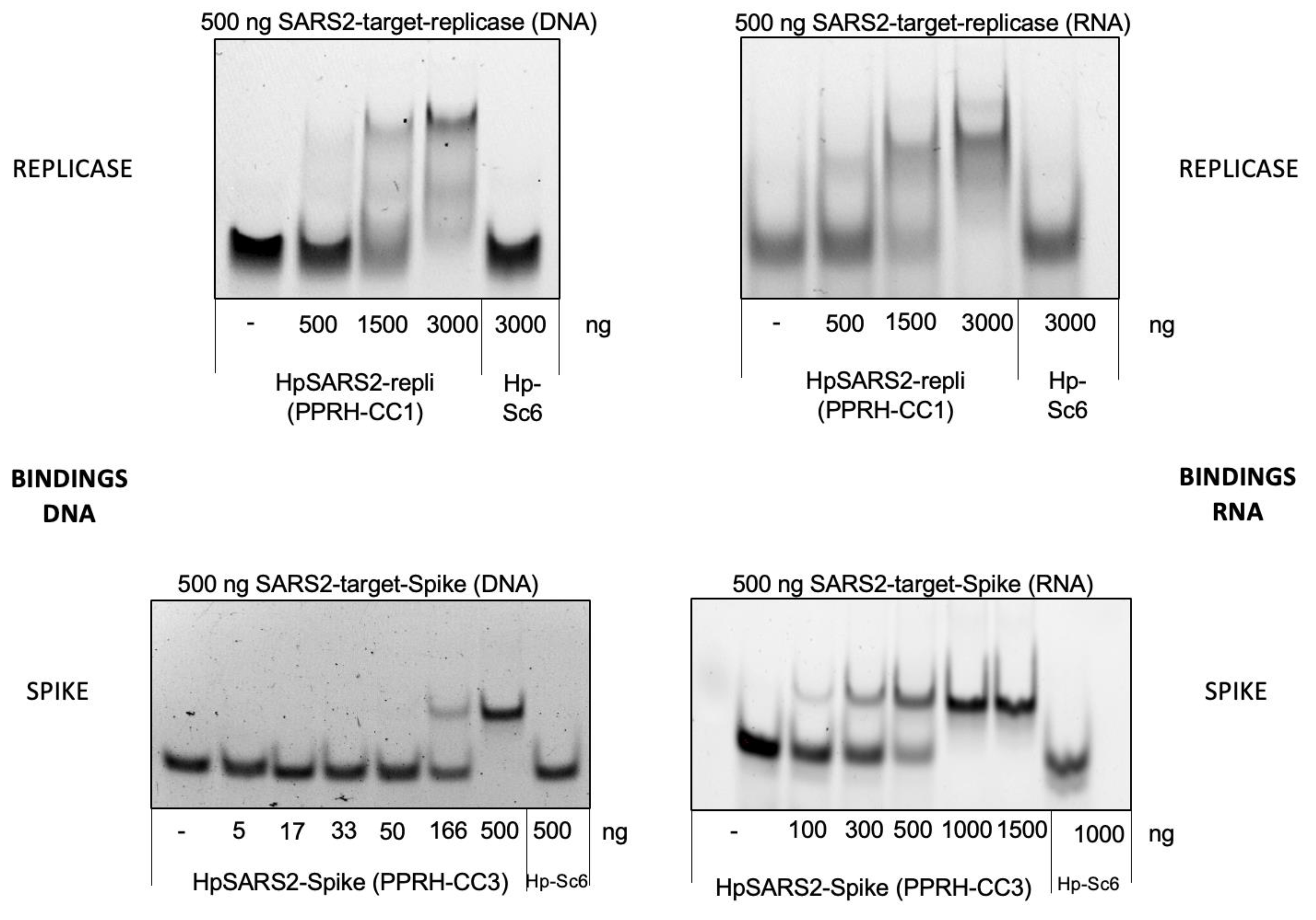
2.2. Gel Shift Binding Assays
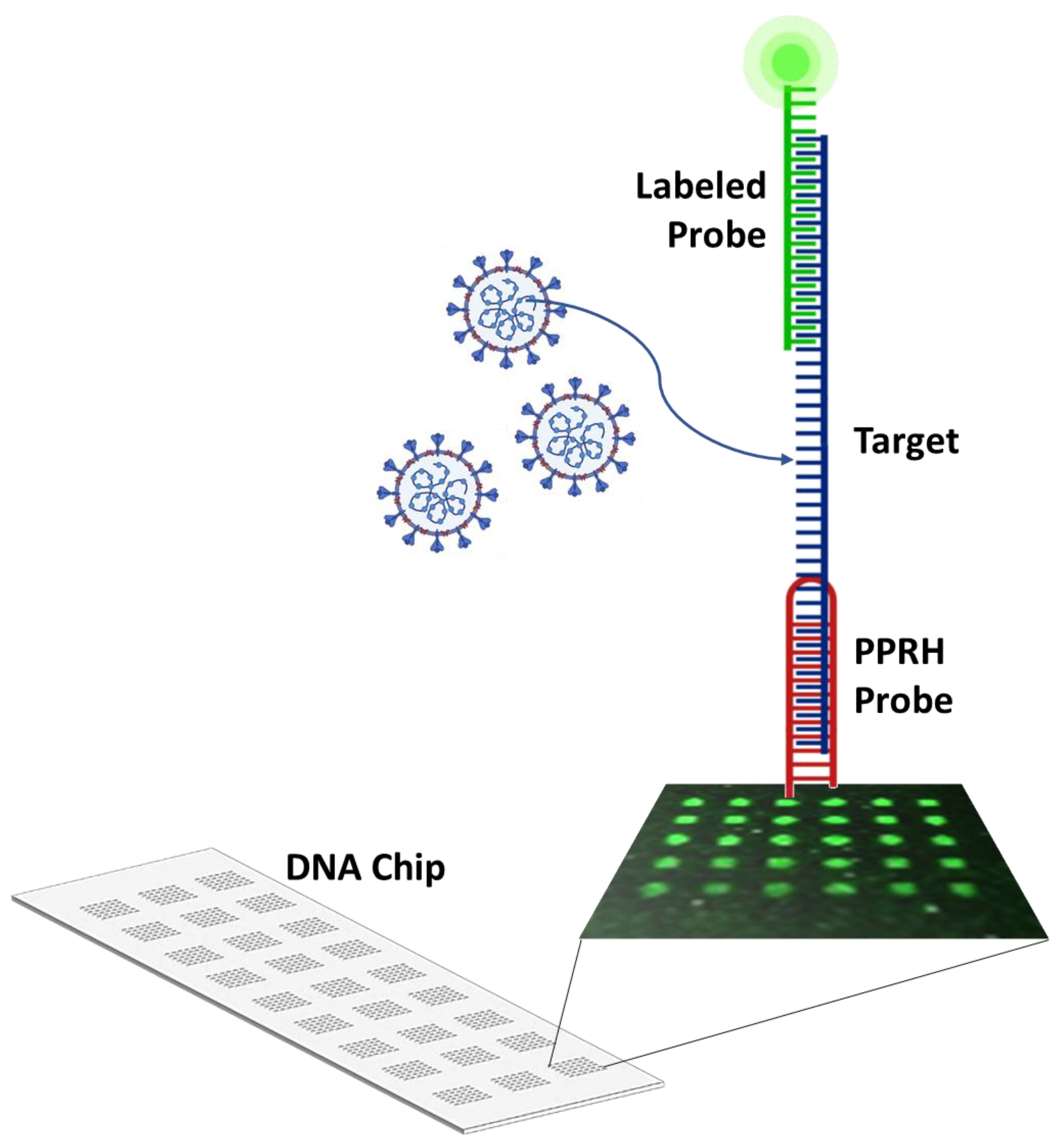
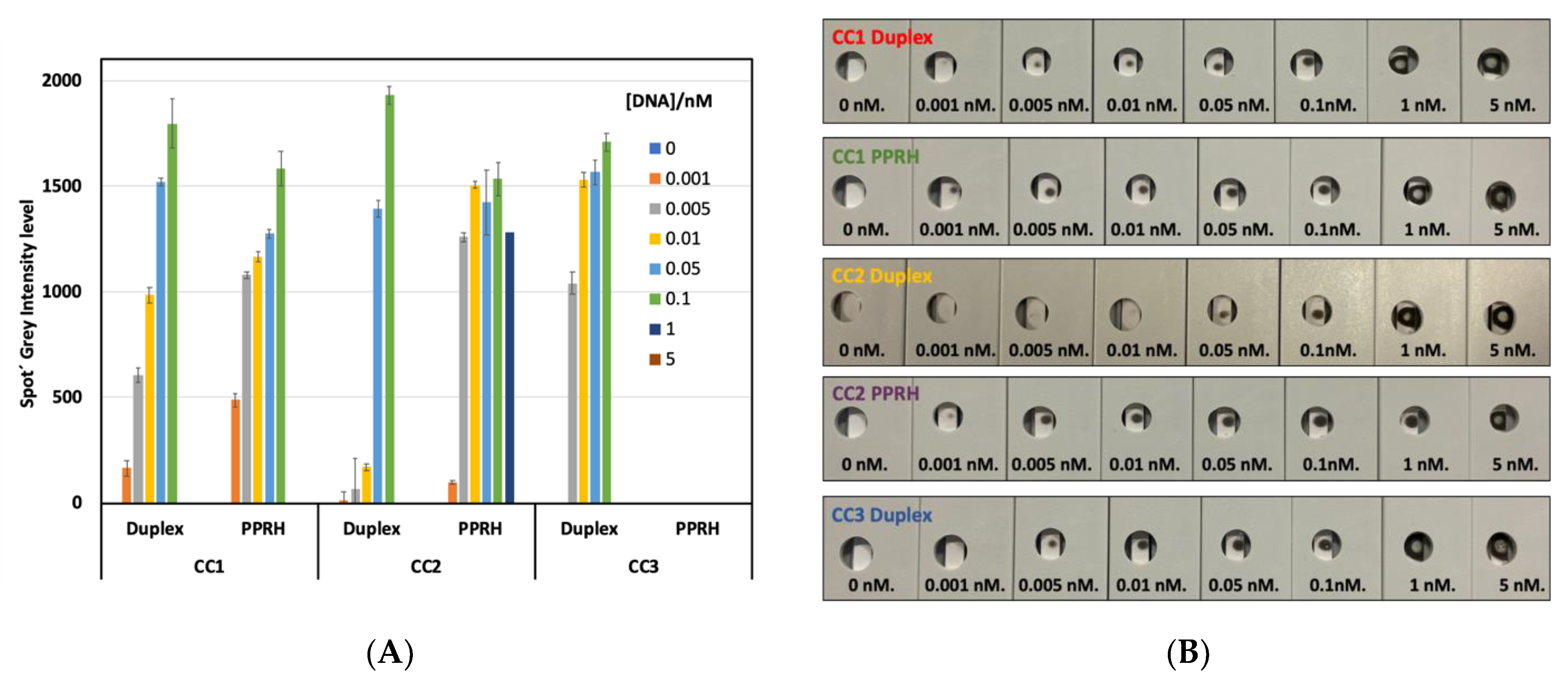
2.3. CC Pair Validation with a Fluorescent DNA Microarray Chip
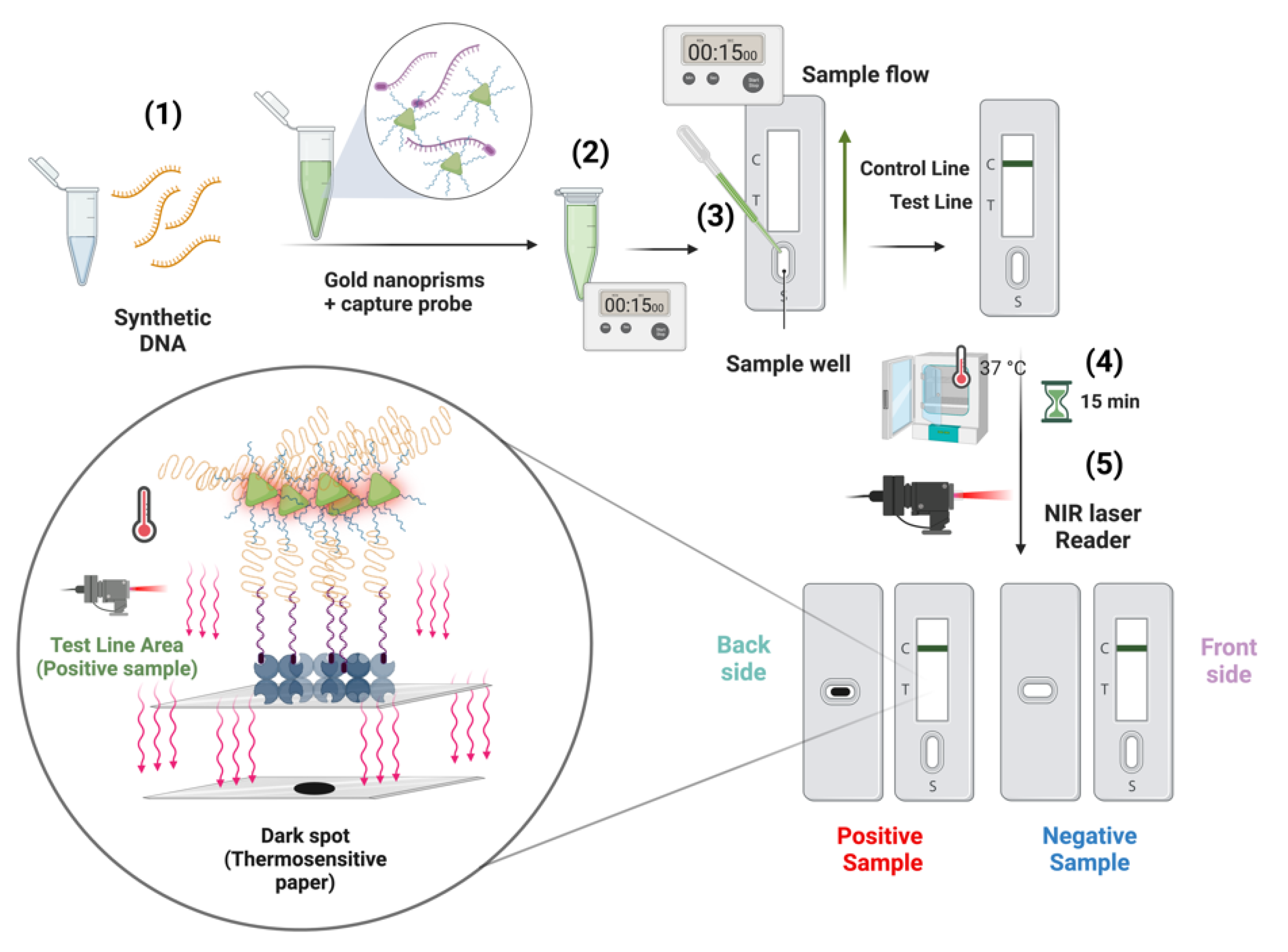
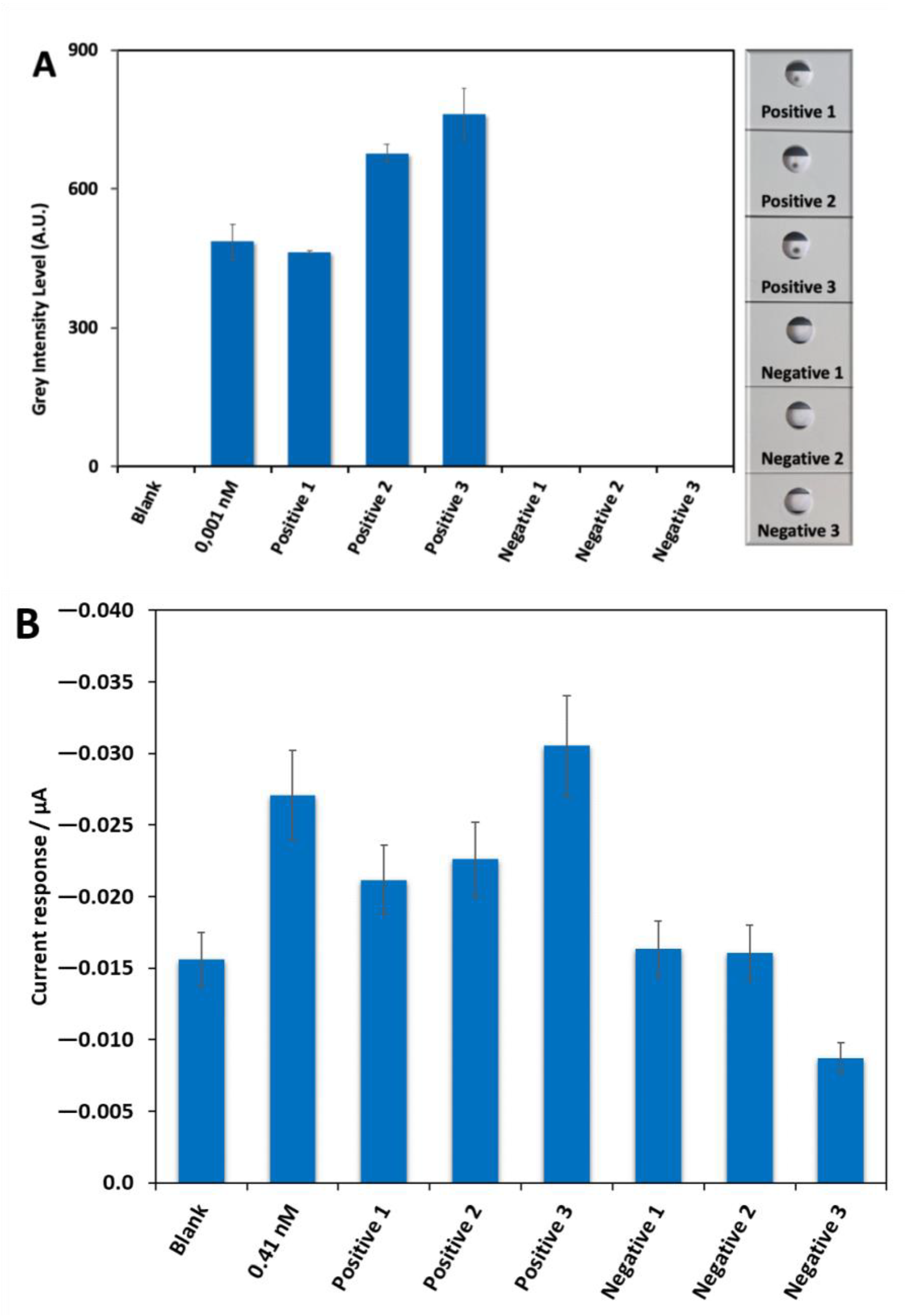
2.4. First Biosensor Device Thermal Lateral Flow System
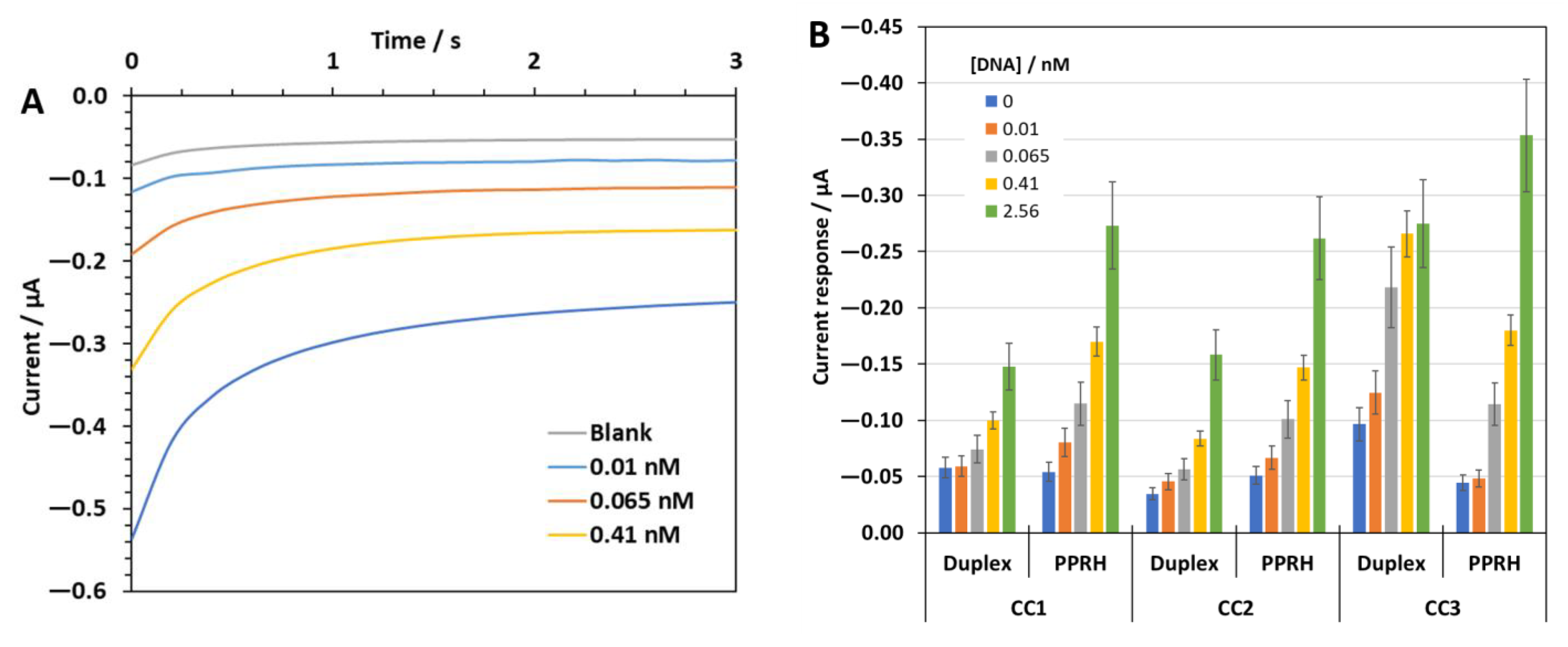
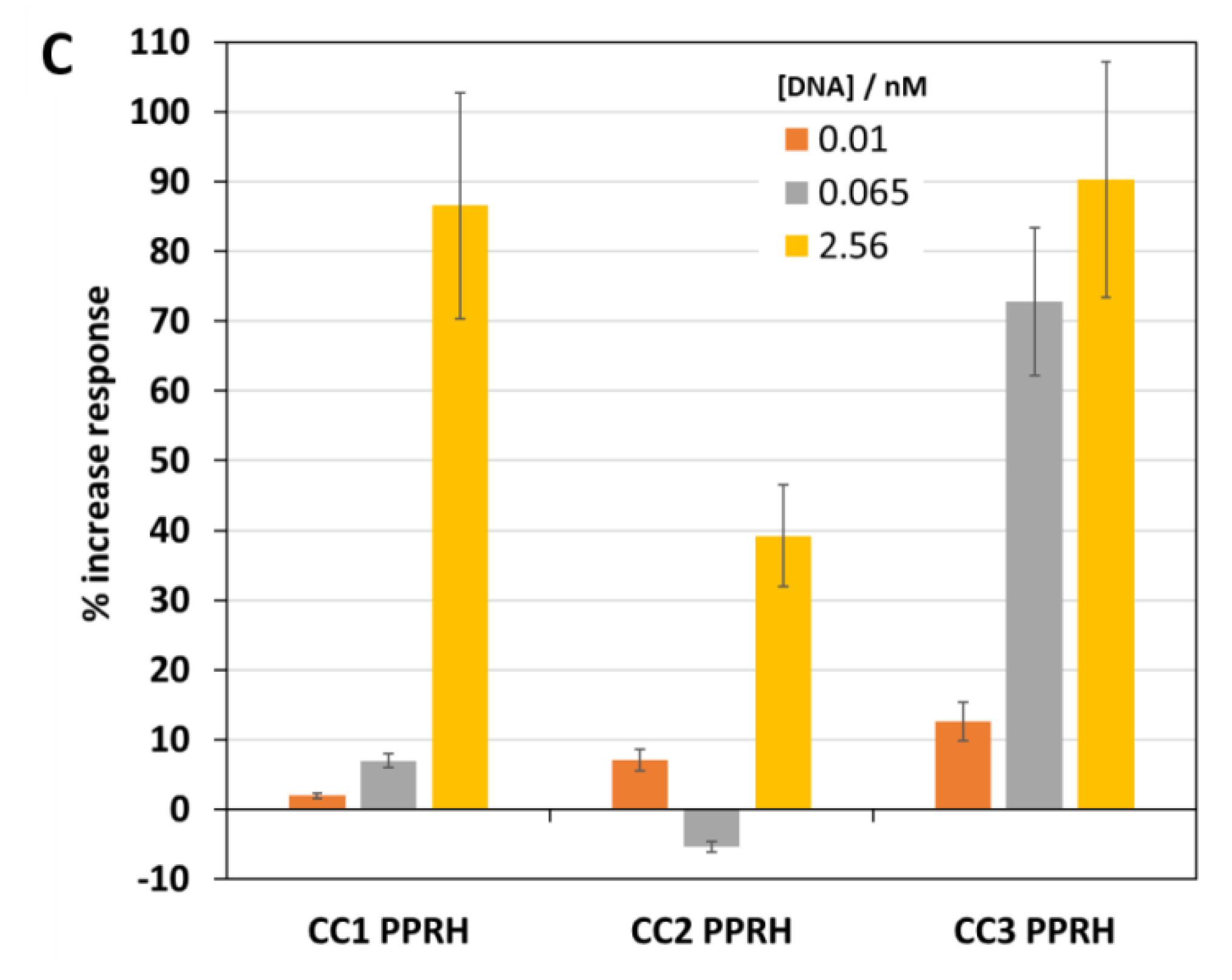
2.5. Second Biosensor Device Compact Electrochemical Biosensor Platform
2.6. Detection of SARS-CoV-2 RNA in Clinically Relevant Samples
3. Materials and Methods
3.1. Design of PPRH
3.2. Synthesis of Oligonucleotides
3.3. Gel-Binding Assays
3.4. Fluorescent DNA Microarray Chip
3.5. Thermal Lateral Flow System Using PPRH as Biosensors Linked to Gold Nanoprisms
3.5.1. Gold Nanoprisms (NPrs) Synthesis
3.5.2. Nanoprisms Biofunctionalization
3.5.3. Capture Molecules (Test and Control Lines) Preparation
3.5.4. Preparation of Lateral Flow Test Strips
3.5.5. Thermal Lateral Flow Assay (TLFA) Methodology
3.6. Electrochemical Biosensor
- A reusable electrochemical cell of two gold thin-film electrodes fabricated by a standard photolithographic/lift-off process on 4-inch silicon wafers at the IMB-CNM Clean Room facilities [27]. Additionally, 8 × 8.3 mm2 silicon chips, each one including a 1 × 1 mm2 working electrode and a 1.5 × 1 mm2 counter/reference electrode were manufactured.
- A disposable fluidic channel made of Whatman® cellulose chromatography paper, Grade 1, cut using a custom-made die cutter (Tecnocut, Barcelona, Spain) sandwiched between two polyvinyl layers patterned using a blade plotter (CAMM-1 Servo Cutter, Roland DG, Barcelona, Spain) to expose the fluidic channels in the sample addition and detection areas.
- A poly(methyl methacrylate) cartridge to integrate and align the cell and the fluidic channel, machined using a CO2-laser printer (Epilog Mini 24, Epilog Laser, Golden, CO, USA). The bottom part of the cartridge included an Nd magnet to trap MNPs inside the platform, as explained below.
3.7. Determination of SARS-CoV-2 RNA in Clinically Relevant Samples
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mercer, T.R.; Salit, M. Testing at the scale during COVID-19 pandemics. Nat. Genet. Rev. 2021, 22, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleickeret, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Wikramaratna, P.; Paton, R.; Ghafari, M.; Lourenco, J. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR 2020. Eur. Surveill. 2020, 25, 2000568. [Google Scholar]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef] [PubMed]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. A DNA detection using recombination proteins. PLoS Biol. 2006, 4, 1115–1121. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Esslerzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Naringer, N.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISP-Cas12 based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; Derby, M.D.L.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.S.; Sathe, L.; Munugala, C.; Jones, E.M.; Gasperini, M.; Lubock, N.B.; Yarza, F.; Thompson, E.M.; Kovary, K.M.; Park, J.; et al. Massively scaled-up testing for SARS-CoV-2 RNA via next-generation sequencing of pooled and barcoded nasal and saliva samples. Nat. Biomed. Eng. 2021, 5, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Lambert-Niclot, S.; Cuffel, A.; Le Pape, S.; Vauloup-Fellous, C.; Morand-Joubert, L.; Roque-Afonso, A.M.; Le Goff, J.; Delaugerre, C. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J. Clin. Microbiol. 2020, 58, e00977-e20. [Google Scholar] [CrossRef] [PubMed]
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef]
- Calvo-Lozano, O.; Aviñó, A.; Friaza, V.; Medina-Escuela, A.; Huertas, C.S.; Calderón, E.J.; Eritja, R.; Lechuga, L.M. Fast and accurate Pneumocystis pneumonia diagnosis in human samples using a label-free plasmonic biosensor. Nanomaterials 2020, 10, 1246. [Google Scholar] [CrossRef]
- Pla, L.; Aviñó, A.; Eritja, R.; Ruiz-Gaitán, A.; Pemán, J.; Friaza, V.; Calderón, E.J.; Aznar, E.; Martínez-Máñez, R.; Santiago-Felipe, S. Triplex hybridization-based nanosystem for the rapid screening of Pneumocystis pneumonia in clinical samples. J. Fungi 2020, 6, 292. [Google Scholar] [CrossRef]
- Noé, V.; Aubets, E.; Félix, A.J.; Ciudad, C.J. Nucleic acids therapeutics using PolyPurine Reverse Hoogsteen hairpins. Biochem. Pharm. 2021, 189, 114371. [Google Scholar] [CrossRef]
- Coma, S.; Noé, V.; Eritja, R.; Ciudad, C.J. Strand displacement of double-stranded DNA by triplex-forming antiparallel purine-hairpins. Oligonucleotides 2005, 15, 269–283. [Google Scholar] [CrossRef]
- Aubets, E.; Chillon, M.; Ciudad, C.J.; Noe, V. PolyPurine Reverse Hoogsteen Hairpins work as RNA species for gene silencing. Int. J. Mol. Sci. 2021, 22, 10025. [Google Scholar] [CrossRef]
- Aviñó, A.; Huertas, C.S.; Lechuga, L.M.; Eritja, R. Sensitive and label-free detection of miRNA-145 by triplex formation. Anal. Bioanal. Chem. 2016, 408, 885–893. [Google Scholar] [CrossRef]
- Ribes, A.; Santiago-Felipe, S.; Aviñó, A.; Candela-Noguera, V.; Eritja, R.; Sancenón, F.; Martínez-Máñez, R.; Aznar, E. Design of oligonucleotide-capped mesoporous silica nanoparticles for the detection of miRNA-145 by duplex and triplex formation. Sens. Actuators B. Chem. 2018, 277, 598–603. [Google Scholar] [CrossRef]
- Huertas, C.S.; Aviñó, A.; Kurachi, C.; Piqué, A.; Sandoval, J.; Eritja, R.; Esteller, M.; Lechuga, L.M. Label-free DNA-methylation detection by direct ds-DNA fragment screening using poly-purine hairpins. Biosens. Bioelectr. 2018, 120, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Ranki, M.; Palva, A.; Virtanen, M.; Laaksonen, M.; Soderlund, H. Sandwich hybridization as a convenient method for the detection of nucleic acids in crude samples. Gene 1983, 21, 77–85. [Google Scholar] [CrossRef]
- Aviñó, A.; Eritja, R.; Ciudad, C.; Noé, V. Parallel clamps and polypurine hairpins (PPRH) for gene silencing and triplex-affinity capture: Design, synthesis and use. Curr. Protoc. Nucleic Acid Chem. 2019, 77, e78. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.U.; Schmithausen, R.M.; Li, D.; Max, L.; Jacobs, M.L.; Hollstein, R.; Blumenstock, K.; Liebing, J.; Słabicki, M.; Ben-Shmuel, A.; et al. LAMP-Seq enables sensitive, multiplexed COVID-19 diagnostics using molecular barcoding. Nat. Biotechnol. 2021, 39, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, B.; Grazu, V.; Ibarra, A.; Magen, C.; del Pino, P.; de la Fuente, J.M. Tailoring the synthesis and heating ability of gold nanoprisms for bioapplications. Langmuir 2012, 28, 8965–8970. [Google Scholar] [CrossRef]
- Gutiérrez-Capitán, M.; Baldi, A.; Merlos, A.; Fernández-Sánchez, C. Array of individually addressable two-electrode electrochemical cells sharing a single counter/reference electrode for multiplexed enzyme activity measurements. Biosens. Bioelectr. 2022, 201, 113952. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aviñó, A.; Cuestas-Ayllón, C.; Gutiérrez-Capitán, M.; Vilaplana, L.; Grazu, V.; Noé, V.; Balada, E.; Baldi, A.; Félix, A.J.; Aubets, E.; et al. Detection of SARS-CoV-2 Virus by Triplex Enhanced Nucleic Acid Detection Assay (TENADA). Int. J. Mol. Sci. 2022, 23, 15258. https://doi.org/10.3390/ijms232315258
Aviñó A, Cuestas-Ayllón C, Gutiérrez-Capitán M, Vilaplana L, Grazu V, Noé V, Balada E, Baldi A, Félix AJ, Aubets E, et al. Detection of SARS-CoV-2 Virus by Triplex Enhanced Nucleic Acid Detection Assay (TENADA). International Journal of Molecular Sciences. 2022; 23(23):15258. https://doi.org/10.3390/ijms232315258
Chicago/Turabian StyleAviñó, Anna, Carlos Cuestas-Ayllón, Manuel Gutiérrez-Capitán, Lluisa Vilaplana, Valeria Grazu, Véronique Noé, Eva Balada, Antonio Baldi, Alex J. Félix, Eva Aubets, and et al. 2022. "Detection of SARS-CoV-2 Virus by Triplex Enhanced Nucleic Acid Detection Assay (TENADA)" International Journal of Molecular Sciences 23, no. 23: 15258. https://doi.org/10.3390/ijms232315258
APA StyleAviñó, A., Cuestas-Ayllón, C., Gutiérrez-Capitán, M., Vilaplana, L., Grazu, V., Noé, V., Balada, E., Baldi, A., Félix, A. J., Aubets, E., Valiuska, S., Domínguez, A., Gargallo, R., Eritja, R., Marco, M.-P., Fernández-Sánchez, C., Martínez de la Fuente, J., & Ciudad, C. J. (2022). Detection of SARS-CoV-2 Virus by Triplex Enhanced Nucleic Acid Detection Assay (TENADA). International Journal of Molecular Sciences, 23(23), 15258. https://doi.org/10.3390/ijms232315258










