Tumor-Derived Small Extracellular Vesicles Involved in Breast Cancer Progression and Drug Resistance
Abstract
1. Introduction
2. Tumor-Derived sEVs in Breast Cancer
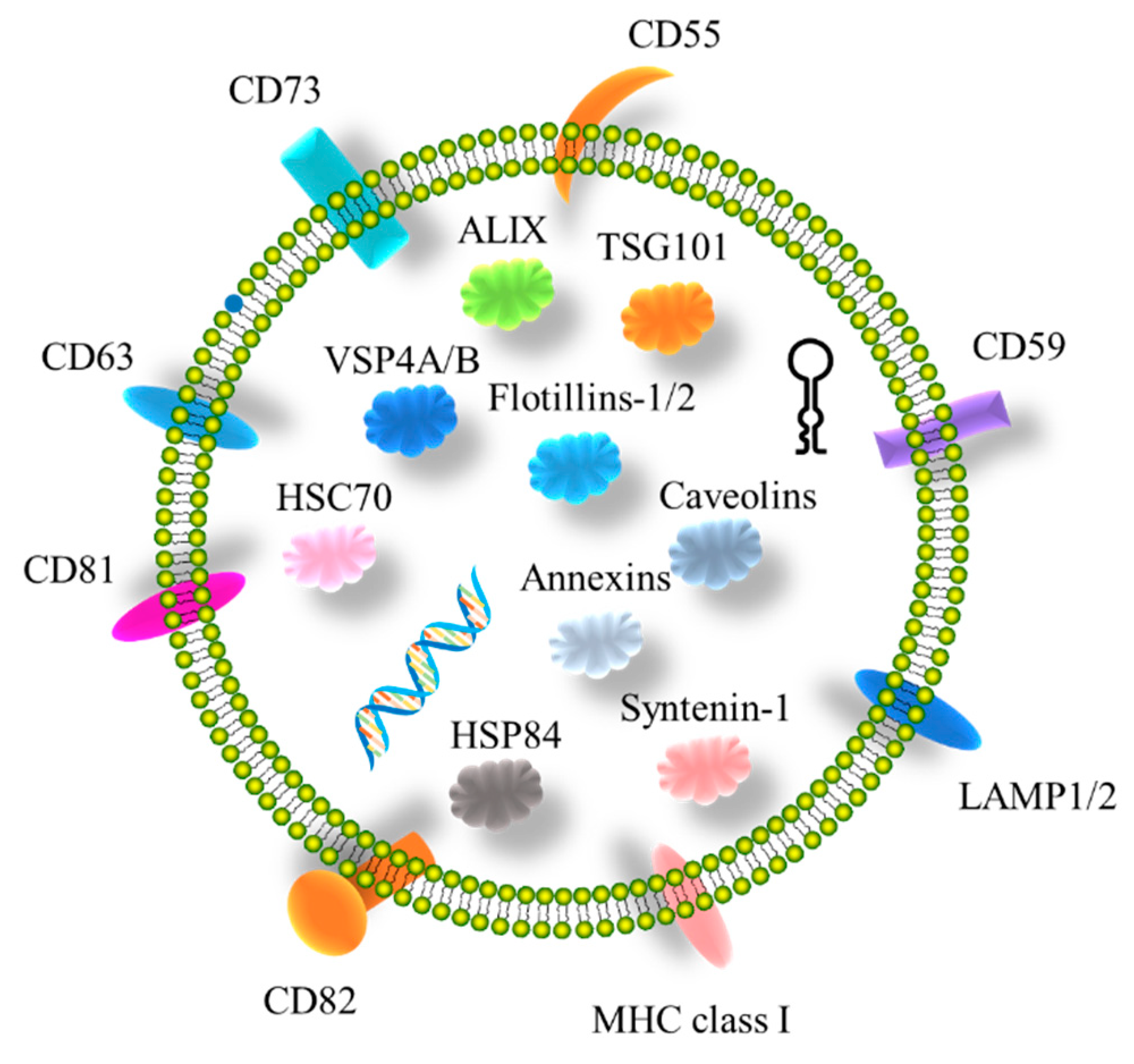
2.1. Characterization of Tumor-Derived sEVs in Breast Cancer
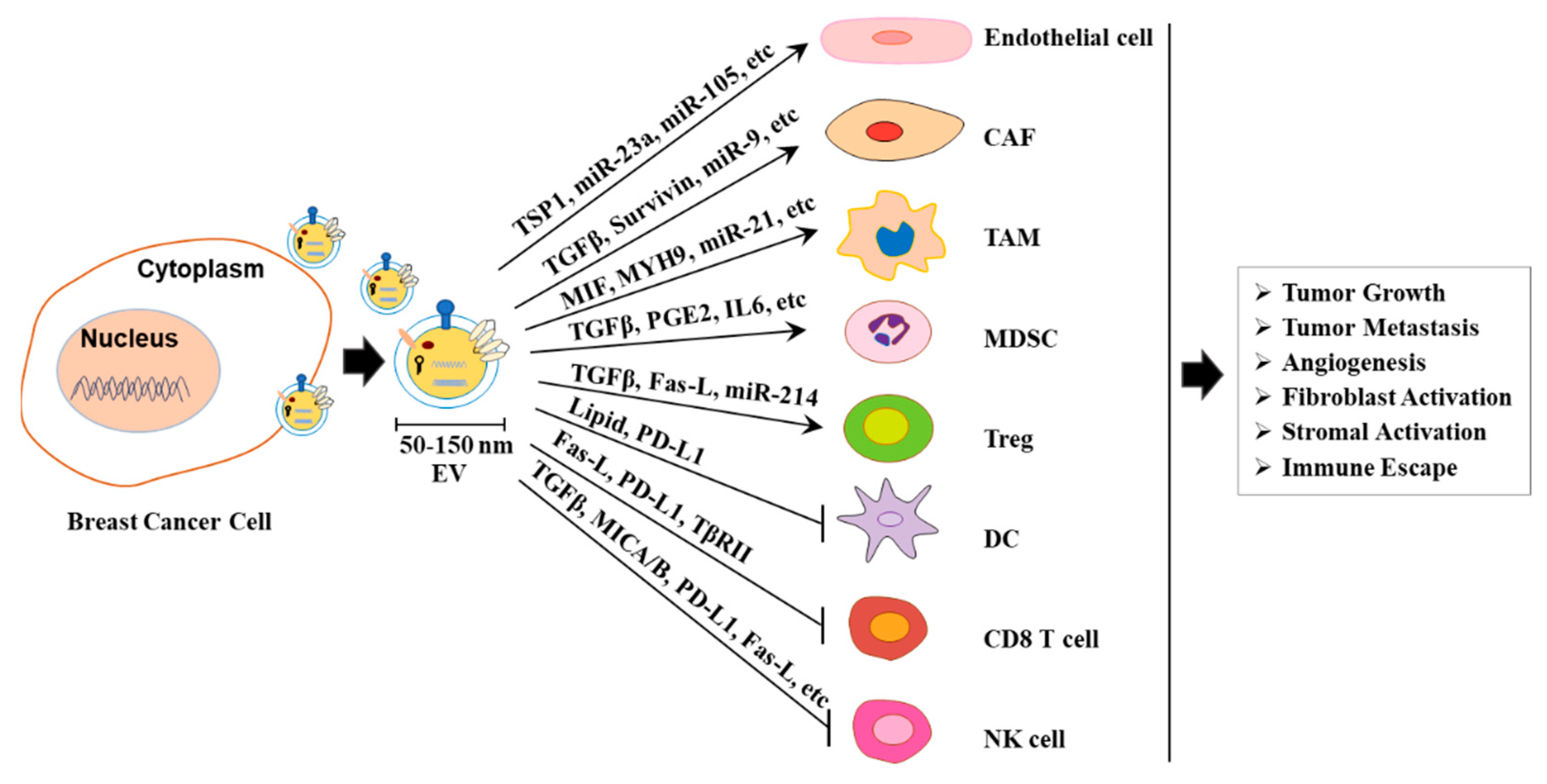
2.2. Tumor-Derived sEVs Facilitate Tumor Development
2.3. Tumor-Derived sEVs Promote Cancer Metastasis
| Molecules in sEVs | Target Cells | Functions | Ref. |
|---|---|---|---|
| TSP1 | Endothelial cells | TSP1-enriched exosomes promoted the transendothelial migration of malignant cells and decreased the expression of intercellular junction proteins. Exosomal TSP1 facilitated the transendothelial migration of breast cancer cells via disrupting the intercellular integrity of endothelial cells. | [48] |
| miR-105 | Endothelial cells | Cancer-secreted miR-105 efficiently destroys tight junctions and the integrity of these natural barriers against metastasis. | [42] |
| CEMIP | Endothelial cells | Promotion of vascular remodeling, leading to the formation of a pre-metastatic niche. | [49] |
| TGF-β | Fibroblast, bone marrow myeloid cells | Transducing SMAD-dependent signaling. Inhibiting either signaling or β-glycan expression attenuated differentiation. | [50,51] |
| Survivin | Fibroblasts | Breast cancer cell-derived Survivin upregulates SOD1 expression in fibroblasts and converts them into myofibroblasts, conversely inducing breast cancer progression in vitro and in vivo. | [52] |
| miR-9 | Fibroblasts | Enhancing the switch to CAF phenotype, thus contributing to tumor growth. | [53] |
| Myosin-9 | Macrophages | SIPA1-high breast cancer can enhance macrophage infiltration through sEVs enriched with myosin-9, which might aggravate the malignancy of breast cancer. | [54] |
| gp130 | Macrophages | Breast cancer cell-derived exosomes transfer gp130 to macrophages in vitro, which alters macrophage polarization by activating the STAT3 pathway. | [55] |
| EGFR | Macrophages | Breast cancer cell-derived exosomes transferred activated EGFR to host macrophages, which inhibited their production of type I interferons and antiviral immunity, resulting in compromised innate immunity. | [56] |
| PGE2 | Bone marrow myeloid cells | MDSC-mediated promotion of tumor progression is dependent on T-exosome prostaglandin E2 (PGE2) and TGF-β molecules. T-exosomes can induce the accumulation of MDSCs expressing COX-2, IL-6, VEGF, and arginase-1. | [51] |
| lncRNA SNHG16 | Tregs | BC-derived exosomal SNHG16/miR-16-5p/SMAD5-regulatory axis potentiates TGF-β1/SMAD5 pathway activation, thus inducing CD73 expression in Vδ1 T cells. | [57] |
| TβRII | CD8 T cells | Malignant breast cancer cells can transfer active TGF-β type II receptor (TβRII) via tumor-derived extracellular vesicles (TEV) and, thereby, stimulate TGF-β signaling in recipient cells. EV-TβRII delivered as a cargo to CD8+ T cells induces the activation of SMAD3, which is associated and cooperated with TCF1 transcription factor to impose CD8+ T cell exhaustion. | [58] |
| Integrins α6β4 and α6β1 | Associated with lung metastasis. | [46] | |
| Integrin αvβ5 | Linked to liver metastasis. | [46] | |
| Palmitoylated proteins | Macrophages | NF-κB activation. | [59] |
| miR-122 | Stromal cells | By preventing glucose uptake in stromal cells via miR-122-mediated inhibition of pyruvate kinase, breast cancer cells create a PMN with greater glucose availability for their own utilization. | [36] |
2.4. Biomarkers of sEVs
3. Tumor-Derived sEVs Play Vital Roles in the BC Microenvironment
3.1. Effects of Tumor-Derived sEVs on Angiogenesis
3.2. Effects of Tumor-Derived sEVs on Fibroblast and Stromal Activation
3.3. Effects of Tumor-Derived sEVs on Immune Cells
3.4. Effects of Microenvironment-Derived sEVs on BC Development
3.5. sEVs Modulate BC Drug Resistance
4. sEV-Based BC Diagnosis and Therapy
| Contents in sEVs | Biomarkers in sEVs | Functions (Mechanism)/Usage | Ref. |
|---|---|---|---|
| Protein | ADAM10, Annexin-1, CD9, metalloprotease, and HSP70 | Activation of RhoA and Notch signaling. Promotion of cell motility and tumor progression. | [138,187] |
| CD82 | Negative correlation between CD82 expression in tissues and CD82 content in exosomes. | [188] | |
| GPC1 | Overexpression of GPC1-mediated epithelial–mesenchymal transition (EMT), which promotes invasion and migration. | [195] | |
| Annexin A2 | Serum exo-AnxA2 is high in AA women with TNBC and promotes angiogenesis. | [84,196] | |
| Developmental endothelial Locus-1 (Del-1) | Plasma Del-1 levels are significantly high (p < 0.0001) in patients with breast cancer, but return to normal after tumor removal. | [191] | |
| Survivin-2B | The protein may be used as a biomarker for patients with early breast cancer. | [193] | |
| miRNA | miR-21, miR-1246 | Circulating exosomal miRNAs, miRNA-21 and miRNA-1246, are abundant in patients with breast cancer. | [123] |
| miR-181d-5p | Targeting CDX2 and downregulation of CDX2 and HOXA5. Enhancement of breast cancer aggressiveness. | [134] | |
| miR-21, miR-378e, miR-143 | Induction of stemness and EMT phenotype of breast cancer. | [100] | |
| miR-500a-5p | Targeting USP28 and downregulation of USP28. Promotion of proliferation and metastasis of breast cancer cells. | [135] | |
| miR-4516 | Targeting FOS such as antigen 1 (FOSL1). Promotion of TNBC development. | [197] | |
| miR-22 | Targeting ESR1 and PTEN and downregulation of ESR1 and PTEN. Promotion of tamoxifen resistance. | [136] | |
| miR-1246 | The exosomal miRNA-1246 in the peripheral blood can distinguish 46 breast cancer patients from 28 healthy controls with 100% sensitivity and 93% specificity. | [61] | |
| lncRNA | SNHG3 | Targeting miR-330-5p and promotion of PKM expression and glycolysis metabolism. | [139] |
| circRNA | circHIF1A | Induction of CD44 expression by targeting and down-regulating miR-580-5p. Promotion of breast cancer cell proliferation and stemness in hypoxic stress. | [137] |
| Payload/Drug Names | Targets | Ref. |
|---|---|---|
| Cell cycle quiescence and chemotherapy-resistant mesenchymal stem cells-derived ssEVs | Facilitating breast cancer cells to progressively differentiate into dormancy in the perivascular region of bone marrow | [223] |
| Tumor-acclimated MSCs-derived ssEVs | Induction of monocytic myeloid-derived suppressor cells | [224] |
| M1-type macrophages-derived ssEVs loaded with Paclitaxel (PTX) | Microtubule | [225] |
| Human fetal lung fibroblast-derived ssEVs loaded with FA and irastatin | Ferroptosis/Folic acid | [217] |
| Hybrid Exosome (HE) loaded with water-soluble doxorubicin | DNA | [226] |
| Generalized exosomal nanobiological conjugate produced by pH-responsive macrophage M1 binding with anti-tumor effect of antibodies | Specific proteins | [214] |
| Exosomes isolated from MSCCXCR4 + TRAIL transduced with CXCR4 and TRAIL (exosome CXCR4 + TRAIL) loaded with carboplatin | Synthesis of DNA | [227] |
| Biomimetic tumor-derived exosomes loaded with (TEX)-Liposome-paclitaxel (PTX) | Microtubule | [228] |
| Folic acid (FA)-functionalized bovine milk-derived ssEVs loaded with Paclitaxel (PAC) and 5-fluorouracil (5-FU) | Microtubule/synthesis of DNA | [229] |
| miR-567 | Cancer-associated ATG5 | [230] |
| let-7a miRNA | EGFR | [231] |
| Ultrasonic sensitizer indocyanine green (ICG) | Folic acid | [232] |
| Deintegrin and metalloproteinase 15 (A15), doxorubicin (Dox) and cholesterol-modified miRNA 159 | DNA/GAMYB | [233] |
| Cationic bovine serum albumin (CBSA) conjugated siS100A4 nanoparticles | Lung PMN | [234] |
| Surface display monoclonal antibodies against human CD3 and human HER2 | Human CD3 and human HER2 | [235] |
| The conjugates of reactive oxygen species (ROS)-responsive sulfide-linked paclitaxel-Linoleic acid conjugates (PTXS-LA) and cucurbitin B (CuB) (EMPC) loaded with CD44 | Circulating tumor cells | [236] |
5. Conclusions and Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Zhao, J.; Liang, J.; Wang, H.; Chen, Y.; Huang, G. Exosomes: A novel insight into traditional Chinese medicine. Front. Pharmacol. 2022, 13, 844782. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Chen, X.; Feng, J.; Chen, W.; Shao, S.; Chen, L.; Wan, H. Small extracellular vesicles: From promoting pre-metastatic niche formation to therapeutic strategies in breast cancer. Cell Commun. Signal. 2022, 20, 141. [Google Scholar] [CrossRef]
- Palazzolo, S.; Canzonieri, V.; Rizzolio, F. The history of small extracellular vesicles and their implication in cancer drug resistance. Front. Oncol. 2022, 12, 948843. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.L.; Wang, S.W.; Sun, W.C.; Shu, C.W.; Kao, Y.C.; Shiao, M.S.; Chen, C.L. Sorafenib suppresses TGF-beta responses by inducing caveolae/lipid raft-mediated internalization/degradation of cell-surface type II TGF-beta receptors: Implications in development of effective adjunctive therapy for hepatocellular carcinoma. Biochem. Pharmacol. 2018, 154, 39–53. [Google Scholar] [CrossRef]
- Johannes, L.; Parton, R.G.; Bassereau, P.; Mayor, S. Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 2015, 16, 311–321. [Google Scholar] [CrossRef] [PubMed]
- King, J.S.; Kay, R.R. The origins and evolution of macropinocytosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180158. [Google Scholar] [CrossRef]
- Gray, M.; Botelho, R.J. Phagocytosis: Hungry, Hungry Cells. Methods Mol. Biol. 2017, 1519, 1–16. [Google Scholar] [CrossRef]
- Lai, R.C.; Chen, T.S.; Lim, S.K. Mesenchymal stem cell exosome: A novel stem cell-based therapy for cardiovascular disease. Regen. Med. 2011, 6, 481–492. [Google Scholar] [CrossRef]
- Timmers, L.; Lim, S.K.; Hoefer, I.E.; Arslan, F.; Lai, R.C.; van Oorschot, A.A.; Goumans, M.J.; Strijder, C.; Sze, S.K.; Choo, A.; et al. Human mesenchymal stem cell-conditioned medium improves cardiac function following myocardial infarction. Stem Cell Res. 2011, 6, 206–214. [Google Scholar] [CrossRef]
- Timmers, L.; Lim, S.K.; Arslan, F.; Armstrong, J.S.; Hoefer, I.E.; Doevendans, P.A.; Piek, J.J.; El Oakley, R.M.; Choo, A.; Lee, C.N.; et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2007, 1, 129–137. [Google Scholar] [CrossRef]
- Salmond, N.; Williams, K.C. Isolation and characterization of extracellular vesicles for clinical applications in cancer—Time for standardization? Nanoscale Adv. 2021, 3, 1830–1852. [Google Scholar] [CrossRef]
- Wahlgren, J.; Karlson, T.D.; Brisslert, M.; Sani, F.V.; Telemo, E.; Sunnerhagen, P.; Valadi, H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012, 40, e130. [Google Scholar] [CrossRef] [PubMed]
- Ojeda-Hernandez, D.D.; Hernandez-Sapiens, M.A.; Reza-Zaldivar, E.E.; Canales-Aguirre, A.; Matias-Guiu, J.A.; Matias-Guiu, J.; Mateos-Diaz, J.C.; Gomez-Pinedo, U.; Sancho-Bielsa, F. Exosomes and Biomaterials: In Search of a New Therapeutic Strategy for Multiple Sclerosis. Life 2022, 12, 1417. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, 516–519. [Google Scholar] [CrossRef]
- Kim, D.K.; Kang, B.; Kim, O.Y.; Choi, D.S.; Lee, J.; Kim, S.R.; Go, G.; Yoon, Y.J.; Kim, J.H.; Jang, S.C.; et al. EVpedia: An integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell. Vesicles 2013, 19, 20384. [Google Scholar] [CrossRef]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 16, 18374. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, Y.; Liang, L.; Wang, Y.; Huang, W.; Zhao, K.; Liu, S.; Deng, G.; Chen, J. Tumor-derived extracellular vesicles shuttle c-Myc to promote gastric cancer growth and metastasis via the KCNQ1OT1/miR-556-3p/CLIC1 axis. Cell Death Dis. 2022, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Ruivo, C.F.; Bastos, N.; Adem, B.; Batista, I.; Duraes, C.; Melo, C.A.; Castaldo, S.A.; Campos-Laborie, F.; Moutinho-Ribeiro, P.; Morao, B.; et al. Extracellular Vesicles from Pancreatic Cancer Stem Cells Lead an Intratumor Communication Network (EVNet) to fuel tumour progression. Gut 2022, 71, 2043–2068. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Rana, S.; Zoller, M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia 2013, 15, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Sedgwick, A.E.; Clancy, J.W.; Olivia Balmert, M.; D’Souza-Schorey, C. Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci. Rep. 2015, 5, 14748. [Google Scholar] [CrossRef]
- Sinha, S.; Hoshino, D.; Hong, N.H.; Kirkbride, K.C.; Grega-Larson, N.E.; Seiki, M.; Tyska, M.J.; Weaver, A.M. Cortactin promotes exosome secretion by controlling branched actin dynamics. J. Cell Biol. 2016, 214, 197–213. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Hon, K.W.; Abu, N.; Ab Mutalib, N.S.; Jamal, R. Exosomes as potential biomarkers and targeted therapy in colorectal cancer: A mini-review. Front. Pharmacol. 2017, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Treps, L.; Perret, R.; Edmond, S.; Ricard, D.; Gavard, J. Glioblastoma stem-like cells secrete the pro-angiogenic VEGF-A factor in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1359479. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Hung, J.Y.; Chang, W.A.; Lin, Y.S.; Pan, Y.C.; Tsai, P.H.; Wu, C.Y.; Kuo, P.L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef]
- Hendrix, A.; Maynard, D.; Pauwels, P.; Braems, G.; Denys, H.; Van den Broecke, R.; Lambert, J.; Van Belle, S.; Cocquyt, V.; Gespach, C.; et al. Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. J. Natl. Cancer Inst 2010, 102, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Xu, Z.; Zhang, Y.; Tan, C.; Hu, W.; Wang, M.; Xu, Y.; Tang, J. Exosome-mediated miR-222 transferring: An insight into NF-kappaB-mediated breast cancer metastasis. Exp. Cell Res. 2018, 369, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Keklikoglou, I.; Cianciaruso, C.; Guc, E.; Squadrito, M.L.; Spring, L.M.; Tazzyman, S.; Lambein, L.; Poissonnier, A.; Ferraro, G.B.; Baer, C.; et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat. Cell Biol. 2019, 21, 190–202. [Google Scholar] [CrossRef]
- Cen, J.; Feng, L.; Ke, H.; Bao, L.; Li, L.Z.; Tanaka, Y.; Weng, J.; Su, L. Exosomal thrombospondin-1 disrupts the integrity of endothelial intercellular junctions to facilitate breast cancer cell metastasis. Cancers 2019, 11, 1946. [Google Scholar] [CrossRef]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- Webber, J.; Steadman, R.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010, 70, 9621–9630. [Google Scholar] [CrossRef]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.B.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.J.; Zhang, L.; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, T.; Chen, J.; Ni, H.; Li, W. Survivin in breast cancer-derived exosomes activates fibroblasts by up-regulating SOD1, whose feedback promotes cancer proliferation and metastasis. J. Biol. Chem. 2020, 295, 13737–13752. [Google Scholar] [CrossRef] [PubMed]
- Baroni, S.; Romero-Cordoba, S.; Plantamura, I.; Dugo, M.; D’Ippolito, E.; Cataldo, A.; Cosentino, G.; Angeloni, V.; Rossini, A.; Daidone, M.G.; et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016, 7, e2312. [Google Scholar] [CrossRef]
- Feng, L.Y.; Weng, J.; Yao, C.G.; Wang, R.Y.; Wang, N.; Zhang, Y.L.; Tanaka, Y.; Su, L. Extracellular Vesicles Derived from SIPA1(high) Breast Cancer Cells Enhance Macrophage Infiltration and Cancer Metastasis through Myosin-9. Biology 2022, 11, 543. [Google Scholar] [CrossRef]
- Ham, S.; Lima, L.G.; Chai, E.P.Z.; Muller, A.; Lobb, R.J.; Krumeich, S.; Wen, S.W.; Wiegmans, A.P.; Moller, A. Breast Cancer-Derived Exosomes Alter Macrophage Polarization via gp130/STAT3 Signaling. Front. Immunol. 2018, 9, 871. [Google Scholar] [CrossRef]
- Gao, L.; Wang, L.; Dai, T.; Jin, K.; Zhang, Z.; Wang, S.; Xie, F.; Fang, P.; Yang, B.; Huang, H.; et al. Tumor-derived exosomes antagonize innate antiviral immunity. Nat. Immunol. 2018, 19, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Fang, Q.Q.; Chen, W.Z.; Jiang, J.X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.B.; Xia, W.J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+gammadelta1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef]
- Xie, F.; Zhou, X.; Su, P.; Li, H.; Tu, Y.; Du, J.; Pan, C.; Wei, X.; Zheng, M.; Jin, K.; et al. Breast cancer cell-derived extracellular vesicles promote CD8(+) T cell exhaustion via TGF-beta type II receptor signaling. Nat. Commun. 2022, 13, 4461. [Google Scholar] [CrossRef]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Van Haute, D.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z.; et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci. Rep. 2014, 4, 5750. [Google Scholar] [CrossRef]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef]
- Zhai, L.Y.; Li, M.X.; Pan, W.L.; Chen, Y.; Li, M.M.; Pang, J.X.; Zheng, L.; Chen, J.X.; Duan, W.J. In Situ Detection of Plasma Exosomal MicroRNA-1246 for Breast Cancer Diagnostics by a Au Nanoflare Probe. ACS Appl. Mater. Interfaces 2018, 10, 39478–39486. [Google Scholar] [CrossRef] [PubMed]
- Stevic, I.; Muller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marme, F.; Schem, C.; Stickeler, E.; Denkert, C.; van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.S.; Kim, D.K.; Kim, Y.K.; Gho, Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 2013, 13, 1554–1571. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Kim, Y.K.; Gho, Y.S. Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrom. Rev. 2015, 34, 474–490. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.M.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 14, 27066. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lasser, C.; Szabo, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzas, E.I.; Lotvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 12, 20677. [Google Scholar] [CrossRef]
- Hanson, P.I.; Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012, 28, 337–362. [Google Scholar] [CrossRef] [PubMed]
- Szwedowicz, U.; Lapinska, Z.; Gajewska-Naryniecka, A.; Choromanska, A. Exosomes and Other Extracellular Vesicles with High Therapeutic Potential: Their Applications in Oncology, Neurology, and Dermatology. Molecules 2022, 27, 1303. [Google Scholar] [CrossRef] [PubMed]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavik, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef]
- Liu, S.; Gonzalez-Prieto, R.; Zhang, M.; Geurink, P.P.; Kooij, R.; Iyengar, P.V.; van Dinther, M.; Bos, E.; Zhang, X.; Le Devedec, S.E.; et al. Deubiquitinase Activity Profiling Identifies UCHL1 as a Candidate Oncoprotein That Promotes TGFbeta-Induced Breast Cancer Metastasis. Clin. Cancer Res. 2020, 26, 1460–1473. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Yang, S.; Pi, H.; Li, Z.; Yao, P.; Zhang, Q.; Wang, Q.; Shen, P.; Li, X.; et al. Proteomic Landscape of Exosomes Reveals the Functional Contributions of CD151 in Triple-Negative Breast Cancer. Mol. Cell. Proteom. 2021, 20, 100121. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, J.N.; Demory Beckler, M.; Gephart, J.D.; Franklin, J.L.; Bogatcheva, G.; Kremers, G.J.; Piston, D.W.; Ayers, G.D.; McConnell, R.E.; Tyska, M.J.; et al. Amphiregulin exosomes increase cancer cell invasion. Curr. Biol. 2011, 21, 779–786. [Google Scholar] [CrossRef]
- Kreger, B.T.; Johansen, E.R.; Cerione, R.A.; Antonyak, M.A. The Enrichment of Survivin in Exosomes from Breast Cancer Cells Treated with Paclitaxel Promotes Cell Survival and Chemoresistance. Cancers 2016, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Schwich, E.; Rebmann, V. The Inner and Outer Qualities of Extracellular Vesicles for Translational Purposes in Breast Cancer. Front. Immunol. 2018, 9, 584. [Google Scholar] [CrossRef]
- Yang, S.J.; Wang, D.D.; Li, J.; Xu, H.Z.; Shen, H.Y.; Chen, X.; Zhou, S.Y.; Zhong, S.L.; Zhao, J.H.; Tang, J.H. Predictive role of GSTP1-containing exosomes in chemotherapy-resistant breast cancer. Gene 2017, 623, 5–14. [Google Scholar] [CrossRef]
- Todorova, D.; Simoncini, S.; Lacroix, R.; Sabatier, F.; Dignat-George, F. Extracellular vesicles in angiogenesis. Circ. Res. 2017, 120, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523. [Google Scholar] [CrossRef]
- Sruthi, T.V.; Edatt, L.; Raji, G.R.; Kunhiraman, H.; Shankar, S.S.; Shankar, V.; Ramachandran, V.; Poyyakkara, A.; Kumar, S.V.B. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J. Cell. Physiol. 2018, 233, 3498–3514. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Mkhobongo, B.; Chandran, R.; Abrahamse, H. The Role of Melanoma Cell-Derived Exosomes (MTEX) and Photodynamic Therapy (PDT) within a Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 9726. [Google Scholar] [CrossRef]
- Maji, S.; Chaudhary, P.; Akopova, I.; Nguyen, P.M.; Hare, R.J.; Gryczynski, I.; Vishwanatha, J.K. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol. Cancer Res. 2017, 15, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, G.S.; Poutahidis, T.; Erdman, S.E.; Kirsch, R.; Riddell, R.H.; Diamandis, E.P. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol. Cancer Res. 2012, 10, 1403–1418. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Zheng, W.; Dong, J.; Wang, Y.; Lai, J.; Liu, X.; Yin, F. Tumor microenvironment in pancreatic ductal adenocarcinoma: Implications in immunotherapy. World J. Gastroenterol. 2022, 28, 3297–3313. [Google Scholar] [CrossRef] [PubMed]
- Cirri, P.; Chiarugi, P. Cancer-associated-fibroblasts and tumour cells: A diabolic liaison driving cancer progression. Cancer Metastasis Rev. 2012, 31, 195–208. [Google Scholar] [CrossRef]
- Jung, D.W.; Che, Z.M.; Kim, J.; Kim, K.; Kim, K.Y.; Williams, D.; Kim, J. Tumor-stromal crosstalk in invasion of oral squamous cell carcinoma: A pivotal role of CCL7. Int. J. Cancer 2010, 127, 332–344. [Google Scholar] [CrossRef]
- Erez, N.; Truitt, M.; Olson, P.; Arron, S.T.; Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 2010, 17, 135–147. [Google Scholar] [CrossRef]
- Olumi, A.F.; Grossfeld, G.D.; Hayward, S.W.; Carroll, P.R.; Tlsty, T.D.; Cunha, G.R. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999, 59, 5002–5011. [Google Scholar] [CrossRef] [PubMed]
- Fotsitzoudis, C.; Koulouridi, A.; Messaritakis, I.; Konstantinidis, T.; Gouvas, N.; Tsiaoussis, J.; Souglakos, J. Cancer-Associated Fibroblasts: The Origin, Biological Characteristics and Role in Cancer-A Glance on Colorectal Cancer. Cancers 2022, 14, 4394. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Kwon, A.; Huh, Y.H.; Rhee, S.; Song, W.K. Tumor-derived miR-130b-3p induces cancer-associated fibroblast activation by targeting SPIN90 in luminal A breast cancer. Oncogenesis 2022, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.T.; Peng, B.; Zhang, D.X.; Ma, V.; Mathey-Andrews, C.A.; Lam, C.K.; Kiomourtzis, T.; Jin, J.; McReynolds, L.; Huang, L.; et al. Tumor-secreted extracellular vesicles promote the activation of cancer-associated fibroblasts via the transfer of microRNA-125b. J. Extracell. Vesicles 2019, 8, 1599680. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: Present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Yang, H.; Zhang, X.; Xu, W. The emerging roles of exosomes in tumor-stroma interaction. J. Cancer Res. Clin. Oncol. 2016, 142, 1897–1907. [Google Scholar] [CrossRef] [PubMed]
- Antonyak, M.A.; Li, B.; Boroughs, L.K.; Johnson, J.L.; Druso, J.E.; Bryant, K.L.; Holowka, D.A.; Cerione, R.A. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4852–4857. [Google Scholar] [CrossRef]
- Cho, J.A.; Park, H.; Lim, E.H.; Lee, K.W. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int. J. Oncol. 2012, 40, 130–138. [Google Scholar] [CrossRef]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhao, H.; Jiang, Y.; Yin, C.; Zhang, J. HCC-Derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells 2019, 8, 558. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Song, Y.; Miao, F.; Chen, G.; Zhu, Y.; Wu, N.; Pang, L.; Chen, Z.; Chen, X. PDL1-positive exosomes suppress antitumor immunity by inducing tumor-specific CD8(+) T cell exhaustion during metastasis. Cancer Sci. 2021, 112, 3437–3454. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef]
- Baig, M.S.; Roy, A.; Rajpoot, S.; Liu, D.; Savai, R.; Banerjee, S.; Kawada, M.; Faisal, S.M.; Saluja, R.; Saqib, U.; et al. Tumor-derived exosomes in the regulation of macrophage polarization. Inflamm. Res. 2020, 69, 435–451. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, H.; Yuan, X.; Jiang, P.; Qian, H.; Xu, W. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol. Cancer 2018, 17, 146. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The cellular and molecular origin of tumor-associated macrophages. Science 2014, 344, 921–925. [Google Scholar] [CrossRef]
- Goudot, C.; Coillard, A.; Villani, A.C.; Gueguen, P.; Cros, A.; Sarkizova, S.; Tang-Huau, T.L.; Bohec, M.; Baulande, S.; Hacohen, N.; et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity 2017, 47, 582–596.e6. [Google Scholar] [CrossRef]
- Cassetta, L.; Fragkogianni, S.; Sims, A.H.; Swierczak, A.; Forrester, L.M.; Zhang, H.; Soong, D.Y.H.; Cotechini, T.; Anur, P.; Lin, E.Y.; et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 2019, 35, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Haderk, F.; Schulz, R.; Iskar, M.; Cid, L.L.; Worst, T.; Willmund, K.V.; Schulz, A.; Warnken, U.; Seiler, J.; Benner, A.; et al. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immunol. 2017, 2, eaah5509. [Google Scholar] [CrossRef] [PubMed]
- Cocozza, F.; Grisard, E.; Martin-Jaular, L.; Mathieu, M.; Thery, C. SnapShot: Extracellular Vesicles. Cell 2020, 182, 262–262.e1. [Google Scholar] [CrossRef] [PubMed]
- Kugeratski, F.G.; Kalluri, R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021, 288, 10–35. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Kowal, J.; Thery, C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160479. [Google Scholar] [CrossRef]
- Tkach, M.; Thalmensi, J.; Timperi, E.; Gueguen, P.; Nevo, N.; Grisard, E.; Sirven, P.; Cocozza, F.; Gouronnec, A.; Martin-Jaular, L.; et al. Extracellular vesicles from triple negative breast cancer promote pro-inflammatory macrophages associated with better clinical outcome. Proc. Natl. Acad. Sci. USA 2022, 119, e2107394119. [Google Scholar] [CrossRef]
- Wen, S.W.; Sceneay, J.; Lima, L.G.; Wong, C.S.; Becker, M.; Krumeich, S.; Lobb, R.J.; Castillo, V.; Wong, K.N.; Ellis, S.; et al. The Biodistribution and Immune Suppressive Effects of Breast Cancer-Derived Exosomes. Cancer Res. 2016, 76, 6816–6827. [Google Scholar] [CrossRef]
- Wieckowski, E.U.; Visus, C.; Szajnik, M.; Szczepanski, M.J.; Storkus, W.J.; Whiteside, T.L. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J. Immunol. 2009, 183, 3720–3730. [Google Scholar] [CrossRef]
- Dieterich, L.C. Mechanisms of extracellular vesicle-mediated immune evasion in melanoma. Front. Immunol. 2022, 13, 1002551. [Google Scholar] [CrossRef]
- Shen, Y.; Guo, D.; Weng, L.; Wang, S.; Ma, Z.; Yang, Y.; Wang, P.; Wang, J.; Cai, Z. Tumor-derived exosomes educate dendritic cells to promote tumor metastasis via HSP72/HSP105-TLR2/TLR4 pathway. Oncoimmunology 2017, 6, e1362527. [Google Scholar] [CrossRef] [PubMed]
- Ashiru, O.; Boutet, P.; Fernandez-Messina, L.; Aguera-Gonzalez, S.; Skepper, J.N.; Vales-Gomez, M.; Reyburn, H.T. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 2010, 70, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. BCR 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Kowal, J.; Zucchetti, A.E.; Enserink, L.; Jouve, M.; Lankar, D.; Saitakis, M.; Martin-Jaular, L.; Thery, C. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017, 36, 3012–3028. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef]
- Denton, A.E.; Roberts, E.W.; Fearon, D.T. Stromal Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2018, 1060, 99–114. [Google Scholar] [CrossRef]
- LeBleu, V.S.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Models Mech. 2018, 11, dmm029447. [Google Scholar] [CrossRef]
- Ziani, L.; Chouaib, S.; Thiery, J. Alteration of the Antitumor Immune Response by Cancer-Associated Fibroblasts. Front. Immunol. 2018, 9, 414. [Google Scholar] [CrossRef]
- Ohlund, D.; Elyada, E.; Tuveson, D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014, 211, 1503–1523. [Google Scholar] [CrossRef]
- Kanzaki, R.; Pietras, K. Heterogeneity of cancer-associated fibroblasts: Opportunities for precision medicine. Cancer Sci. 2020, 111, 2708–2717. [Google Scholar] [CrossRef]
- Brechbuhl, H.M.; Finlay-Schultz, J.; Yamamoto, T.M.; Gillen, A.E.; Cittelly, D.M.; Tan, A.C.; Sams, S.B.; Pillai, M.M.; Elias, A.D.; Robinson, W.A.; et al. Fibroblast Subtypes Regulate Responsiveness of Luminal Breast Cancer to Estrogen. Clin. Cancer Res. 2017, 23, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jia, H.H.; Xu, Y.Q.; Zhou, X.; Zhao, X.H.; Wang, Y.F.; Song, X.; Zhu, Z.Y.; Sun, T.; Dou, Y.; et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ss1 secretion. Mol. Cancer 2018, 17, 5. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, C.H.; Tan, L.D.; Wang, Q.S.; Li, X.Q.; Feng, Y.M. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br. J. Cancer 2014, 110, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wei, H.; Wang, J.; Li, L.; Chen, A.; Li, Z. MicroRNA-181d-5p-Containing Exosomes Derived from CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol. Ther. Nucleic Acids 2020, 19, 654–667. [Google Scholar] [CrossRef]
- Chen, B.; Sang, Y.; Song, X.; Zhang, D.; Wang, L.; Zhao, W.; Liang, Y.; Zhang, N.; Yang, Q. Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics 2021, 11, 3932–3947. [Google Scholar] [CrossRef]
- Gao, Y.; Li, X.; Zeng, C.; Liu, C.; Hao, Q.; Li, W.; Zhang, K.; Zhang, W.; Wang, S.; Zhao, H.; et al. CD63(+) Cancer-Associated Fibroblasts Confer Tamoxifen Resistance to Breast Cancer Cells through Exosomal miR-22. Adv. Sci. 2020, 7, 2002518. [Google Scholar] [CrossRef]
- Zhan, Y.; Du, J.; Min, Z.; Ma, L.; Zhang, W.; Zhu, W.; Liu, Y. Carcinoma-associated fibroblasts derived exosomes modulate breast cancer cell stemness through exonic circHIF1A by miR-580-5p in hypoxic stress. Cell Death Discov. 2021, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, M.; Principe, S.; Jackson, H.W.; Luga, V.; Fang, H.; Molyneux, S.D.; Shao, Y.W.; Aiken, A.; Waterhouse, P.D.; Karamboulas, C.; et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 2014, 16, 889–901. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Liu, W.; Li, X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl. Biochem. Biotechnol. 2020, 191, 1084–1099. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Lux, M.P.; Fasching, P.A.; Schrauder, M.G.; Hein, A.; Jud, S.M.; Rauh, C.; Beckmann, M.W. The PI3K Pathway: Background and Treatment Approaches. Breast Care 2016, 11, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X. Cancer-Associated Fibroblasts-Derived Exosomes Suppress Immune Cell Function in Breast Cancer via the miR-92/PD-L1 Pathway. Front. Immunol. 2020, 11, 2026. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kim, C.G.; Kim, S.K.; Shin, S.J.; Choe, E.A.; Park, S.H.; Shin, E.C.; Kim, J. YAP-Induced PD-L1 Expression Drives Immune Evasion in BRAFi-Resistant Melanoma. Cancer Immunol. Res. 2018, 6, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhou, X.; Wang, J.; Liu, Z.; Han, S.; Wan, L.; Sun, X.; Chen, H. Adipose-derived mesenchymal stem cells and extracellular vesicles confer antitumor activity in preclinical treatment of breast cancer. Pharmacol. Res. 2020, 157, 104843. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Guo, J.; Yu, L.; Guo, T.; Wang, J.; Wang, X.; Chen, Y. PD-L1(+) exosomes from bone marrow-derived cells of tumor-bearing mice inhibit antitumor immunity. Cell. Mol. Immunol. 2021, 18, 2402–2409. [Google Scholar] [CrossRef]
- Romagnoli, G.G.; Zelante, B.B.; Toniolo, P.A.; Migliori, I.K.; Barbuto, J.A. Dendritic Cell-Derived Exosomes may be a Tool for Cancer Immunotherapy by Converting Tumor Cells into Immunogenic Targets. Front. Immunol. 2014, 5, 692. [Google Scholar] [CrossRef]
- Kaban, K.; Hinterleitner, C.; Zhou, Y.; Salva, E.; Kantarci, A.G.; Salih, H.R.; Marklin, M. Therapeutic Silencing of BCL-2 Using NK Cell-Derived Exosomes as a Novel Therapeutic Approach in Breast Cancer. Cancers 2021, 13, 2397. [Google Scholar] [CrossRef]
- Dallavalle, S.; Dobricic, V.; Lazzarato, L.; Gazzano, E.; Machuqueiro, M.; Pajeva, I.; Tsakovska, I.; Zidar, N.; Fruttero, R. Improvement of conventional anti-cancer drugs as new tools against multidrug resistant tumors. Drug Resist. Updates 2020, 50, 100682. [Google Scholar] [CrossRef]
- Battista, T.; Fiorillo, A.; Chiarini, V.; Genovese, I.; Ilari, A.; Colotti, G. Roles of Sorcin in Drug Resistance in Cancer: One Protein, Many Mechanisms, for a Novel Potential Anticancer Drug Target. Cancers 2020, 12, 887. [Google Scholar] [CrossRef]
- Navas, T.; Kinders, R.J.; Lawrence, S.M.; Ferry-Galow, K.V.; Borgel, S.; Hollingshead, M.G.; Srivastava, A.K.; Alcoser, S.Y.; Makhlouf, H.R.; Chuaqui, R.; et al. Clinical Evolution of Epithelial-Mesenchymal Transition in Human Carcinomas. Cancer Res. 2020, 80, 304–318. [Google Scholar] [CrossRef]
- Garcia-Mayea, Y.; Mir, C.; Masson, F.; Paciucci, R.; ME, L.L. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Semin. Cancer Biol. 2020, 60, 166–180. [Google Scholar] [CrossRef]
- Gong, J.; Jaiswal, R.; Dalla, P.; Luk, F.; Bebawy, M. Microparticles in cancer: A review of recent developments and the potential for clinical application. Semin. Cell Dev. Biol. 2015, 40, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Fremder, E.; Munster, M.; Aharon, A.; Miller, V.; Gingis-Velitski, S.; Voloshin, T.; Alishekevitz, D.; Bril, R.; Scherer, S.J.; Loven, D.; et al. Tumor-derived microparticles induce bone marrow-derived cell mobilization and tumor homing: A process regulated by osteopontin. Int. J. Cancer 2014, 135, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Barenholz-Cohen, T.; Merkher, Y.; Haj, J.; Shechter, D.; Kirchmeier, D.; Shaked, Y.; Weihs, D. Lung mechanics modifications facilitating metastasis are mediated in part by breast cancer-derived extracellular vesicles. Int. J. Cancer 2020, 147, 2924–2933. [Google Scholar] [CrossRef] [PubMed]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of small molecules in vesicles shed by cancer cells: Association with gene expression and chemosensitivity profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar] [PubMed]
- Locher, K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016, 23, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Nedeljkovic, M.; Damjanovic, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Ambudkar, S.V.; Kimchi-Sarfaty, C.; Sauna, Z.E.; Gottesman, M.M. P-glycoprotein: From genomics to mechanism. Oncogene 2003, 22, 7468–7485. [Google Scholar] [CrossRef] [PubMed]
- Bebawy, M.; Combes, V.; Lee, E.; Jaiswal, R.; Gong, J.; Bonhoure, A.; Grau, G.E. Membrane microparticles mediate transfer of P-glycoprotein to drug sensitive cancer cells. Leukemia 2009, 23, 1643–1649. [Google Scholar] [CrossRef]
- Levchenko, A.; Mehta, B.M.; Niu, X.; Kang, G.; Villafania, L.; Way, D.; Polycarpe, D.; Sadelain, M.; Larson, S.M. Intercellular transfer of P-glycoprotein mediates acquired multidrug resistance in tumor cells. Proc. Natl. Acad. Sci. USA 2005, 102, 1933–1938. [Google Scholar] [CrossRef]
- Sousa, D.; Lima, R.T.; Vasconcelos, M.H. Intercellular Transfer of Cancer Drug Resistance Traits by Extracellular Vesicles. Trends Mol. Med. 2015, 21, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.M.; Zhu, X.Y.; Chen, W.X.; Zhong, S.L.; Hu, Q.; Ma, T.F.; Zhang, J.; Chen, L.; Tang, J.H.; Zhao, J.H. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumour. Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Cai, Y.; He, D.; Zou, C.; Zhang, P.; Lo, C.Y.; Xu, Z.; Chan, F.L.; Yu, S.; Chen, Y.; et al. Transient receptor potential channel TRPC5 is essential for P-glycoprotein induction in drug-resistant cancer cells. Proc. Natl. Acad. Sci. USA 2012, 109, 16282–16287. [Google Scholar] [CrossRef]
- Wang, T.; Ning, K.; Lu, T.X.; Sun, X.; Jin, L.; Qi, X.; Jin, J.; Hua, D. Increasing circulating exosomes-carrying TRPC5 predicts chemoresistance in metastatic breast cancer patients. Cancer Sci. 2017, 108, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, Z.; Hua, D.; He, D.; Wang, L.; Zhang, P.; Wang, J.; Cai, Y.; Gao, C.; Zhang, X.; et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 6389–6394. [Google Scholar] [CrossRef]
- Santos, J.C.; Lima, N.D.S.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef]
- Coles, B.F.; Kadlubar, F.F. Detoxification of electrophilic compounds by glutathione S-transferase catalysis: Determinants of individual response to chemical carcinogens and chemotherapeutic drugs? Biofactors 2003, 17, 115–130. [Google Scholar] [CrossRef]
- Safaei, R.; Larson, B.J.; Cheng, T.C.; Gibson, M.A.; Otani, S.; Naerdemann, W.; Howell, S.B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol. Cancer Ther. 2005, 4, 1595–1604. [Google Scholar] [CrossRef]
- Pompili, S.; Vetuschi, A.; Sferra, R.; Cappariello, A. Extracellular Vesicles and Resistance to Anticancer Drugs: A Tumor Skeleton Key for Unhinging Chemotherapies. Front. Oncol. 2022, 12, 933675. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben David, G.; Shlomi, T.; Gil, Z. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.R.; Johnston, P.G.; Longley, D.B. Anti-apoptotic mechanisms of drug resistance in cancer. Curr. Cancer Drug Targets 2009, 9, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.Y.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.J.; Ye, M.L.; He, X.H.; Zhang, F.; Han, J. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018, 53, 1013–1026. [Google Scholar] [CrossRef]
- Hellwinkel, J.E.; Redzic, J.S.; Harland, T.A.; Gunaydin, D.; Anchordoquy, T.J.; Graner, M.W. Glioma-derived extracellular vesicles selectively suppress immune responses. Neuro Oncol. 2016, 18, 497–506. [Google Scholar] [CrossRef]
- Battke, C.; Ruiss, R.; Welsch, U.; Wimberger, P.; Lang, S.; Jochum, S.; Zeidler, R. Tumour exosomes inhibit binding of tumour-reactive antibodies to tumour cells and reduce ADCC. Cancer Immunol. Immunother. 2011, 60, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Ciravolo, V.; Huber, V.; Ghedini, G.C.; Venturelli, E.; Bianchi, F.; Campiglio, M.; Morelli, D.; Villa, A.; Della Mina, P.; Menard, S.; et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol. 2012, 227, 658–667. [Google Scholar] [CrossRef]
- Martinez, V.G.; O’Neill, S.; Salimu, J.; Breslin, S.; Clayton, A.; Crown, J.; O’Driscoll, L. Resistance to HER2-targeted anti-cancer drugs is associated with immune evasion in cancer cells and their derived extracellular vesicles. Oncoimmunology 2017, 6, e1362530. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef]
- Van den Boorn, J.G.; Schlee, M.; Coch, C.; Hartmann, G. SiRNA delivery with exosome nanoparticles. Nat. Biotechnol. 2011, 29, 325–326. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Li, Z.; Zhu, J.; Wang, P.; Fan, G.; Dai, Y.; Zheng, Z.; Liu, Y. Serum expression levels of microRNA-382-3p, -598-3p, -1246 and -184 in breast cancer patients. Oncol. Lett. 2016, 12, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef]
- Detchokul, S.; Williams, E.D.; Parker, M.W.; Frauman, A.G. Tetraspanins as regulators of the tumour microenvironment: Implications for metastasis and therapeutic strategies. Br. J. Pharmacol. 2014, 171, 5462–5490. [Google Scholar] [CrossRef]
- Galindo-Hernandez, O.; Villegas-Comonfort, S.; Candanedo, F.; Gonzalez-Vazquez, M.C.; Chavez-Ocana, S.; Jimenez-Villanueva, X.; Sierra-Martinez, M.; Salazar, E.P. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch. Med. Res. 2013, 44, 208–214. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, W.; Bu, J.; Li, Y.; Li, R.; Nie, R.; Xiao, C.; Ma, K.; Huang, X.; Li, Y. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol. Carcinog. 2019, 58, 674–685. [Google Scholar] [CrossRef]
- Ando, W.; Kikuchi, K.; Uematsu, T.; Yokomori, H.; Takaki, T.; Sogabe, M.; Kohgo, Y.; Otori, K.; Ishikawa, S.; Okazaki, I. Novel breast cancer screening: Combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci. Rep. 2019, 9, 13595. [Google Scholar] [CrossRef]
- Rupp, A.K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marme, F.; Sultmann, H.; Altevogt, P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011, 122, 437–446. [Google Scholar] [CrossRef]
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Jung, J.H.; Chae, Y.S.; Bae, H.I.; Kim, Y.B.; Kim, I.S.; Park, H.Y.; et al. Identification of Developmental Endothelial Locus-1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin. Cancer Res. 2016, 22, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Chae, Y.S.; Jung, J.H.; Kim, I.S.; Park, H.Y.; Baek, M.C. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget 2016, 7, 40189–40199. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Bennit, H.F.; Turay, D.; Perez, M.; Mirshahidi, S.; Yuan, Y.; Wall, N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; McGee, A.R.; Kaur, K.; Thundat, T. Nanomechanical sandwich assay for multiple cancer biomarkers in breast cancer cell-derived exosomes. Nanoscale 2016, 8, 15137–15141. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Gibbs, L.D.; Maji, S.; Lewis, C.M.; Suzuki, S.; Vishwanatha, J.K. Serum exosomal-annexin A2 is associated with African-American triple-negative breast cancer and promotes angiogenesis. Breast Cancer Res. 2020, 22, 11. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, B.G.; Jang, Y.; Kang, S.; Lee, J.H.; Cho, N.H. The stromal loss of miR-4516 promotes the FOSL1-dependent proliferation and malignancy of triple negative breast cancer. Cancer Lett. 2020, 469, 256–265. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef]
- Batrakova, E.V.; Kim, M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control. Release 2015, 219, 396–405. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, K.E.; Dwyer, R.M. Engineering Exosomes for Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 1122. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Amreddy, N.; Razaq, M.; Towner, R.; Zhao, Y.D.; Ahmed, R.A.; Munshi, A.; Ramesh, R. Exosomes as Theranostics for Lung Cancer. Adv. Cancer Res. 2018, 139, 1–33. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Vader, P.; Mol, E.A.; Pasterkamp, G.; Schiffelers, R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. [Google Scholar] [CrossRef]
- Toffoli, G.; Hadla, M.; Corona, G.; Caligiuri, I.; Palazzolo, S.; Semeraro, S.; Gamini, A.; Canzonieri, V.; Rizzolio, F. Exosomal doxorubicin reduces the cardiac toxicity of doxorubicin. Nanomedicine 2015, 10, 2963–2971. [Google Scholar] [CrossRef]
- Yerneni, S.S.; Lathwal, S.; Shrestha, P.; Shirwan, H.; Matyjaszewski, K.; Weiss, L.; Yolcu, E.S.; Campbell, P.G.; Das, S.R. Rapid On-Demand Extracellular Vesicle Augmentation with Versatile Oligonucleotide Tethers. ACS Nano 2019, 13, 10555–10565. [Google Scholar] [CrossRef]
- Raghav, K.P.; Wang, W.; Liu, S.; Chavez-MacGregor, M.; Meng, X.; Hortobagyi, G.N.; Mills, G.B.; Meric-Bernstam, F.; Blumenschein, G.R., Jr.; Gonzalez-Angulo, A.M. cMET and phospho-cMET protein levels in breast cancers and survival outcomes. Clin. Cancer Res. 2012, 18, 2269–2277. [Google Scholar] [CrossRef]
- Li, S.; Wu, Y.; Ding, F.; Yang, J.; Li, J.; Gao, X.; Zhang, C.; Feng, J. Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer. Nanoscale 2020, 12, 10854–10862. [Google Scholar] [CrossRef]
- Bahrami, A.; Aledavood, A.; Anvari, K.; Hassanian, S.M.; Maftouh, M.; Yaghobzade, A.; Salarzaee, O.; ShahidSales, S.; Avan, A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J. Cell. Physiol. 2018, 233, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.S.; Kang, C.W.; Kang, S.; Jang, Y.; Chae, Y.C.; Kim, B.G.; Cho, N.H. ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 2020, 39, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Pando, A.; Reagan, J.L.; Quesenberry, P.; Fast, L.D. Extracellular vesicles in leukemia. Leuk. Res. 2018, 64, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Wu, G.; Zhang, J.; Huang, L.L.; Ding, J.; Jiang, A.; Zhang, Y.; Liu, Y.; Li, J.; Pu, K.; et al. Responsive Exosome Nano-bioconjugates for Synergistic Cancer Therapy. Angew. Chem. Int. Ed. Engl. 2020, 59, 2018–2022. [Google Scholar] [CrossRef] [PubMed]
- Gorji-Bahri, G.; Hashemi, A.; Moghimi, H.R. ExomiRs: A Novel Strategy in Cancer Diagnosis and Therapy. Curr. Gene Ther. 2018, 18, 336–350. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Michael, I.J.; Park, J.; Granick, S.; Cho, Y.K. Cloaked Exosomes: Biocompatible, Durable, and Degradable Encapsulation. Small 2018, 14, e1802052. [Google Scholar] [CrossRef]
- Yu, M.; Gai, C.; Li, Z.; Ding, D.; Zheng, J.; Zhang, W.; Lv, S.; Li, W. Targeted exosome-encapsulated erastin induced ferroptosis in triple negative breast cancer cells. Cancer Sci. 2019, 110, 3173–3182. [Google Scholar] [CrossRef]
- Barok, M.; Puhka, M.; Vereb, G.; Szollosi, J.; Isola, J.; Joensuu, H. Cancer-derived exosomes from HER2-positive cancer cells carry trastuzumab-emtansine into cancer cells leading to growth inhibition and caspase activation. BMC Cancer 2018, 18, 504. [Google Scholar] [CrossRef]
- Shen, M.; Ren, X. New insights into the biological impacts of immune cell-derived exosomes within the tumor environment. Cancer Lett. 2018, 431, 115–122. [Google Scholar] [CrossRef]
- Viaud, S.; Terme, M.; Flament, C.; Taieb, J.; Andre, F.; Novault, S.; Escudier, B.; Robert, C.; Caillat-Zucman, S.; Tursz, T.; et al. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: A role for NKG2D ligands and IL-15Ralpha. PLoS ONE 2009, 4, e4942. [Google Scholar] [CrossRef]
- Bell, B.M.; Kirk, I.D.; Hiltbrunner, S.; Gabrielsson, S.; Bultema, J.J. Designer exosomes as next-generation cancer immunotherapy. Nanomedicine 2016, 12, 163–169. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Sandiford, O.A.; Donnelly, R.J.; El-Far, M.H.; Burgmeyer, L.M.; Sinha, G.; Pamarthi, S.H.; Sherman, L.S.; Ferrer, A.I.; DeVore, D.E.; Patel, S.A.; et al. Mesenchymal Stem Cell-Secreted Extracellular Vesicles Instruct Stepwise Dedifferentiation of Breast Cancer Cells into Dormancy at the Bone Marrow Perivascular Region. Cancer Res. 2021, 81, 1567–1582. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Mandal, G.; Roy Chowdhury, S.; Purohit, S.; Payne, K.K.; Anadon, C.; Gupta, A.; Swanson, P.; Yu, X.; Conejo-Garcia, J.R.; et al. Exosomes Produced by Mesenchymal Stem Cells Drive Differentiation of Myeloid Cells into Immunosuppressive M2-Polarized Macrophages in Breast Cancer. J. Immunol. 2019, 203, 3447–3460. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Huang, Q.; Peng, C.; Yao, L.; Chen, H.; Qiu, Z.; Wu, Y.; Wang, L.; Chen, W. Exosomes from M1-Polarized Macrophages Enhance Paclitaxel Antitumor Activity by Activating Macrophages-Mediated Inflammation. Theranostics 2019, 9, 1714–1727. [Google Scholar] [CrossRef]
- Rayamajhi, S.; Nguyen, T.D.T.; Marasini, R.; Aryal, S. Macrophage-derived exosome-mimetic hybrid vesicles for tumor targeted drug delivery. Acta Biomater. 2019, 94, 482–494. [Google Scholar] [CrossRef]
- Liu, M.; Hu, Y.; Chen, G. The Antitumor Effect of Gene-Engineered Exosomes in the Treatment of Brain Metastasis of Breast Cancer. Front. Oncol. 2020, 10, 1453. [Google Scholar] [CrossRef]
- Huang, H.; Shao, L.; Chen, Y.; Tang, L.; Liu, T.; Li, J.; Zhu, H. Synergistic strategy with hyperthermia therapy based immunotherapy and engineered exosomes-liposomes targeted chemotherapy prevents tumor recurrence and metastasis in advanced breast cancer. Bioeng. Transl. Med. 2022, 7, e10284. [Google Scholar] [CrossRef]
- Kumar, D.N.; Chaudhuri, A.; Dehari, D.; Shekher, A.; Gupta, S.C.; Majumdar, S.; Krishnamurthy, S.; Singh, S.; Kumar, D.; Agrawal, A.K. Combination Therapy Comprising Paclitaxel and 5-Fluorouracil by Using Folic Acid Functionalized Bovine Milk Exosomes Improves the Therapeutic Efficacy against Breast Cancer. Life 2022, 12, 1143. [Google Scholar] [CrossRef]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. [Google Scholar] [CrossRef]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013, 21, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Cao, T.G.; Kang, J.H.; You, J.Y.; Kang, H.C.; Rhee, W.J.; Ko, Y.T.; Shim, M.S. Safe and Targeted Sonodynamic Cancer Therapy Using Biocompatible Exosome-Based Nanosonosensitizers. ACS Appl. Mater. Interfaces 2021, 13, 25575–25588. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.P.; Holme, M.N.; Stevens, M.M. Re-Engineering Extracellular Vesicles as Smart Nanoscale Therapeutics. ACS Nano 2017, 11, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Gu, C.; Gan, Y.; Shao, L.; Chen, H.; Zhu, H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control. Release 2020, 318, 1–15. [Google Scholar] [CrossRef]
- Shi, X.; Cheng, Q.; Hou, T.; Han, M.; Smbatyan, G.; Lang, J.E.; Epstein, A.L.; Lenz, H.J.; Zhang, Y. Genetically Engineered Cell-Derived Nanoparticles for Targeted Breast Cancer Immunotherapy. Mol. Ther. 2020, 28, 536–547. [Google Scholar] [CrossRef]
- Wang, K.; Ye, H.; Zhang, X.; Wang, X.; Yang, B.; Luo, C.; Zhao, Z.; Zhao, J.; Lu, Q.; Zhang, H.; et al. An exosome-like programmable-bioactivating paclitaxel prodrug nanoplatform for enhanced breast cancer metastasis inhibition. Biomaterials 2020, 257, 120224. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.; Guo, L.; Tanaka, Y.; Su, L. Tumor-Derived Small Extracellular Vesicles Involved in Breast Cancer Progression and Drug Resistance. Int. J. Mol. Sci. 2022, 23, 15236. https://doi.org/10.3390/ijms232315236
Feng L, Guo L, Tanaka Y, Su L. Tumor-Derived Small Extracellular Vesicles Involved in Breast Cancer Progression and Drug Resistance. International Journal of Molecular Sciences. 2022; 23(23):15236. https://doi.org/10.3390/ijms232315236
Chicago/Turabian StyleFeng, Lingyun, Lijuan Guo, Yoshimasa Tanaka, and Li Su. 2022. "Tumor-Derived Small Extracellular Vesicles Involved in Breast Cancer Progression and Drug Resistance" International Journal of Molecular Sciences 23, no. 23: 15236. https://doi.org/10.3390/ijms232315236
APA StyleFeng, L., Guo, L., Tanaka, Y., & Su, L. (2022). Tumor-Derived Small Extracellular Vesicles Involved in Breast Cancer Progression and Drug Resistance. International Journal of Molecular Sciences, 23(23), 15236. https://doi.org/10.3390/ijms232315236






