Mulberry Leaf Supplements Effecting Anti-Inflammatory Genes and Improving Obesity in Elderly Overweight Dogs
Abstract
:1. Introduction
2. Results
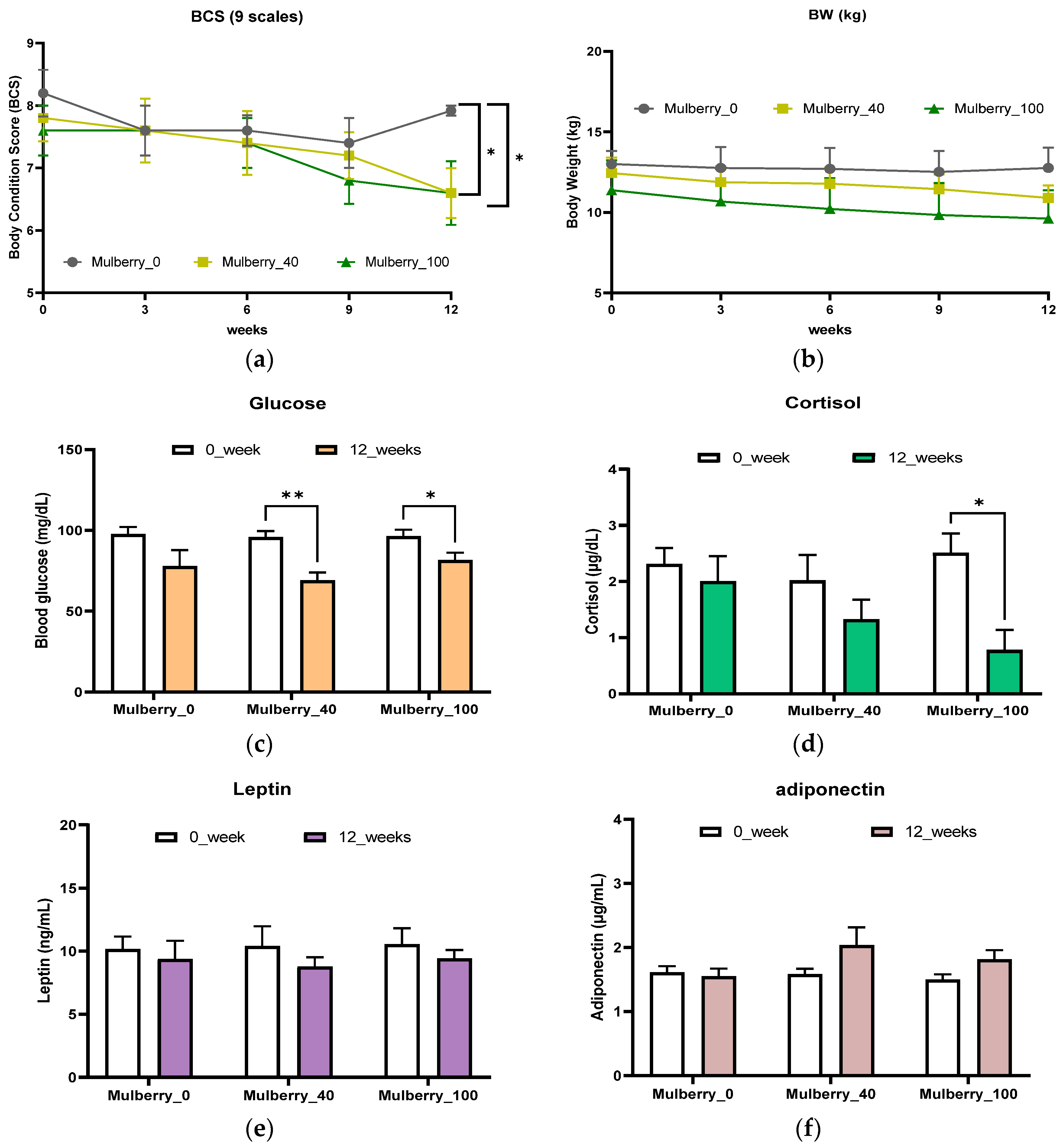
2.1. Body Condition Score (BCS) and Body Weight (BW)
2.2. Obesity-Related Factors in the Blood
2.3. Preprocessing and Alignment of Reads
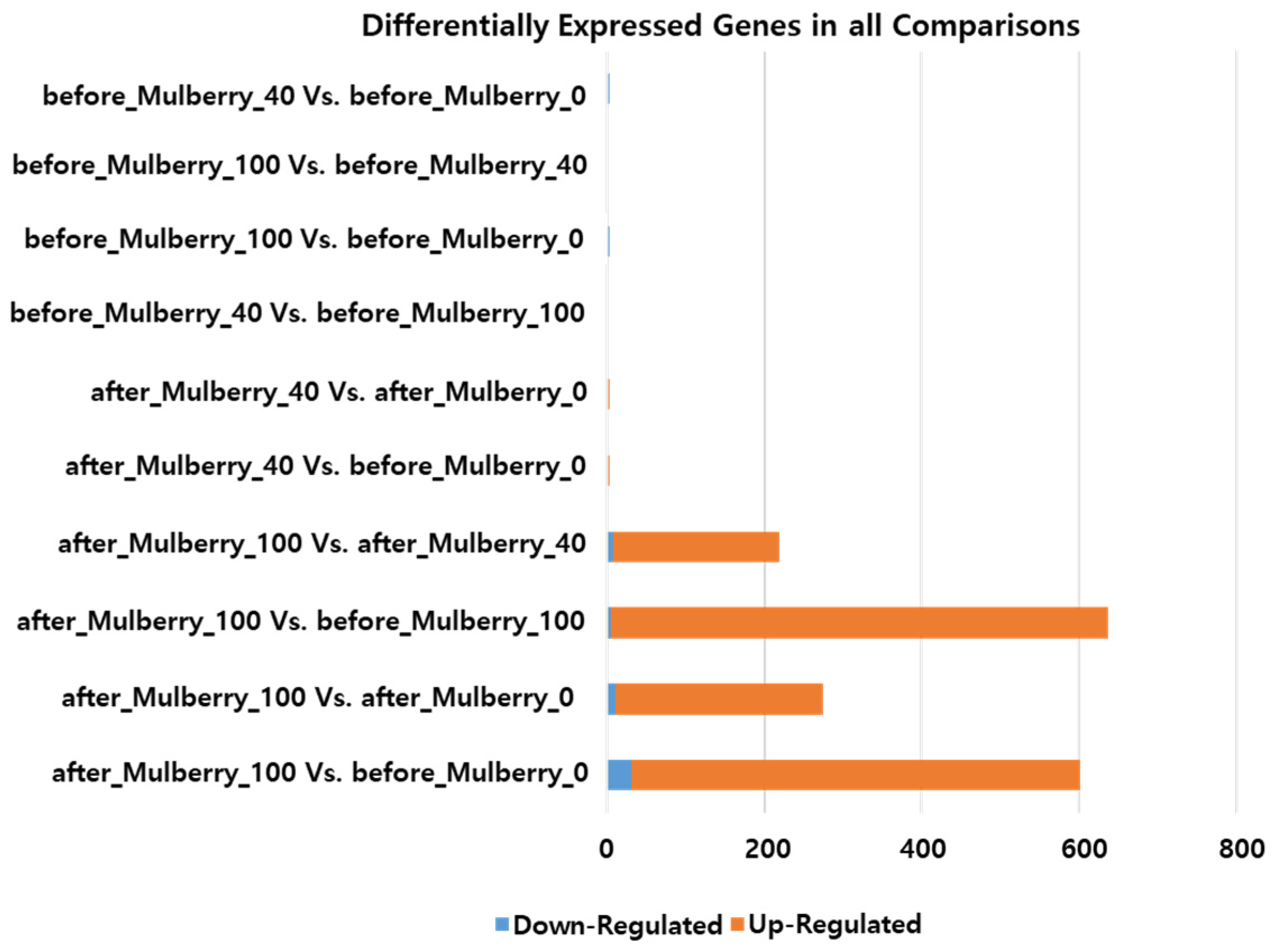
2.4. Assembly and Expression Analysis
2.5. Functional Enrichment Analysis and Obesity-Related Genes
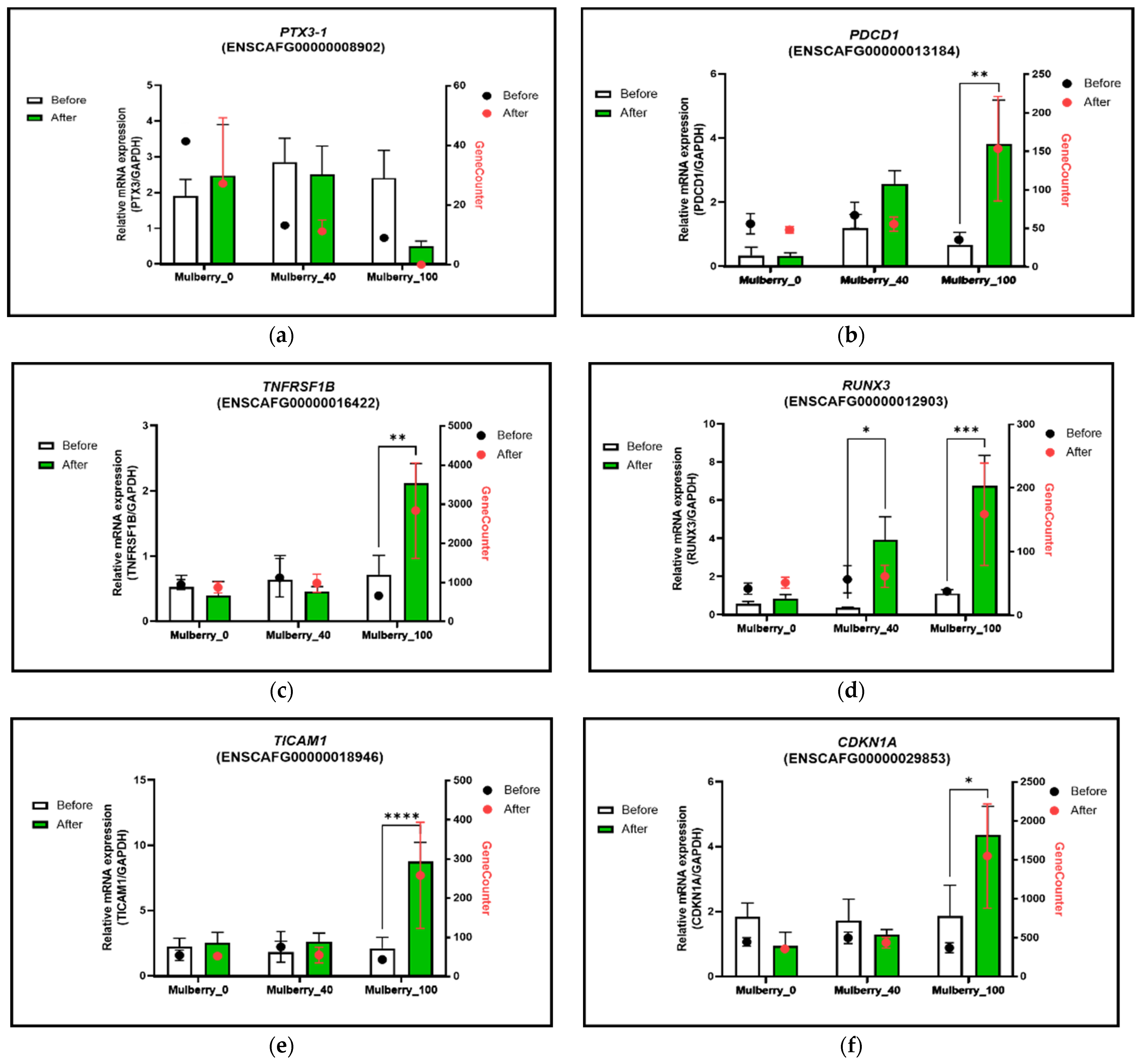
2.6. Real-Time PCR Analysis for Validation
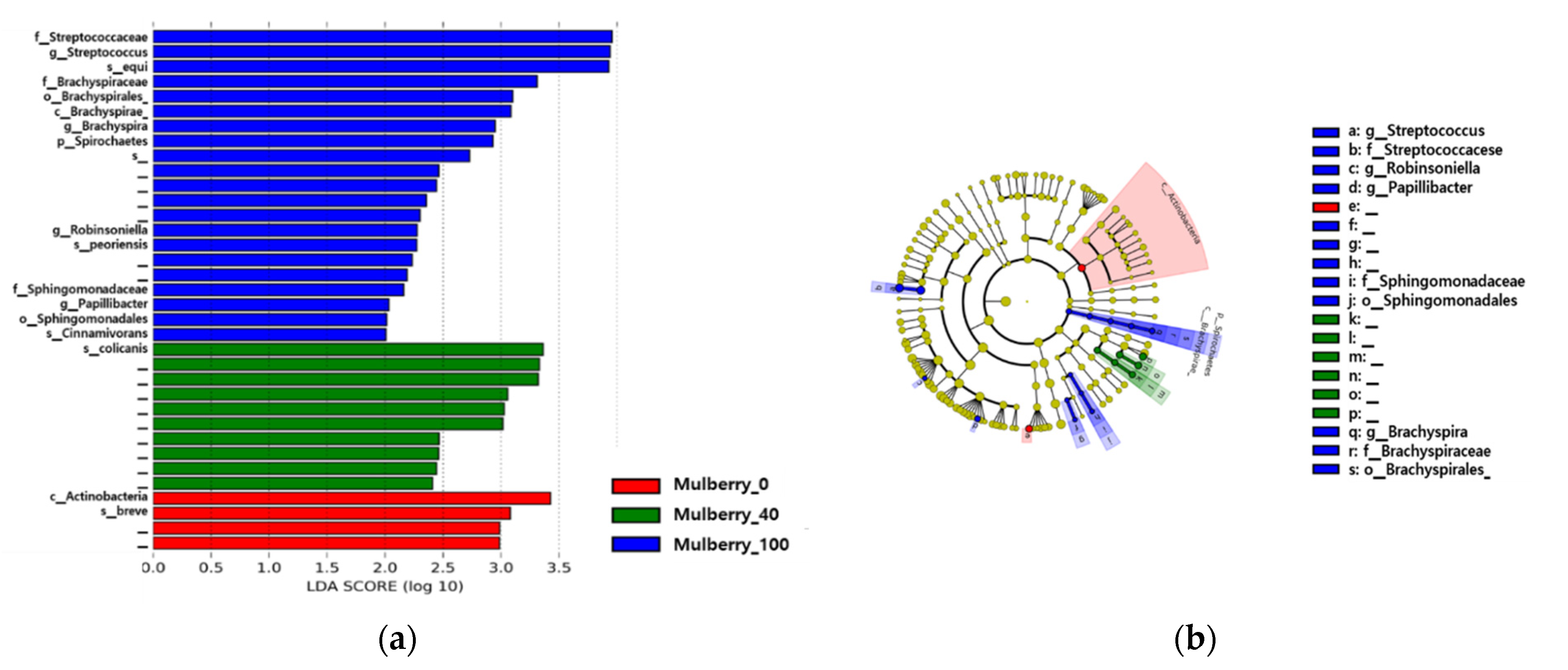
2.7. Gut Microbiome and Diversity Analysis
2.8. Taxonomy Analysis of the Samples
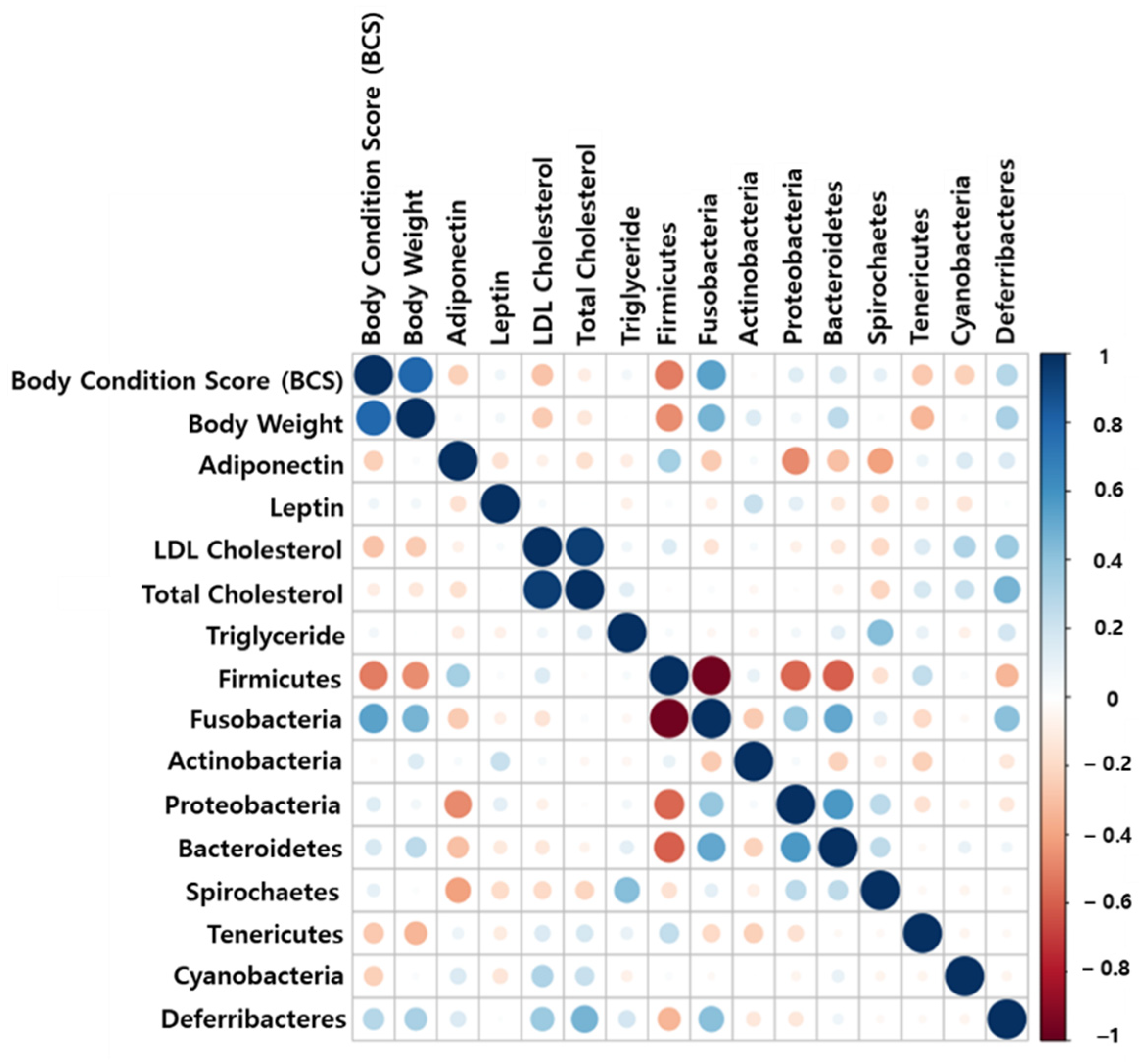
2.9. Correlation Analysis between Obesity-Related Factors and Gut Microbiome
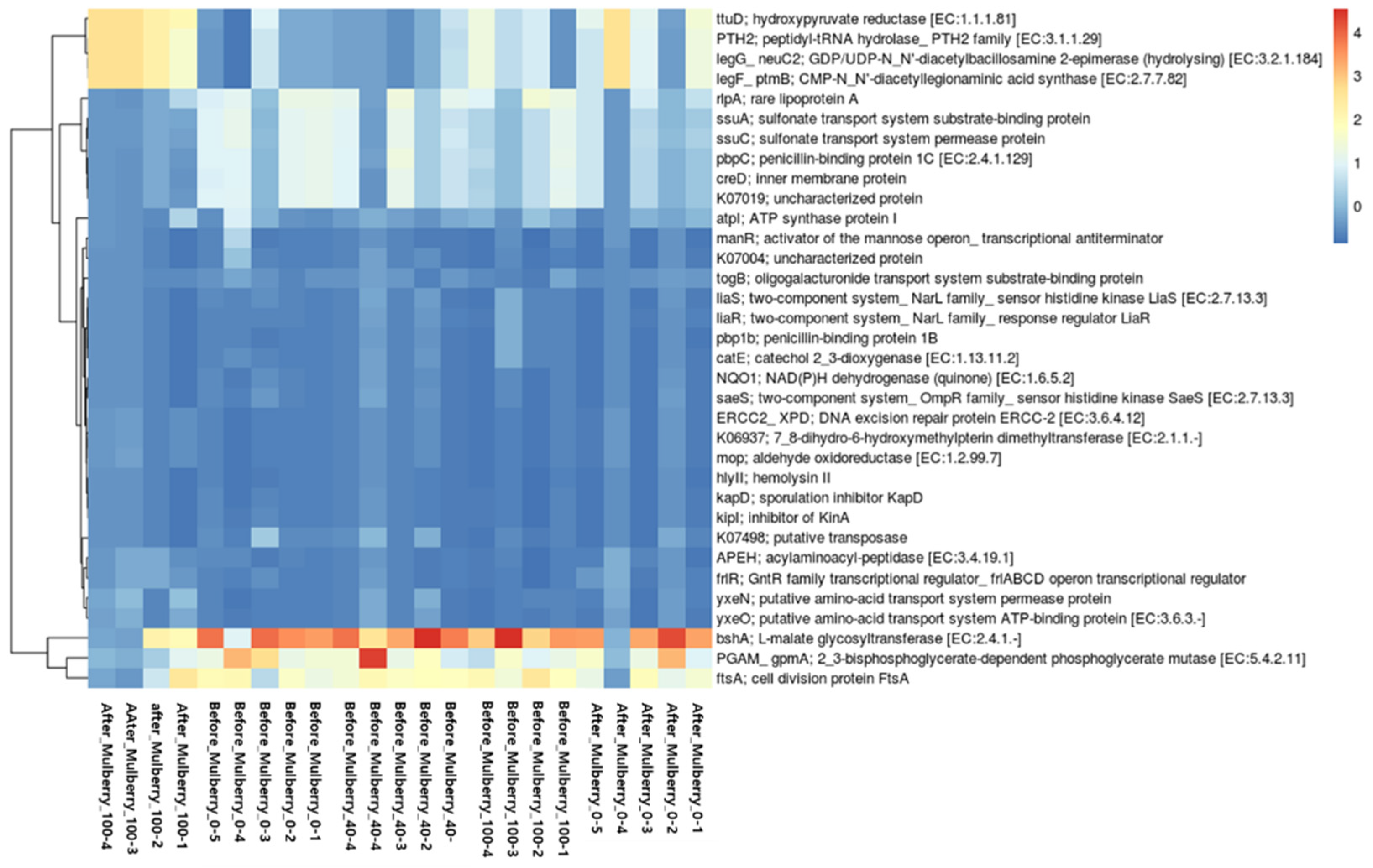
2.10. Functional Potential of Bacterial Community
3. Discussion
4. Materials and Methods
4.1. Animals and Diet
4.2. Blood Sampling, Serum Chemistry, and Fecal Sampling
4.3. RNA-Seq Pipeline Used for Assembly and Differential Expression Analysis
4.4. Comparison of DEGs in Different Groups
4.5. Functional Enrichment of DEGs and Identification of Obesity-Related Genes
4.6. Blood Gene Expression Using Real-Time PCR Analysis
4.7. Gut Microbiome Sequence Analysis
4.8. Taxonomic Annotation
4.9. Diversity Analysis
4.10. Differential Abundance of Taxa
4.11. Correlation Analysis between Obesity-Related Factors and Gut Microbiome
4.12. Prediction of Functional Potential of Bacterial Community with Amplicon Sequences
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Endalifer, M.L.; Diress, G. Epidemiology, Predisposing Factors, Biomarkers, and Prevention Mechanism of Obesity: A Systematic Review. J. Obes. 2020, 2020, 6134362. [Google Scholar] [CrossRef]
- Rosengren, A. Obesity and cardiovascular health: The size of the problem. Eur. Heart J. 2021, 42, 3404–3406. [Google Scholar] [CrossRef]
- Khwatenge, C.N.; Pate, M.; Miller, L.C.; Sang, Y. Immunometabolic Dysregulation at the Intersection of Obesity and COVID-19. Front. Immunol. 2021, 12, 732913. [Google Scholar] [CrossRef]
- Dhurandhar, N.V.; Bailey, D.; Thomas, D. Interaction of obesity and infections. Obes. Rev. 2015, 16, 1017–1029. [Google Scholar] [CrossRef]
- McGreevy, P.D.; Thomson, P.C.; Pride, C.; Fawcett, A.; Grassi, T.; Jones, B. Prevalence of obesity in dogs examined by Australian veterinary practices and the risk factors involved. Vet. Rec. 2005, 156, 695–702. [Google Scholar] [CrossRef]
- Park, M.; Kim, K.H.; Jaiswal, V.; Choi, J.; Chun, J.L.; Seo, K.M.; Lee, M.J.; Lee, H.J. Effect of black ginseng and silkworm supplementation on obesity, the transcriptome, and the gut microbiome of diet-induced overweight dogs. Sci. Rep. 2021, 11, 16334. [Google Scholar] [CrossRef]
- Cortese, L.; Terrazzano, G.; Pelagalli, A. Leptin and Immunological Profile in Obesity and Its Associated Diseases in Dogs. Int. J. Mol. Sci. 2019, 20, 2392. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Ballantyne, C.M. Metabolic Inflammation and Insulin Resistance in Obesity. Circ. Res. 2020, 126, 1549–1564. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Saltiel, A.R. Inflammatory links between obesity and metabolic disease. J. Clin. Investig. 2011, 121, 2111–2117. [Google Scholar] [CrossRef] [Green Version]
- Osborn, O.; Olefsky, J.M. The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 2012, 18, 363–374. [Google Scholar] [CrossRef]
- Berbudi, A.; Rahmadika, N.; Tjahjadi, A.I.; Ruslami, R. Type 2 Diabetes and its Impact on the Immune System. Curr. Diabetes Rev. 2020, 16, 442–449. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, L.; Huang, X.; Ding, X. Effect of 1-deoxynojirimycin water extract from mulberry leaves on lipid catabolism in high-fat fed mice. Acta Nutr. Sin. 2018, 40, 376–380. [Google Scholar]
- Zhang, R.; Zhang, Q.; Zhu, S.; Liu, B.; Liu, F.; Xu, Y. Mulberry leaf (Morus alba L.): A review of its potential influences in mechanisms of action on metabolic diseases. Pharmacol. Res. 2022, 175, 106029. [Google Scholar] [CrossRef]
- Li, Y.G.; Ji, D.F.; Zhong, S.; Lin, T.B.; Lv, Z.Q.; Hu, G.Y.; Wang, X. 1-deoxynojirimycin inhibits glucose absorption and accelerates glucose metabolism in streptozotocin-induced diabetic mice. Sci. Rep. 2013, 3, 1377. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yu, Z.; Jiang, J.; Li, Y.; Yu, S. Mulberry leaf attenuates atherosclerotic lesions in patients with coronary heart disease possibly via 1-Deoxynojirimycin: A placebo-controlled, double-blind clinical trial. J. Food Biochem. 2021, 45, e13573. [Google Scholar] [CrossRef]
- Wang, R.J.; Yang, C.H.; Hu, M.L. 1-Deoxynojirimycin inhibits metastasis of B16F10 melanoma cells by attenuating the activity and expression of matrix metalloproteinases-2 and -9 and altering cell surface glycosylation. J. Agric. Food Chem. 2010, 58, 8988–8993. [Google Scholar] [CrossRef]
- Kong, W.H.; Oh, S.H.; Ahn, Y.R.; Kim, K.W.; Kim, J.H.; Seo, S.W. Antiobesity effects and improvement of insulin sensitivity by 1-deoxynojirimycin in animal models. J. Agric. Food Chem. 2008, 56, 2613–2619. [Google Scholar] [CrossRef]
- Bharani, S.E.; Asad, M.; Dhamanigi, S.S.; Chandrakala, G.K. Immunomodulatory activity of methanolic extract of Morus alba Linn. (mulberry) leaves. Pak. J. Pharm. Sci. 2010, 23, 63–68. [Google Scholar]
- Xue, M.; Sun, H.; Cao, Y.; Wang, G.; Meng, Y.; Wang, D.; Hong, Y. Mulberry leaf polysaccharides modulate murine bone-marrow-derived dendritic cell maturation. Hum. Vaccines Immunother. 2015, 11, 946–950. [Google Scholar] [CrossRef] [Green Version]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef] [Green Version]
- Forrest, M.S.; Lan, Q.; Hubbard, A.E.; Zhang, L.; Vermeulen, R.; Zhao, X.; Li, G.; Wu, Y.Y.; Shen, M.; Yin, S.; et al. Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers. Environ. Health Perspect. 2005, 113, 801–807. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of diet on the gut microbiota: Rethinking intervention duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef]
- Liu, B.-N.; Liu, X.-T.; Liang, Z.-H.; Wang, J.-H. Gut microbiota in obesity. World J. Gastroenterol. 2021, 27, 3837. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [Green Version]
- Davis, C.D. The gut microbiome and its role in obesity. Nutr. Today 2016, 51, 167. [Google Scholar] [CrossRef] [Green Version]
- Stępień, M.; Stępień, A.; Wlazeł, R.N.; Paradowski, M.; Banach, M.; Rysz, J. Obesity indices and inflammatory markers in obese non-diabetic normo-and hypertensive patients: A comparative pilot study. Lipids Health Dis. 2014, 13, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lafontan, M. Fat cells: Afferent and efferent messages define new approaches to treat obesity. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 119–146. [Google Scholar] [CrossRef]
- Vendramini, T.H.A.; Macedo, H.T.; Amaral, A.R.; Rentas, M.F.; Macegoza, M.V.; Zafalon, R.V.A.; Pedrinelli, V.; Mesquita, L.G.; de Carvalho Balieiro, J.C.; Pfrimer, K. Gene expression of the immunoinflammatory and immunological status of obese dogs before and after weight loss. PLoS ONE 2020, 15, e0238638. [Google Scholar] [CrossRef]
- Quarta, C.; Clemmensen, C.; Zhu, Z.; Yang, B.; Joseph, S.S.; Lutter, D.; Yi, C.-X.; Graf, E.; García-Cáceres, C.; Legutko, B. Molecular integration of incretin and glucocorticoid action reverses immunometabolic dysfunction and obesity. Cell Metab. 2017, 26, 620–632.e6. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.-Y.; Huang, C.-C.; Hu, F.B.; Chavarro, J.E. Vegetarian diets and weight reduction: A meta-analysis of randomized controlled trials. J. Gen. Intern. Med. 2016, 31, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Kjeldsen-Kragh, J. Rheumatoid arthritis treated with vegetarian diets. Am. J. Clin. Nutr. 1999, 70, 594s–600s. [Google Scholar] [CrossRef] [Green Version]
- Menzel, J.; Jabakhanji, A.; Biemann, R.; Mai, K.; Abraham, K.; Weikert, C. Systematic review and meta-analysis of the associations of vegan and vegetarian diets with inflammatory biomarkers. Sci. Rep. 2020, 10, 21736. [Google Scholar] [CrossRef]
- Nigro, E.; Scudiero, O.; Monaco, M.L.; Palmieri, A.; Mazzarella, G.; Costagliola, C.; Bianco, A.; Daniele, A. New insight into adiponectin role in obesity and obesity-related diseases. BioMed Res. Int. 2014, 2014, 658913. [Google Scholar] [CrossRef]
- Obradovic, M.; Sudar-Milovanovic, E.; Soskic, S.; Essack, M.; Arya, S.; Stewart, A.J.; Gojobori, T.; Isenovic, E.R. Leptin and obesity: Role and clinical implication. Front. Endocrinol. 2021, 12, 585887. [Google Scholar] [CrossRef]
- Van der Valk, E.S.; Savas, M.; van Rossum, E.F. Stress and obesity: Are there more susceptible individuals? Curr. Obes. Rep. 2018, 7, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Chawla, A.; Stobdan, T.; Srivastava, R.B.; Jaiswal, V.; Chauhan, R.S.; Kant, A. Sex-biased temporal gene expression in male and female floral buds of seabuckthorn (Hippophae rhamnoides). PLoS ONE 2015, 10, e0124890. [Google Scholar] [CrossRef] [Green Version]
- Guleria, V.; Jaiswal, V. Comparative transcriptome analysis of different stages of Plasmodium falciparum to explore vaccine and drug candidates. Genomics 2020, 112, 796–804. [Google Scholar] [CrossRef]
- Jaiswal, V.; Park, M.; Lee, H.-J. Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-O-Glucoside-Treated Alzheimer’s Mouse Model. Antioxidants 2021, 10, 1435. [Google Scholar] [CrossRef]
- Jaiswal, V.; Cho, Y.-I.; Lee, H.-J. Preliminary study to explore the immune-enhancement mechanism of platycodon grandiflorus extract through comparative transcriptome analysis. Appl. Sci. 2020, 11, 226. [Google Scholar] [CrossRef]
- Syn, N.L.; Teng, M.W.; Mok, T.S.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Zhang, J.; Webster, J.D.; Dugger, D.L.; Goncharov, T.; Roose-Girma, M.; Hung, J.; Kwon, Y.C.; Vucic, D.; Newton, K.; Dixit, V.M. Ubiquitin ligases cIAP1 and cIAP2 limit cell death to prevent inflammation. Cell Rep. 2019, 27, 2679–2689.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Cui, H.; Xu, C.; Sun, Z.; Tang, Z.; Chen, H. RUNX3 protects against acute lung injury by inhibiting the JAK2/STAT3 pathway in rats with severe acute pancreatitis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5382–5391. [Google Scholar] [PubMed]
- Miyashita, Y.; Kouwaki, T.; Tsukamoto, H.; Okamoto, M.; Nakamura, K.; Oshiumi, H. TICAM-1/TRIF associates with Act1 and suppresses IL-17 receptor–mediated inflammatory responses. Life Sci. Alliance 2022, 5, e202101181. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Hayashi, S.; Fukuda, K.; Maeda, T.; Tsubosaka, M.; Kamenaga, T.; Kikuchi, K.; Fujita, M.; Kuroda, Y.; Hashimoto, S. Susceptibility of cyclin-dependent kinase inhibitor 1-deficient mice to rheumatoid arthritis arising from interleukin-1β-induced inflammation. Sci. Rep. 2021, 11, 12516. [Google Scholar] [CrossRef]
- Inoue, K.; Kodama, T.; Daida, H. Pentraxin 3: A novel biomarker for inflammatory cardiovascular disease. Int. J. Vasc. Med. 2012, 2012. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, W.-D.; Wang, Y.-D. The relationship between gut microbiota and inflammatory diseases: The role of macrophages. Front. Microbiol. 2020, 11, 1065. [Google Scholar] [CrossRef]
- Lay, C.; Rigottier-Gois, L.; Holmstrøm, K.; Rajilic, M.; Vaughan, E.E.; de Vos, W.M.; Collins, M.D.; Thiel, R.; Namsolleck, P.; Blaut, M. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 2005, 71, 4153–4155. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, A.; Ben-Jacob, N.; Tayem, H.; Halperin, E.; Iraqi, F.A.; Gophna, U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb. Ecol. 2011, 61, 423–428. [Google Scholar] [CrossRef]
- Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [CrossRef] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Reddy, K.E.; Kim, H.-R.; Jeong, J.Y.; So, K.-M.; Lee, S.; Ji, S.Y.; Kim, M.; Lee, H.-J.; Lee, S.; Kim, K.-H. Impact of breed on the fecal microbiome of dogs under the same dietary condition. J. Microbiol. Biotechnol. 2019, 29, 1947–1956. [Google Scholar] [CrossRef]
- Wang, D.D.; Qi, Q.; Wang, Z.; Usyk, M.; Sotres-Alvarez, D.; Mattei, J.; Tamez, M.; Gellman, M.D.; Daviglus, M.; Hu, F.B. The Gut Microbiome Modifies the Association Between a Mediterranean Diet and Diabetes in USA Hispanic/Latino Population. J. Clin. Endocrinol. Metab. 2022, 107, e924–e934. [Google Scholar] [CrossRef]
- Tsigalou, C.; Konstantinidis, T.; Paraschaki, A.; Stavropoulou, E.; Voidarou, C.; Bezirtzoglou, E. Mediterranean diet as a tool to combat inflammation and chronic diseases. An overview. Biomedicines 2020, 8, 201. [Google Scholar] [CrossRef]
- Laflamme, D. Development and validation of a body condition score system for dogs. Canine Pract. 1997, 22, 10–15. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [Green Version]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
| Group | Species | Gender | Age (y) | BW (kg) | BCS (9-Scale) |
|---|---|---|---|---|---|
| Mulberry_0 | Cocker Spaniels | F | 7 | 11.5 | 8 |
| Beagle | F | 9 | 16.5 | 9 | |
| Cocker Spaniels | F | 10 | 17.4 | 9 | |
| Cocker Spaniels | M | 10 | 12.1 | 8 | |
| Shetland Sheepdog | F | 6 | 11.5 | 7 | |
| Mulberry_40 | Beagle | M | 8 | 15.8 | 9 |
| Cocker Spaniels | M | 6 | 11.7 | 8 | |
| Cocker Spaniels | F | 9 | 10.0 | 7 | |
| Cocker Spaniels | M | 9 | 12.9 | 8 | |
| Shetland Sheepdog | F | 7 | 11.8 | 7 | |
| Mulberry_100 | Beagle | F | 9 | 17.9 | 9 |
| Cocker Spaniels | M | 8 | 12.2 | 8 | |
| Shetland Sheepdog | F | 6 | 10.8 | 7 | |
| Cocker Spaniels | F | 8 | 9.0 | 7 | |
| Cocker Spaniels | F | 9 | 11.5 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, M.; Jaiswal, V.; Kim, K.; Chun, J.; Lee, M.-J.; Shin, J.-H.; Lee, H.-J. Mulberry Leaf Supplements Effecting Anti-Inflammatory Genes and Improving Obesity in Elderly Overweight Dogs. Int. J. Mol. Sci. 2022, 23, 15215. https://doi.org/10.3390/ijms232315215
Park M, Jaiswal V, Kim K, Chun J, Lee M-J, Shin J-H, Lee H-J. Mulberry Leaf Supplements Effecting Anti-Inflammatory Genes and Improving Obesity in Elderly Overweight Dogs. International Journal of Molecular Sciences. 2022; 23(23):15215. https://doi.org/10.3390/ijms232315215
Chicago/Turabian StylePark, Miey, Varun Jaiswal, Kihyun Kim, Julan Chun, Mi-Jin Lee, Jae-Ho Shin, and Hae-Jeung Lee. 2022. "Mulberry Leaf Supplements Effecting Anti-Inflammatory Genes and Improving Obesity in Elderly Overweight Dogs" International Journal of Molecular Sciences 23, no. 23: 15215. https://doi.org/10.3390/ijms232315215
APA StylePark, M., Jaiswal, V., Kim, K., Chun, J., Lee, M.-J., Shin, J.-H., & Lee, H.-J. (2022). Mulberry Leaf Supplements Effecting Anti-Inflammatory Genes and Improving Obesity in Elderly Overweight Dogs. International Journal of Molecular Sciences, 23(23), 15215. https://doi.org/10.3390/ijms232315215







