The Role of Circular RNAs in the Physiology and Pathology of the Mammalian Ovary
Abstract
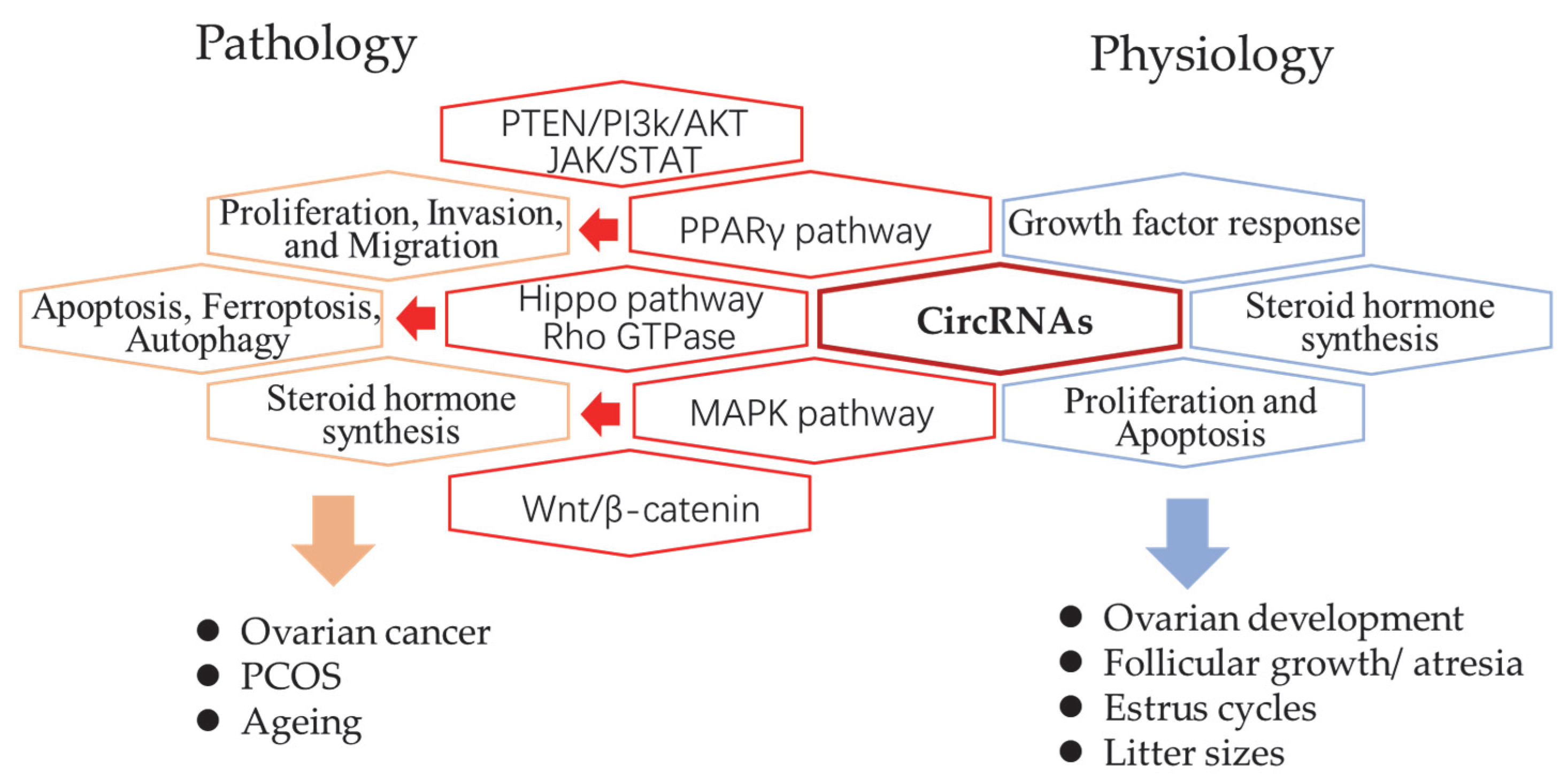
:1. Introduction
2. Characteristics, Functions and Detection of CircRNA
2.1. Stability, Abundance, Conservation, and Specificity of CircRNAs
2.2. Classification, Biogenesis, and Degradation of CircRNAs
2.3. Molecular Functions of CircRNAs
2.4. Detection and Confirmation of CircRNAs
3. CircRNAs in Mammalian Ovaries
3.1. CircRNAs in Ovarian Cancer
3.2. CircRNAs in PCOS
3.3. CircRNAs during Maternal Ageing
3.4. CircRNAs and Ovary Development
3.5. CircRNAs and Follicular Atresia
3.6. CircRNA and High Reproductive Traits
| Species | Tissue | CircRNA | Target miRNA/Gene/Protein | Function | Ref. |
|---|---|---|---|---|---|
| human | OC | Cdr1as | miR-1270/SCAI | sensitizes ovarian cancer to cisplatin | [54] |
| circ-ITCH | miR-145/RASA1 | inhibit tumour progression | [75] | ||
| has_circ_0051240 | miR-637/KLK4 | suppresses cell proliferation, migration, and invasion | [76] | ||
| circEPSTI1 | miR-942 | inhibit cell growth and invasion, induces apoptosis | [77] | ||
| circRNA CDR1 | miR-135b-5p/HIF1AN | decreasing the occurrence and progression of ovarian cancer | [78] | ||
| circLARP4 | down-regulated in cancerous ovarian cells | [79] | |||
| hsa_circ_0007444 | miR-23a-3p/DICER1 | [80] | |||
| circPLEKHM3 | miR-320a/SMG1 | exacerbated the effect of curcumin on ovarian cancer cell proliferation and apoptosis, as well as the anti-tumour effect | [81] | ||
| circABCB10 | miR-1271 | promotes cell proliferation and invasion but inhibits apoptosis | [82] | ||
| circRNA1656 | miR-1301-3p/miR-4660-SIRT3 | down-regulated in HGSOC | [53] | ||
| circ-CSPP1 | miR-1236-3p | promotes proliferation, invasion, and migration | [83] | ||
| has-circ-001567 | promotes cell proliferation and invasion | [84] | |||
| circ-SMAD7 | KLF6 | promotes cell proliferation and invasion | [85] | ||
| circ_0025033 | miR-184/LSM4 | promotes the progression of ovarian cancer | [86] | ||
| circHIPK3 | related to cell growth, migration, and apoptosis | [52] | |||
| PCOS | circ_0023942 | CDK-4 | inhibit granulosa cell proliferation | [87] | |
| circ_0043533 | miR-1179 | related to Bcl-2, CDK2, and Cyclin D1 | [88] | ||
| circ_RANBP9 | miRNA-136-5p/XIAP | exacerbates POS | [89] | ||
| circASPH | miR-375/MAP2K6 | promotes cells proliferation | [90] | ||
| circRHBG | miR-515/SLC7A11 | knockdown of circRHBG promotes ferroptosis in PCOS | [91] | ||
| circ_0005925 | miR-324-3p/MAP2K6 | Promotes Granulosa Cell Growth | [92] | ||
| circ_0043532 | miR-182/SGK3 | promote cell proliferation | [93] | ||
| ovary | circDDX10 | ovarian aging | [58] | ||
| KGN | circUSP36 | PTBP1/NEDD4L | enhance autophagic granulosa cell death | [94] | |
| GCs | circDDX10 | affecting the proliferation and apoptosis and steroid hormone synthesis | [95] | ||
| Pig | ovary | circ-TCP11 | miR-183 | associated with swine litter size | [70] |
| ovary | circSCIN | miR-133, miR-148a/b | affecting estrogen secretion | [71] | |
| GCs | ssc-circINHA-001 | miR-214-5p, miR-7144-3p, miR-9830-5p/INHBA | mediated Inhibin–Activin balance | [66] | |
| GCs | circSLC41A1 | miR-9820-5p/SRSF1 | resists porcine granulosa cell apoptosis and follicular atresia | [67] | |
| GCs | circ-ANKHD1 | miR-27a-3p/SFRP1 | decreased the cell apoptosis rates | [96] | |
| Bovine | GCs | circ_n/a_75 | miR-339a | growth factor response | [64] |
| circ_n/a_303 | miR-2400 and miR-30c | [64] | |||
| Goat | follicles | chi_circ_0008219 | miR-34c-5p, miR-483, miR-1468-3p | higher fecundity rate | [72] |
| Mouse | GCs | circEGFR | miR-125a-3p/CYP19A1 | promoted granulosa cell apoptosis | [59] |
4. Conclusions
- (1)
- The detection and validation of circRNAs are less straightforward and accurate quantification and manipulation of circRNAs are more time-consuming and are more likely to be affected by the linear mRNA encoded by the same gene;
- (2)
- The conservation of circRNA among species is relatively low, which makes it difficult to make comparisons and transfer discovery of one species to another;
- (3)
- The subcellular location and molecular function of circRNAs are varied, which adds complexity to reveal the functional networks of circRNAs. However, precisely because of their versatile and unique structure, circRNAs became a research hotspot of great potential.
- (1)
- circRNAs involved regulatory networks in critical pathways such as steroid hormone biosynthesis, programmed cell death (apoptosis, autophagy, ferroptosis), and oxidative stress response during ovarian physiological processes;
- (2)
- circRNAs involved in tumorigenic or suppressive pathways which can be used as therapeutic targets;
- (3)
- circRNA serving as stable diagnostic and prognostic biomarkers to assess the reproductive status, disease, and applications in assisted reproductive technology;
- (4)
- The roles of exosomal circRNAs in oocyte-granulosa-thecal cell communication;
- (5)
- Identification and functional studies of proteins or peptides products of circRNAs;
- (6)
- Development of practical and effective techniques for quantitative detection of circRNAs. Although our understanding and application of circRNAs in the mammalian ovary are still in the initial stage, these unique molecules hold immense potential for further research and will pave new avenues for the field.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [Green Version]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circ RNA s. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, R.; Gao, Y.; Gao, Y.; Wang, R.; Kadash-Edmondson, K.E.; Liu, B.; Wang, Y.; Lin, L.; Xing, Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021, 12, 266. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Wang, Y.; Fan, Y.; Fang, N.; Wang, T.; Xu, T.; Shu, Y. CircRNAs in cancer metabolism: A review. J. Hematol. Oncol. 2019, 12, 90. [Google Scholar] [CrossRef] [Green Version]
- Orisaka, M.; Tajima, K.; Tsang, B.K.; Kotsuji, F. Oocyte-granulosa-theca cell interactions during preantral follicular development. J. Ovarian Res. 2009, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Quirk, S.M.; Cowan, R.G.; Harman, R.M.; Hu, C.L.; Porter, D.A. Ovarian Follicular Growth and Atresia: The Relationship between Cell Proliferation and Survival. J. Anim. Sci. 2004, 82, E40–E52. [Google Scholar] [CrossRef]
- Baker, T.G. A Quantitative and Cytological Study of Germ Cells in Human Ovaries. Proc. R. Soc. Lond. Ser. B Boil. Sci. 1963, 158, 417–433. [Google Scholar] [CrossRef]
- Umekage, S.; Uehara, T.; Fujita, Y.; Suzuki, H.; Kikuchi, Y. In vivo circular RNA expression by the permuted intron-exon method. Innov. Biotechnol. 2012, 4, 76–90. [Google Scholar]
- Li, H.M.; Ma, X.L.; Li, H.G. Intriguing circles: Conflicts and controversies in circular RNA research. Wiley Interdiscip. Rev. RNA 2019, 10, e1538. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmayr-Heyda, A.; Reiner, A.T.; Auer, K.; Sukhbaatar, N.; Aust, S.; Bachleitner-Hofmann, T.; Mesteri, I.; Grunt, T.W.; Zeillinger, R.; Pils, D. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis and normal human tissues. Sci. Rep. 2015, 5, 8057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Balool, H.H.; Weber, D.; Liu, Y.; Wade, M.; Guleria, K.; Nam, P.; Clayton, J.; Rowe, W.; Coxhead, J.; Irving, J. Post-transcriptional exon shuffling events in humans can be evolutionarily conserved and abundant. Genome Res. 2011, 21, 1788–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.L.; Bao, Y.; Muh-Ching, Y.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Julia, S.; Thomas, P. Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [Green Version]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef]
- Zhang, H.-D.; Jiang, L.-H.; Sun, D.-W.; Hou, J.-C.; Ji, Z.-L. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018, 25, 1–7. [Google Scholar] [CrossRef]
- Altesha, M.A.; Ni, T.; Khan, A.; Liu, K.; Zheng, X. Circular RNA in cardiovascular disease. J. Cell. Physiol. 2019, 234, 5588–5600. [Google Scholar] [CrossRef]
- Ren, S.; Lin, P.; Wang, J.; Yu, H.; Lv, T.; Sun, L.; Du, G. Circular RNAs: Promising molecular biomarkers of human aging-related diseases via functioning as an miRNA sponge. Mol. Ther.-Methods Clin. Dev. 2020, 18, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Noto, J.J.; Schmidt, C.A.; Matera, A.G. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017, 14, 978–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Hu, Z.; Zhou, J.; Tian, C.; Tian, G.; He, M.; Gao, L.; Chen, L.; Li, T.; Peng, H. Interior circular RNA. RNA Biol. 2020, 17, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Du, W.W.; Zeng, K.; Wu, N.; Fang, L.; Lyu, J.; Yee, A.J.; Yang, B.B. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol. Ther. 2021, 29, 2754–2768. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Y.; Zhang, C.; Sun, X. Biological functions of circRNAs and their progress in livestock and poultry. Reprod. Domest. Anim. 2020, 55, 1667–1677. [Google Scholar] [CrossRef]
- Tran, A.M.; Chalbatani, G.M.; Berland, L.; Cruz De los Santos, M.; Raj, P.; Jalali, S.A.; Gharagouzloo, E.; Ivan, C.; Dragomir, M.P.; Calin, G.A. A new world of biomarkers and therapeutics for female reproductive system and breast cancers: Circular RNAs. Front. Cell Dev. Biol. 2020, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef]
- Preußer, C.; Hung, L.-H.; Schneider, T.; Schreiner, S.; Hardt, M.; Moebus, A.; Santoso, S.; Bindereif, A. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1424473. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Li, T.-C.; Wu, Y.-Y.; Yeh, C.-H.; Chiang, W.; Chuang, C.-Y.; Kuo, H.-C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017, 8, 1149. [Google Scholar] [CrossRef] [Green Version]
- Panda, A.C. Circular RNAs act as miRNA sponges. Circ. RNAs 2018, 1087, 67–79. [Google Scholar]
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and characterizing circRNA-protein interaction. Theranostics 2017, 7, 4183. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI J. Natl. Cancer Inst. 2018, 110, 304–315. [Google Scholar] [CrossRef] [Green Version]
- Prats, A.C.; David, F.; Diallo, L.; Roussel, E.; Lacazette, E. Circular RNA, the Key for Translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.-K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2022, 1–11. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhu, Q. Mechanisms regulating abnormal circular RNA biogenesis in cancer. Cancers 2021, 13, 4185. [Google Scholar] [CrossRef]
- Shao, T.; Pan, Y.-H.; Xiong, X.-D. Circular RNA: An important player with multiple facets to regulate its parental gene expression. Mol. Ther.-Nucleic Acids 2021, 23, 369–376. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Y.; Cao, M.; Pantaleo, V.; Burgyan, J.; Li, W.X.; Ding, S.W. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. USA 2012, 109, 3938–3943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Singh, D.; Zeng, Z.; Coleman, S.J.; Huang, Y.; Savich, G.L.; He, X.; Mieczkowski, P.; Grimm, S.A.; Perou, C.M. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010, 38, e178. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, R.; Ma, X.-K.; Chen, L.-L.; Yang, L. Genome-wide annotation of circRNAs and their alternative back-splicing/splicing with CIRCexplorer pipeline. In Epitranscriptomics: Methods and Protocols, Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 137–149. [Google Scholar]
- Schneider, T.; Schreiner, S.; Preußer, C.; Bindereif, A.; Rossbach, O. Northern blot analysis of circular RNAs. In Circular RNAs; Springer: Berlin/Heidelberg, Germany, 2018; pp. 119–133. [Google Scholar]
- Lim, A.; Lim, T.H. Fluorescence In Situ Hybridization on Tissue Sections. Methods Mol. Biol. 2017, 1541, 119. [Google Scholar] [PubMed]
- Liu, W.; Zhang, J.; Zou, C.; Xie, X.; Wang, Y.; Wang, B.; Zhao, Z.; Tu, J.; Wang, X.; Li, H. Microarray Expression Profile and Functional Analysis of Circular RNAs in Osteosarcoma. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 43, 969. [Google Scholar] [CrossRef]
- Zhang, M.A.; Chen, Z.B.; Jiang, C.C.; Liu, Y.A.; Wu, J.A.; Liu, L.A. Circular RNA detection methods: A minireview—ScienceDirect. Talanta 2021, 238, 123066. [Google Scholar]
- Naarmann-de Vries, I.S.; Eschenbach, J.; Schudy, S.; Meder, B.; Dieterich, C. Targeted Analysis of circRNA Expression in Patient Samples by Lexo-circSeq. Front. Mol. Biosci. 2022, 9, 875805. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.-L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Ning, L.; Long, B.; Zhang, W.; Yu, M.; Wang, S.; Cao, D.; Yang, J.; Shen, K.; Huang, Y.; Lang, J. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int. J. Oncol. 2018, 53, 2637–2646. [Google Scholar] [CrossRef] [Green Version]
- Teng, F.; Xu, J.; Zhang, M.; Liu, S.; Gu, Y.; Zhang, M.; Wang, X.; Ni, J.; Qian, B.; Shen, R. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int. J. Biochem. Cell Biol. 2019, 112, 8–17. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, C.; Liu, Y.; Wang, M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci. Trends 2019, 13, 204–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Ji, M.; Wang, Q.; He, N.; Li, Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol. Ther.-Nucleic Acids 2019, 18, 24–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, Q.; Liu, M.; Xu, J.; Liu, Y.; Cao, X.; Dong, X.; Liu, S. Characterization of circular RNA expression profiles in cumulus cells from patients with polycystic ovary syndrome. Fertil. Steril. 2019, 111, 1243–1251.e1241. [Google Scholar] [CrossRef]
- Li, M.; Zeng, Z.; Zhang, A.; Ye, Q.; Su, S.; Xia, T. WGCNA analysis identifies polycystic ovary syndrome-associated circular RNAs that interact with RNA-binding proteins and sponge miRNAs. Int. J. Gen. Med. 2021, 14, 8737. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-P.; Peng, X.-Y.; Lv, X.-Q.; Liu, L.; Li, X.-L.; He, X.; Lv, F.; Pan, Y.; Wang, L.; Liu, K.-F. High throughput circRNAs sequencing profile of follicle fluid exosomes of polycystic ovary syndrome patients. J. Cell. Physiol. 2019, 234, 15537–15547. [Google Scholar] [CrossRef]
- Cai, H.; Li, Y.; Li, H.; Niringiyumukiza, J.D.; Zhang, M.; Chen, L.; Chen, G.; Xiang, W. Identification and characterization of human ovary-derived circular RNAs and their potential roles in ovarian aging. Aging 2018, 10, 2511. [Google Scholar] [CrossRef]
- Jia, W.; Xu, B.; Wu, J. Circular RNA expression profiles of mouse ovaries during postnatal development and the function of circular RNA epidermal growth factor receptor in granulosa cells. Metabolism 2018, 85, 192–204. [Google Scholar] [CrossRef]
- Liang, G.; Yang, Y.; Niu, G.; Tang, Z.; Li, K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017, 24, 523–535. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Gong, W.; He, Y.; Li, N.; Yuan, X. Ovary-derived circular RNAs profile analysis during the onset of puberty in gilts. BMC Genom. 2021, 22, 445. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Z.; He, X.; Jiang, Y.; Ouyang, Y.; Hong, Q.; Chu, M. Differentially Expressed Circular RNA Profile Signatures Identified in Prolificacy Trait of Yunshang Black Goat Ovary at Estrus Cycle. Front. Physiol. 2022, 13, 576. [Google Scholar] [CrossRef]
- Xu, L.; Liu, C.; Na, R.; Zhang, W.; He, Y.; Yuan, Y.; Zhang, H.; Han, Y.; Zeng, Y.; Si, W. Genetic Basis of Follicle Development in Dazu Black Goat by Whole-Transcriptome Sequencing. Animals 2021, 11, 3536. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jiang, H.; Liu, J.B.; Sun, X.L.; Zhang, Z.; Li, S.; Gao, Y.; Yuan, B.; Zhang, J.B. Genome-wide analysis of circular RNAs in bovine cumulus cells treated with BMP15 and GDF9. Sci. Rep. 2018, 8, 7944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, T.; Zhang, J.; Yao, W.; Du, X.; Pan, Z. CircINHA resists granulosa cell apoptosis by upregulating CTGF as a ceRNA of miR-10a-5p in pig ovarian follicles. Biochim. Biophys. Acta 2019, 1862, 194420. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Wang, H.; Zhang, Y.; Zhang, J.; Liu, J.; Pan, Z. circRNA-Mediated Inhibin–Activin Balance Regulation in Ovarian Granulosa Cell Apoptosis and Follicular Atresia. Int. J. Mol. Sci. 2021, 22, 9113. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Zhang, J.; Du, X.; Pan, Z. circSLC41A1 Resists Porcine Granulosa Cell Apoptosis and Follicular Atresia by Promoting SRSF1 through miR-9820-5p Sponging. Int. J. Mol. Sci. 2022, 23, 1509. [Google Scholar] [CrossRef]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Teerds, K.; Tao, J.; Wei, H.; Jaklofsky, M.; Zhao, Z.; Liang, Y.; Li, L.; Wang, C.C.; Zhang, S. Characteristics of Circular RNA Expression Profiles of Porcine Granulosa Cells in Healthy and Atretic Antral Follicles. Int. J. Mol. Sci. 2020, 21, 5217. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, H.; Li, X.; Hu, J.; Sun, S. Genome-Wide Differential Expression Profiling of Ovarian circRNAs Associated With Litter Size in Pigs. Front. Genet. 2019, 10, 1010. [Google Scholar] [CrossRef]
- Liang, G.; Yan, J.; Guo, J.; Tang, Z. Identification of Ovarian Circular RNAs and Differential Expression Analysis between MeiShan and Large White Pigs. Animals 2020, 10, 1114. [Google Scholar] [CrossRef]
- Hu, T.; Qi, X.; Feng, Z.; Zhang, N.; Yang, L.; Suo, X.; Li, X.; Yang, Q.; Chen, M. Circular RNA profiling reveals chi_circ_0008219 function as microRNA sponges in pre-ovulatory ovarian follicles of goats (Capra hircus). Genomics 2018, 110, 257–266. [Google Scholar]
- Liu, A.; Chen, X.; Liu, M.; Zhang, L.; Ma, X.; Tian, S. Differential expression and functional analysis of CircRNA in the ovaries of low and high fecundity hanper sheep. Animals 2021, 11, 1863. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, G.; Tan, Y.; Shuai, J. Expression profile of circular RNAs in continuous light-induced ovarian dysfunction. Ecotoxicol. Environ. Saf. 2022, 242, 113861. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, L.; Chen, J.; Gao, H.; Zhao, W.; Huang, Y.; Jiang, T.; Zhou, J.; Chen, Y. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem. Biophys. Res. Commun. 2018, 505, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xia, B.; Xu, Y.; Zhang, Y.; Xu, J.; Lou, G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wang, S.; Li, G.; Zhao, X.; Jiang, F.; Liu, J.; Tan, W. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J. Cell. Mol. Med. 2019, 23, 3597–3602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Mao, M.; Jiang, J.; Zhu, D.; Li, P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019, 12, 3869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, I.; Karedath, T.; Andrews, S.S.; Al, I.K.; Mohamoud, Y.A.; Querleu, D.; Rafii, A.; Malek, J.A. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 2016, 7, 36366. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Sun, Y.; Xu, H.; Shi, Y.; Shen, R.; Teng, F.; Xu, J.; Jia, X. Circular RNA Hsa_Circ_0007444 Inhibits Tumor Progression Through MiR-23a-3p/DICER1 in Ovarian Cancer. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Sun, S.; Fang, H. Curcumin inhibits ovarian cancer progression by regulating circ-PLEKHM3/miR-320a/SMG1 axis. J. Ovarian Res. 2021, 14, 158. [Google Scholar] [CrossRef]
- Lin, X.; Chen, Y.; Ye, X.; Xia, X. Circular RNA ABCB10 promotes cell proliferation and invasion, but inhibits apoptosis via regulating themicroRNA1271mediated Capn4/Wnt/βcatenin signaling pathway in epithelial ovarian cancer. Mol. Med. Rep. 2021, 23, 387. [Google Scholar] [CrossRef]
- Li, Q.-H.; Liu, Y.; Chen, S.; Zong, Z.-h.; Du, Y.-P.; Sheng, X.-J.; Zhao, Y. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed. Pharmacother. 2019, 114, 108832. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Zhong, J.; Pang, L. Upregulation of Circular RNA VPS13C-has-circ-001567 Promotes Ovarian Cancer Cell Proliferation and Invasion. Cancer Biother. Radiopharm. 2019, 34, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qin, X.P.; Lang, Y.P.; Kou, D.; Shao, Z.W. Circular RNA circ-SMAD7 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5603–5610. [Google Scholar] [PubMed]
- Hou, W.; Zhang, Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol.—Res. Pract. 2021, 217, 153275. [Google Scholar] [CrossRef]
- Zhao, C.; Zhou, Y.; Shen, X.; Gong, M.; Lu, Y.; Fang, C.; Chen, J.; Ju, R. Circular RNA expression profiling in the fetal side of placenta from maternal polycystic ovary syndrome and circ_0023942 inhibits the proliferation of human ovarian granulosa cell. Arch. Gynecol. Obstet. 2020, 301, 963–971. [Google Scholar] [CrossRef]
- Chen, A.-X.; Jin, R.-Y.; Zhou, W.-M.; Ye, Y.-J.; Lu, J.-L.; Ren, Y.-F.; Xuan, F.-L. CircRNA circ_0043533 facilitates cell growth in polycystic ovary syndrome by targeting miR-1179. Reprod. Biol. 2022, 22, 100637. [Google Scholar] [CrossRef]
- Lu, X.; Gao, H.; Zhu, B.; Lin, G. Circular RNA circ_RANBP9 exacerbates polycystic ovary syndrome via microRNA-136-5p/XIAP axis. Bioengineered 2021, 12, 6748–6758. [Google Scholar] [CrossRef]
- Wu, G.; Xia, J.; Yang, Z.; Chen, Y.; Jiang, W.; Yin, T.; Yang, J. CircASPH promotes KGN cells proliferation through miR-375/MAP2K6 axis in Polycystic Ovary Syndrome. J. Cell. Mol. Med. 2022, 26, 1817–1825. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, S.; Cai, B.; Wang, Z.; Chen, M.; Zheng, Z.; Zhou, C. Involvement of ferroptosis in the granulosa cells proliferation of PCOS through the circRHBG/miR-515/SLC7A11 axis. Ann. Transl. Med. 2021, 9, 1348. [Google Scholar] [CrossRef]
- Tu, P.; Yan, S.; Zhang, F. Circ_0005925 Promotes Granulosa Cell Growth by Targeting MiR-324-3p to Upregulate MAP2K6 in Polycystic Ovary Syndrome. Biochem. Genet. 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xiong, F.; Bai, Y.; Xiao, J.; Zhang, Y.; Chen, J.; Li, Q. Circ_0043532 regulates miR-182/SGK3 axis to promote granulosa cell progression in polycystic ovary syndrome. Reprod. Biol. Endocrinol. 2021, 19, 103681. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, J.; Li, Y.-Y.; Li, M.-Q.; Liao, H.-Q. CircRNA circUSP36 impairs the stability of NEDD4L mRNA through recruiting PTBP1 to enhance ULK1-mediated autophagic granulosa cell death. J. Reprod. Immunol. 2022, 153, 103681. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Chang, T.; Li, Y.; Jia, Y.; Li, H.; Zhang, M.; Su, P.; Zhang, L.; Xiang, W. Circular DDX10 is associated with ovarian function and assisted reproductive technology outcomes through modulating the proliferation and steroidogenesis of granulosa cells. Aging 2021, 13, 9592. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, F.; Fan, Y.; Xie, S.; Li, C.; Meng, L.; Li, L.; Zhang, S.; Wei, H. A novel identified circ-ANKHD1 targets the miR-27a-3p/SFRP1 signaling pathway and modulates the apoptosis of granulosa cells. Environ. Sci. Pollut. Res. 2021, 28, 57459–57469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, Y.; Liu, H.; Pan, Z. MicroRNAs in ovarian follicular atresia and granulosa cell apoptosis. Reprod. Biol. Endocrinol. 2019, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-J.; Chen, X.; Li, C.-P.; Li, X.-M.; Liu, C.; Liu, B.-H.; Shan, K.; Jiang, Q.; Zhao, C.; Yan, B. Identification and Characterization of Circular Rnas as a New Class of Putative Biomarkers in Diabetes Retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6500–6509. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, C.; Jia, C.; Zhang, Y.; Qing, X.; Zhang, Y.; Liu, J.; Xu, S.; Pan, Z. The Role of Circular RNAs in the Physiology and Pathology of the Mammalian Ovary. Int. J. Mol. Sci. 2022, 23, 15204. https://doi.org/10.3390/ijms232315204
Zhang J, Wang C, Jia C, Zhang Y, Qing X, Zhang Y, Liu J, Xu S, Pan Z. The Role of Circular RNAs in the Physiology and Pathology of the Mammalian Ovary. International Journal of Molecular Sciences. 2022; 23(23):15204. https://doi.org/10.3390/ijms232315204
Chicago/Turabian StyleZhang, Jinbi, Caixia Wang, Chao Jia, Yi Zhang, Xinxin Qing, Yuge Zhang, Jingge Liu, Shiyong Xu, and Zengxiang Pan. 2022. "The Role of Circular RNAs in the Physiology and Pathology of the Mammalian Ovary" International Journal of Molecular Sciences 23, no. 23: 15204. https://doi.org/10.3390/ijms232315204
APA StyleZhang, J., Wang, C., Jia, C., Zhang, Y., Qing, X., Zhang, Y., Liu, J., Xu, S., & Pan, Z. (2022). The Role of Circular RNAs in the Physiology and Pathology of the Mammalian Ovary. International Journal of Molecular Sciences, 23(23), 15204. https://doi.org/10.3390/ijms232315204





