Kruppel-like Factors in Skeletal Physiology and Pathologies
Abstract
1. Skeletal Development and Maintenance
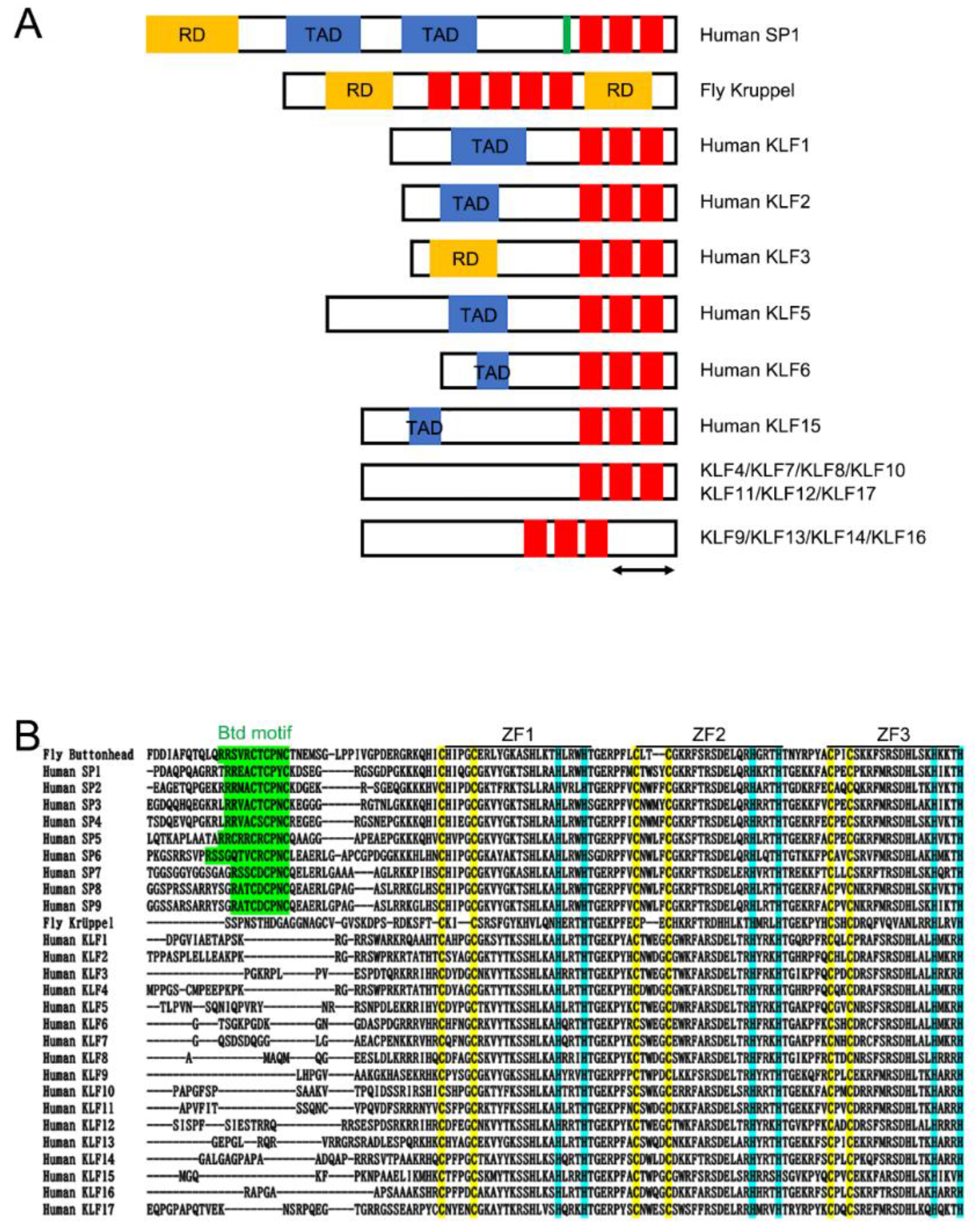
2. Krüppel in Drosophila
3. Kruppel-like Factors in Vertebrates
4. Roles of KLFs in Normal Bone Physiology (Table 1)
4.1. KLF1
4.2. KLF2
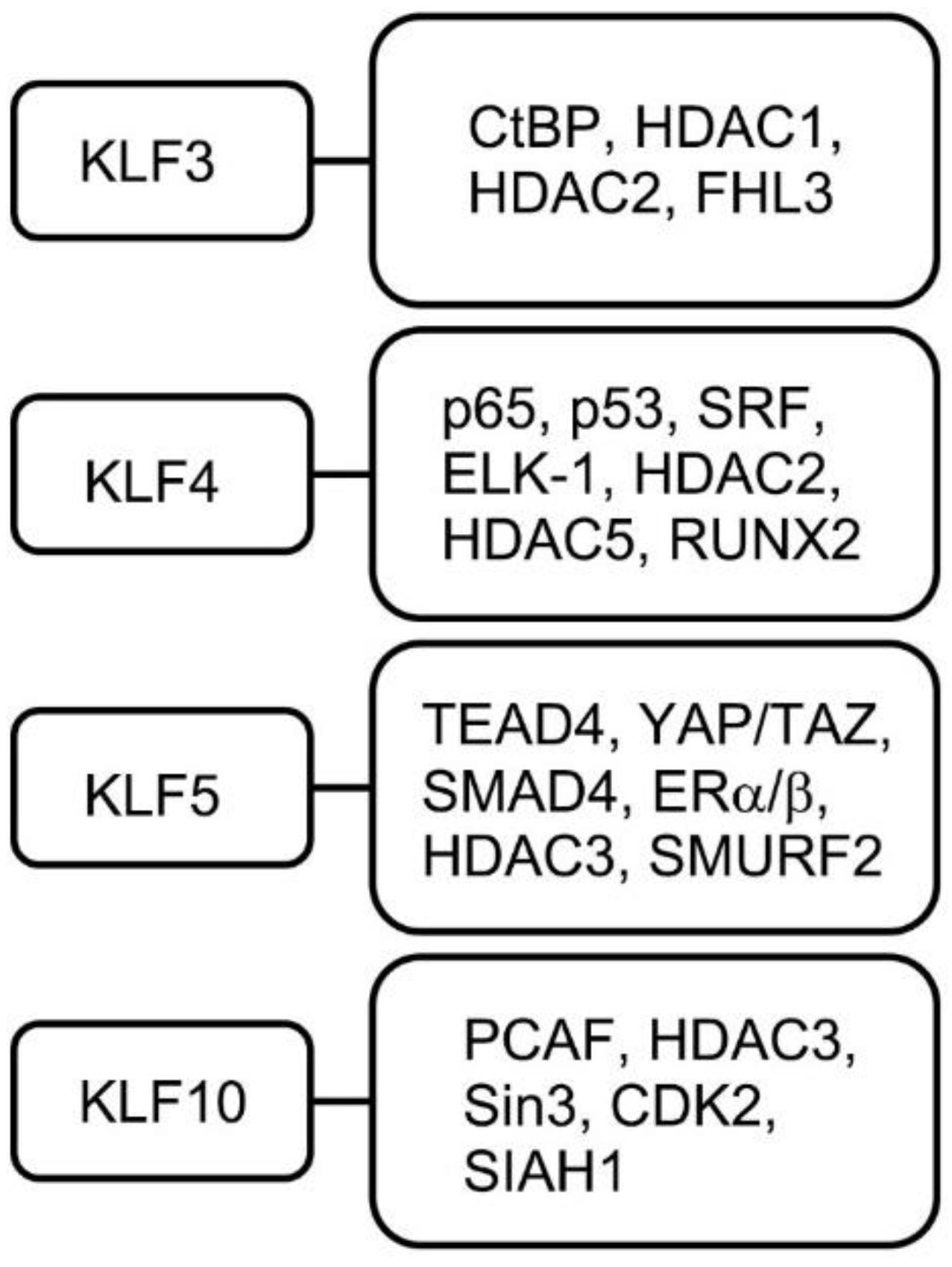
4.3. KLF3
4.4. KLF4
4.5. KLF5
4.6. KLF6
4.7. KLF7
4.8. KLF8
4.9. KLF9
4.10. KLF10
4.11. KLF11
| Factors | Phenotypes | Studies |
|---|---|---|
| KLF1 | Growth defects (LOF in human; unlikely cell-autonomous actions) | [36,37,38,39] |
| KLF2 | Abnormal craniofacial morphogenesis (LOF in mouse) | [42] |
| KLF3 | High bone mass (LOF in human; by mutated lncRNA) | [46] |
| KLF4 | High bone mass with increased osteoclast differentiation (osteoblast-specific LOF in mouse) Repressed osteoclast maturation (osteoblast-specific GOF in mouse) | [55] [57] |
| KLF5 | Retarded skeletal growth with delayed primary ossification center formation (LOF in mouse) | [71] |
| KLF6 | Remain to be investigated | – |
| KLF7 | High bone mineral density associated with its low expression (in human) | [81] |
| KLF8 | Remain to be investigated | – |
| KLF9 | Remain to be investigated | – |
| KLF10 | Growth defect with suppressed cartilage maturation and primary ossification center formation | [102,103] |
| KLF11 | None | [114] |
| KLF12 | Remain to be investigated (in vitro studies exist) | – |
| KLF13 | Remain to be investigated (in vitro studies exist) | – |
| KLF14 | Remain to be investigated (in vitro studies exist) | – |
| KLF15 | Remain to be investigated (in vitro studies exist) | – |
| KLF16 | Remain to be investigated (in vitro studies exist) | – |
| KLF17 | Remain to be investigated | – |
4.12. KLF12
4.13. KLF13
4.14. KLF14
4.15. KLF15
4.16. KLF16
4.17. KLF17
5. Role of KLFs in Osteoarthritis, Osteoporosis, and Osteosarcoma
5.1. Osteoarthritis
5.2. Osteoporosis
5.3. Bone Tumors
| Factors | Conclusion | Studies | |
|---|---|---|---|
| Osteoarthritis | KLF2 KLF4 KLF5 KLF15 | Downregulated in OA patients Represses IL1β-induced MMP13 expression Suppresses ROS production Downregulated in human OA samples Induces PRG4 and collagen genes Downregulated in human OA samples Downregulated in human OA samples | [133,134,135,136,137,138,139] [140,141] [136,141] [135,136] |
| Osteoporosis | KLF3 KLF4 KLF10 | Endothelial cell-specific KO mice show high bone mass Osteoblast-specific KO mice show high bone mass Osteoprogenitor-specific KO mice show low bone mass Downregulated in osteoporotic patients Downregulated in ovariectomy rats Female Klf10 KO mice show reduced bone strength | [46] [54,60] [103,166,167] |
| Osteosarcoma | KLF2 KLF4 KLF5 KLF6 KLF8 KLF9 KLF11 | Downregulated in OS cells in vitro Induces cancer stemness Highly expressed in OS cells ML264 (KLF5 inhibitor) induces OS cell activation Downregulated in OS cells Highly expressed in OS samples Induces cancer stemness Downregulated in OS cells Downregulated in OS samples Suppresses cancer stem cell induction | [181] [183,184] [192,193] [194] [185,186] [194] [186,191] |
6. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2011, 13, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Le Douarin, N.M.; Smith, J. Development of the peripheral nervous system from the neural crest. Annu. Rev. Cell Biol. 1988, 4, 375–404. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, A.D.; Olsen, B.R. Bone development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Salhotra, A.; Shah, H.N.; Levi, B.; Longaker, M.T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020, 21, 696–711. [Google Scholar] [CrossRef]
- Chan, C.K.; Seo, E.Y.; Chen, J.Y.; Lo, D.; McArdle, A.; Sinha, R.; Tevlin, R.; Seita, J.; Vincent-Tompkins, J.; Wearda, T.; et al. Identification and specification of the mouse skeletal stem cell. Cell 2015, 160, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.K.F.; Gulati, G.S.; Sinha, R.; Tompkins, J.V.; Lopez, M.; Carter, A.C.; Ransom, R.C.; Reinisch, A.; Wearda, T.; Murphy, M.; et al. Identification of the Human Skeletal Stem Cell. Cell 2018, 175, 43–56.e21. [Google Scholar] [CrossRef]
- Schuh, R.; Aicher, W.; Gaul, U.; Cote, S.; Preiss, A.; Maier, D.; Seifert, E.; Nauber, U.; Schroder, C.; Kemler, R.; et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell 1986, 47, 1025–1032. [Google Scholar] [CrossRef]
- Knipple, D.C.; Seifert, E.; Rosenberg, U.B.; Preiss, A.; Jackle, H. Spatial and temporal patterns of Kruppel gene expression in early Drosophila embryos. Nature 1985, 317, 40–44. [Google Scholar] [CrossRef]
- Preiss, A.; Rosenberg, U.B.; Kienlin, A.; Seifert, E.; Jackle, H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature 1985, 313, 27–32. [Google Scholar] [CrossRef]
- Rosenberg, U.B.; Preiss, A.; Seifert, E.; Jackle, H.; Knipple, D.C. Production of phenocopies by Kruppel antisense RNA injection into Drosophila embryos. Nature 1985, 313, 703–706. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Ruppert, J.M.; Bigner, S.H.; Vogelstein, B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature 1988, 332, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, J.M.; Kinzler, K.W.; Wong, A.J.; Bigner, S.H.; Kao, F.T.; Law, M.L.; Seuanez, H.N.; O’Brien, S.J.; Vogelstein, B. The GLI-Kruppel family of human genes. Mol. Cell Biol. 1988, 8, 3104–3113. [Google Scholar] [CrossRef]
- Dang, D.T.; Pevsner, J.; Yang, V.W. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 2000, 32, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef]
- Nusslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Surkova, S.; Myasnikova, E.; Reinitz, J.; Samsonova, M. Modeling of gap gene expression in Drosophila Kruppel mutants. PLoS Comput. Biol. 2012, 8, e1002635. [Google Scholar] [CrossRef]
- Bieker, J.J. Kruppel-like factors: Three fingers in many pies. J. Biol. Chem. 2001, 276, 34355–34358. [Google Scholar] [CrossRef]
- Lomberk, G.; Urrutia, R. The family feud: Turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 2005, 392, 1–11. [Google Scholar] [CrossRef]
- McConnell, B.B.; Ghaleb, A.M.; Nandan, M.O.; Yang, V.W. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays News Rev. Mol. Cell Dev. Biol. 2007, 29, 549–557. [Google Scholar] [CrossRef]
- McConnell, B.B.; Yang, V.W. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef]
- Iuchi, S. Three classes of C2H2 zinc finger proteins. Cell Mol. Life Sci. CMLS 2001, 58, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Presnell, J.S.; Schnitzler, C.E.; Browne, W.E. KLF/SP Transcription Factor Family Evolution: Expansion, Diversification, and Innovation in Eukaryotes. Genome Biol. Evol. 2015, 7, 2289–2309. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.R.; Kadonaga, J.T.; Bell, S.P.; Tjian, R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 1986, 234, 47–52. [Google Scholar] [CrossRef]
- Kadonaga, J.T.; Carner, K.R.; Masiarz, F.R.; Tjian, R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 1987, 51, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, C.; Winoto, A. Cloning of GT box-binding proteins: A novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell Biol. 1992, 12, 4251–4261. [Google Scholar] [CrossRef] [PubMed]
- Pesce, M.; Marin Gomez, M.; Philipsen, S.; Scholer, H.R. Binding of Sp1 and Sp3 transcription factors to the Oct-4 gene promoter. Cell Mol. Biol. 1999, 45, 709–716. [Google Scholar]
- Suske, G. The Sp-family of transcription factors. Gene 1999, 238, 291–300. [Google Scholar] [CrossRef]
- Cook, T.; Gebelein, B.; Urrutia, R. Sp1 and its likes: Biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci. 1999, 880, 94–102. [Google Scholar] [CrossRef]
- Hardeman, J. The First Permanent Molar and Its Early Management. Am. J. Dent. Sci. 1884, 18, 59–63. [Google Scholar]
- Wimmer, E.A.; Jackle, H.; Pfeifle, C.; Cohen, S.M. A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature 1993, 366, 690–694. [Google Scholar] [CrossRef]
- Suske, G.; Bruford, E.; Philipsen, S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics 2005, 85, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, E.A.; Frommer, G.; Purnell, B.A.; Jackle, H. buttonhead and D-Sp1: A novel Drosophila gene pair. Mech. Dev. 1996, 59, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Borg, J.; Papadopoulos, P.; Georgitsi, M.; Gutierrez, L.; Grech, G.; Fanis, P.; Phylactides, M.; Verkerk, A.J.; van der Spek, P.J.; Scerri, C.A.; et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 2010, 42, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Nuez, B.; Michalovich, D.; Bygrave, A.; Ploemacher, R.; Grosveld, F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 1995, 375, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Perkins, A.C.; Sharpe, A.H.; Orkin, S.H. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 1995, 375, 318–322. [Google Scholar] [CrossRef]
- Wickramasinghe, S.N.; Illum, N.; Wimberley, P.D. Congenital dyserythropoietic anaemia with novel intra-erythroblastic and intra-erythrocytic inclusions. Br. J. Haematol. 1991, 79, 322–330. [Google Scholar] [CrossRef]
- Arnaud, L.; Saison, C.; Helias, V.; Lucien, N.; Steschenko, D.; Giarratana, M.C.; Prehu, C.; Foliguet, B.; Montout, L.; de Brevern, A.G.; et al. A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am. J. Hum. Genet. 2010, 87, 721–727. [Google Scholar] [CrossRef]
- Tamary, H.; Dgany, O.; Proust, A.; Krasnov, T.; Avidan, N.; Eidelitz-Markus, T.; Tchernia, G.; Genevieve, D.; Cormier-Daire, V.; Bader-Meunier, B.; et al. Clinical and molecular variability in congenital dyserythropoietic anaemia type I. Br. J. Haematol. 2005, 130, 628–634. [Google Scholar] [CrossRef]
- Heimpel, H.; Schwarz, K.; Ebnother, M.; Goede, J.S.; Heydrich, D.; Kamp, T.; Plaumann, L.; Rath, B.; Roessler, J.; Schildknecht, O.; et al. Congenital dyserythropoietic anemia type I (CDA I): Molecular genetics, clinical appearance, and prognosis based on long-term observation. Blood 2006, 107, 334–340. [Google Scholar] [CrossRef]
- Tallack, M.R.; Perkins, A.C. Three fingers on the switch: Kruppel-like factor 1 regulation of gamma-globin to beta-globin gene switching. Curr. Opin. Hematol. 2013, 20, 193–200. [Google Scholar] [CrossRef]
- Wani, M.A.; Conkright, M.D.; Jeffries, S.; Hughes, M.J.; Lingrel, J.B. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Kruppel-like factor. Genomics 1999, 60, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.A.; Means, R.T., Jr.; Lingrel, J.B. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998, 7, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Wang, Z.; Tao, Y.; Bai, J.; Yu, B.; Shen, J.; Sun, H.; Xiao, L.; Xu, Y.; Zhou, J.; et al. KLF2 regulates osteoblast differentiation by targeting of Runx2. Lab. Investig. A J. Tech. Methods Pathol. 2019, 99, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Kim, J.H.; Kim, K.; Seong, S.; Kim, N. The IRF2BP2-KLF2 axis regulates osteoclast and osteoblast differentiation. BMB Rep. 2019, 52, 469–474. [Google Scholar] [CrossRef]
- Das, H.; Kumar, A.; Lin, Z.; Patino, W.D.; Hwang, P.M.; Feinberg, M.W.; Majumder, P.K.; Jain, M.K. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc. Natl. Acad. Sci. USA 2006, 103, 6653–6658. [Google Scholar] [CrossRef]
- Yang, M.; Guo, Q.; Peng, H.; Xiao, Y.Z.; Xiao, Y.; Huang, Y.; Li, C.J.; Su, T.; Zhang, Y.L.; Lei, M.X.; et al. Kruppel-like factor 3 inhibition by mutated lncRNA Reg1cp results in human high bone mass syndrome. J. Exp. Med. 2019, 216, 1944–1964. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Ai, Z.; Zeng, J.; Fu, Y.; Zhang, L.; Wu, X. Bone mesenchymal stem cells (BMSCs)-derived exosomal microRNA-21-5p regulates Kruppel-like factor 3 (KLF3) to promote osteoblast proliferation in vitro. Bioengineered 2022, 13, 11933–11944. [Google Scholar] [CrossRef]
- Zhu, E.; Zhang, J.; Zhou, J.; Yuan, H.; Zhao, W.; Wang, B. miR-20a-5p promotes adipogenic differentiation of murine bone marrow stromal cells via targeting Kruppel-like factor 3. J. Mol. Endocrinol. 2018, 60, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Crossley, M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998, 17, 5129–5140. [Google Scholar] [CrossRef]
- Shi, Y.; Sawada, J.; Sui, G.; Affar el, B.; Whetstine, J.R.; Lan, F.; Ogawa, H.; Luke, M.P.; Nakatani, Y.; Shi, Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003, 422, 735–738. [Google Scholar] [CrossRef]
- Turner, J.; Nicholas, H.; Bishop, D.; Matthews, J.M.; Crossley, M. The LIM protein FHL3 binds basic Kruppel-like factor/Kruppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J. Biol. Chem. 2003, 278, 12786–12795. [Google Scholar] [CrossRef]
- Shields, J.M.; Christy, R.J.; Yang, V.W. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J. Biol. Chem. 1996, 271, 20009–20017. [Google Scholar] [CrossRef] [PubMed]
- Segre, J.A.; Bauer, C.; Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999, 22, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Nakashima, K.; de Crombrugghe, B. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: A new tool to examine physiology and disease of postnatal bone and tooth. Am. J. Pathol. 2004, 165, 1875–1882. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, K.; Youn, B.U.; Lee, J.; Kim, I.; Shin, H.I.; Akiyama, H.; Choi, Y.; Kim, N. Kruppel-like factor 4 attenuates osteoblast formation, function, and cross talk with osteoclasts. J. Cell Biol. 2014, 204, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Michikami, I.; Fukushi, T.; Tanaka, M.; Egusa, H.; Maeda, Y.; Ooshima, T.; Wakisaka, S.; Abe, M. Kruppel-like factor 4 regulates membranous and endochondral ossification. Exp. Cell Res. 2012, 318, 311–325. [Google Scholar] [CrossRef]
- Fujikawa, J.; Tanaka, M.; Itoh, S.; Fukushi, T.; Kurisu, K.; Takeuchi, Y.; Morisaki, I.; Wakisaka, S.; Abe, M. Kruppel-like factor 4 expression in osteoblasts represses osteoblast-dependent osteoclast maturation. Cell Tissue Res. 2014, 358, 177–187. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Kito, A.; Itoh, S.; Naruse, H.; Fujikawa, J.; Sadek, K.M.; Akiyama, S.; Yamashiro, T.; Wakisaka, S.; Abe, M. Kruppel-Like Factor 4 represses osteoblast differentiation via ciliary Hedgehog signaling. Exp. Cell Res. 2018, 371, 417–425. [Google Scholar] [CrossRef]
- Saeki, N.; Itoh, Y.; Kanai, R.; Itoh, S.; Inububishi, T.; Akiyama, S.; Inui-Yamamoto, C.; Abe, M. Pregnane X receptor (PXR) represses osteoblast differentiation through repression of the Hedgehog signaling pathway. Exp. Cell Res. 2022, 416, 113156. [Google Scholar] [CrossRef]
- Yu, S.; Guo, J.; Sun, Z.; Lin, C.; Tao, H.; Zhang, Q.; Cui, Y.; Zuo, H.; Lin, Y.; Chen, S.; et al. BMP2-dependent gene regulatory network analysis reveals Klf4 as a novel transcription factor of osteoblast differentiation. Cell Death Dis. 2021, 12, 197. [Google Scholar] [CrossRef]
- Zhang, W.; Geiman, D.E.; Shields, J.M.; Dang, D.T.; Mahatan, C.S.; Kaestner, K.H.; Biggs, J.R.; Kraft, A.S.; Yang, V.W. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 2000, 275, 18391–18398. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, M.W.; Cao, Z.; Wara, A.K.; Lebedeva, M.A.; Senbanerjee, S.; Jain, M.K. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J. Biol. Chem. 2005, 280, 38247–38258. [Google Scholar] [CrossRef]
- Liu, Y.; Sinha, S.; McDonald, O.G.; Shang, Y.; Hoofnagle, M.H.; Owens, G.K. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J. Biol. Chem. 2005, 280, 9719–9727. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Gan, Q.; Owens, G.K. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am. J. Physiol. Cell Physiol. 2008, 295, C1175–C1182. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Yamashita, M.; Horimai, C.; Hayashi, M. Deletion of Kruppel-like factor 4 in endothelial and hematopoietic cells enhances neointimal formation following vascular injury. J. Am. Heart Assoc. 2014, 3, e000622. [Google Scholar] [CrossRef]
- Lin, H.; Liu, H.; Sun, Q.; Yuan, G.; Zhang, L.; Chen, Z. KLF4 promoted odontoblastic differentiation of mouse dental papilla cells via regulation of DMP1. J. Cell. Physiol. 2013, 228, 2076–2085. [Google Scholar] [CrossRef]
- Liu, H.; Lin, H.; Zhang, L.; Sun, Q.; Yuan, G.; Zhang, L.; Chen, S.; Chen, Z. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J. Biol. Chem. 2013, 288, 9261–9271. [Google Scholar] [CrossRef]
- Tao, H.; Lin, H.; Sun, Z.; Pei, F.; Zhang, J.; Chen, S.; Liu, H.; Chen, Z. Klf4 Promotes Dentinogenesis and Odontoblastic Differentiation via Modulation of TGF-beta Signaling Pathway and Interaction With Histone Acetylation. J. Bone Miner. Res. 2019, 34, 1502–1516. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. 2021, 112, 2097–2117. [Google Scholar] [CrossRef]
- Shindo, T.; Manabe, I.; Fukushima, Y.; Tobe, K.; Aizawa, K.; Miyamoto, S.; Kawai-Kowase, K.; Moriyama, N.; Imai, Y.; Kawakami, H.; et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat. Med. 2002, 8, 856–863. [Google Scholar] [CrossRef]
- Shinoda, Y.; Ogata, N.; Higashikawa, A.; Manabe, I.; Shindo, T.; Yamada, T.; Kugimiya, F.; Ikeda, T.; Kawamura, N.; Kawasaki, Y.; et al. Kruppel-like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. J. Biol. Chem. 2008, 283, 24682–24689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, Y.; Wu, Q.; Xie, L.; Barwick, B.; Fu, C.; Li, X.; Wu, D.; Xia, S.; Chen, J.; et al. Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat. Commun. 2021, 12, 1714. [Google Scholar] [CrossRef]
- Guo, P.; Dong, X.Y.; Zhao, K.; Sun, X.; Li, Q.; Dong, J.T. Opposing effects of KLF5 on the transcription of MYC in epithelial proliferation in the context of transforming growth factor beta. J. Biol. Chem. 2009, 284, 28243–28252. [Google Scholar] [CrossRef]
- Du, J.X.; Hagos, E.G.; Nandan, M.O.; Bialkowska, A.B.; Yu, B.; Yang, V.W. The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Kruppel-like factor 5 protein. J. Biol. Chem. 2011, 286, 40354–40364. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Akaogi, K.; Suzuki, T.; Osakabe, A.; Yamaguchi, C.; Sunahara, N.; Ishida, J.; Kako, K.; Ogawa, S.; Fujimura, T.; et al. Estrogen regulates tumor growth through a nonclassical pathway that includes the transcription factors ERbeta and KLF5. Sci. Signal. 2011, 4, ra22. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shi, Q.; Xu, S.; Du, C.; Liang, L.; Wu, K.; Wang, K.; Wang, X.; Chang, L.S.; He, D.; et al. Curcumin promotes KLF5 proteasome degradation through downregulating YAP/TAZ in bladder cancer cells. Int. J. Mol. Sci. 2014, 15, 15173–15187. [Google Scholar] [CrossRef]
- Wang, C.; Nie, Z.; Zhou, Z.; Zhang, H.; Liu, R.; Wu, J.; Qin, J.; Ma, Y.; Chen, L.; Li, S.; et al. The interplay between TEAD4 and KLF5 promotes breast cancer partially through inhibiting the transcription of p27Kip1. Oncotarget 2015, 6, 17685–17697. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, H.B.; Shi, Q.; Yang, C.; Ma, J.B.; Jin, B.; Wang, X.; He, D.; Guo, P. KLF5 downregulation desensitizes castration-resistant prostate cancer cells to docetaxel by increasing BECN1 expression and inducing cell autophagy. Theranostics 2019, 9, 5464–5477. [Google Scholar] [CrossRef]
- Narla, G.; Heath, K.E.; Reeves, H.L.; Li, D.; Giono, L.E.; Kimmelman, A.C.; Glucksman, M.J.; Narla, J.; Eng, F.J.; Chan, A.M.; et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 2001, 294, 2563–2566. [Google Scholar] [CrossRef]
- Matsumoto, N.; Kubo, A.; Liu, H.; Akita, K.; Laub, F.; Ramirez, F.; Keller, G.; Friedman, S.L. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 2006, 107, 1357–1365. [Google Scholar] [CrossRef]
- Chen, C.; Hu, F.; Miao, S.; Sun, L.; Jiao, Y.; Xu, M.; Huang, X.; Yang, Y.; Zhou, R. Transcription Factor KLF7 Promotes Osteoclast Differentiation by Suppressing HO-1. Front. Genet. 2022, 13, 798433. [Google Scholar] [CrossRef]
- Chen, Y.J.; Chang, W.A.; Huang, M.S.; Chen, C.H.; Wang, K.Y.; Hsu, Y.L.; Kuo, P.L. Identification of novel genes in aging osteoblasts using next-generation sequencing and bioinformatics. Oncotarget 2017, 8, 113598–113613. [Google Scholar] [CrossRef] [PubMed]
- Ke, K.; Safder, M.A.; Sul, O.J.; Kim, W.K.; Suh, J.H.; Joe, Y.; Chung, H.T.; Choi, H.S. Hemeoxygenase-1 maintains bone mass via attenuating a redox imbalance in osteoclast. Mol. Cell Endocrinol. 2015, 409, 11–20. [Google Scholar] [CrossRef]
- Florczyk-Soluch, U.; Jozefczuk, E.; Stepniewski, J.; Bukowska-Strakova, K.; Mendel, M.; Viscardi, M.; Nowak, W.N.; Jozkowicz, A.; Dulak, J. Various roles of heme oxygenase-1 in response of bone marrow macrophages to RANKL and in the early stage of osteoclastogenesis. Sci. Rep. 2018, 8, 10797. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Gu, X.; Gao, J.; Wang, Z.; Liu, G.; Barkema, H.W.; Han, B. Chlorogenic acid promotes the Nrf2/HO-1 anti-oxidative pathway by activating p21(Waf1/Cip1) to resist dexamethasone-induced apoptosis in osteoblastic cells. Free Radic. Biol. Med. 2019, 137, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, J. KLF8 transcription factor participates in oncogenic transformation. Oncogene 2007, 26, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Hu, L.; Li, T.; Lahiri, S.; Shen, C.; Wason, M.S.; Mukherjee, D.; Xie, H.; Yu, L.; Zhao, J. A novel role of Kruppel-like factor 8 in DNA repair in breast cancer cells. J. Biol. Chem. 2012, 287, 43720–43729. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Wang, Y.; Zhao, G.; Xie, R.; Liu, C.; Xiao, X.; Wu, K.; Nie, Y.; Zhang, H.; et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J. Cancer Res. Clin. Oncol. 2013, 139, 1033–1042. [Google Scholar] [CrossRef]
- Lu, H.; Hu, L.; Yu, L.; Wang, X.; Urvalek, A.M.; Li, T.; Shen, C.; Mukherjee, D.; Lahiri, S.K.; Wason, M.S.; et al. KLF8 and FAK cooperatively enrich the active MMP14 on the cell surface required for the metastatic progression of breast cancer. Oncogene 2014, 33, 2909–2917. [Google Scholar] [CrossRef]
- Kumar, S.; Behera, A.; Saha, P.; Kumar Srivastava, A. The role of Kruppel-like factor 8 in cancer biology: Current research and its clinical relevance. Biochem. Pharmacol. 2021, 183, 114351. [Google Scholar] [CrossRef]
- Jin, Y.; Yang, L.; Li, X. MicroRNA-652 promotes cell proliferation and osteosarcoma invasion by directly targeting KLF9. Exp. Ther. Med. 2020, 20, 2953–2960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, T.; He, F.; Chen, A.C.; Yang, H.; Zhu, X. Identification of Key Genes and Pathways in Osteoarthritis via Bioinformatic Tools: An Updated Analysis. Cartilage 2021, 13, 1457S–1464S. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhou, H.; Rong, W. Circular RNA_0078767 upregulates Kruppel-like factor 9 expression by targeting microRNA-889, thereby inhibiting the progression of osteosarcoma. Bioengineered 2022, 13, 14313–14328. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, K.; Cao, Y.; Hu, Y.; Shao, Z.; Tong, W.; Han, Y.; Liu, Y. KLF9 regulates miR-338-3p/NRCAM axis to block the progression of osteosarcoma cells. J. Cancer 2022, 13, 2029–2039. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Zhou, Y.; Qu, Y.; Hou, T.; Ge, W.; Zhang, S. KLF9 and EPYC acting as feature genes for osteoarthritis and their association with immune infiltration. J. Orthop. Surg. Res. 2022, 17, 365. [Google Scholar] [CrossRef]
- Subramaniam, M.; Harris, S.A.; Oursler, M.J.; Rasmussen, K.; Riggs, B.L.; Spelsberg, T.C. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995, 23, 4907–4912. [Google Scholar] [CrossRef]
- Hefferan, T.E.; Reinholz, G.G.; Rickard, D.J.; Johnsen, S.A.; Waters, K.M.; Subramaniam, M.; Spelsberg, T.C. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J. Biol. Chem. 2000, 275, 20255–20259. [Google Scholar] [CrossRef]
- Johnsen, S.A.; Subramaniam, M.; Janknecht, R.; Spelsberg, T.C. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 2002, 21, 5783–5790. [Google Scholar] [CrossRef]
- Johnsen, S.A.; Subramaniam, M.; Katagiri, T.; Janknecht, R.; Spelsberg, T.C. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J. Cell. Biochem. 2002, 87, 233–241. [Google Scholar] [CrossRef]
- Johnsen, S.A.; Subramaniam, M.; Monroe, D.G.; Janknecht, R.; Spelsberg, T.C. Modulation of transforming growth factor beta (TGFbeta)/Smad transcriptional responses through targeted degradation of TGFbeta-inducible early gene-1 by human seven in absentia homologue. J. Biol. Chem. 2002, 277, 30754–30759. [Google Scholar] [CrossRef]
- Hawse, J.R.; Subramaniam, M.; Monroe, D.G.; Hemmingsen, A.H.; Ingle, J.N.; Khosla, S.; Oursler, M.J.; Spelsberg, T.C. Estrogen receptor beta isoform-specific induction of transforming growth factor beta-inducible early gene-1 in human osteoblast cells: An essential role for the activation function 1 domain. Mol. Endocrinol. 2008, 22, 1579–1595. [Google Scholar] [CrossRef]
- Lee, J.M.; Ko, J.Y.; Park, J.W.; Lee, W.K.; Song, S.U.; Im, G.I. KLF10 is a modulatory factor of chondrocyte hypertrophy in developing skeleton. J. Orthop. Res. 2020, 38, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Hawse, J.R.; Iwaniec, U.T.; Bensamoun, S.F.; Monroe, D.G.; Peters, K.D.; Ilharreborde, B.; Rajamannan, N.M.; Oursler, M.J.; Turner, R.T.; Spelsberg, T.C.; et al. TIEG-null mice display an osteopenic gender-specific phenotype. Bone 2008, 42, 1025–1031. [Google Scholar] [CrossRef]
- Bensamoun, S.F.; Hawse, J.R.; Subramaniam, M.; Ilharreborde, B.; Bassillais, A.; Benhamou, C.L.; Fraser, D.G.; Oursler, M.J.; Amadio, P.C.; An, K.N.; et al. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone 2006, 39, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Gingery, A.; Subramaniam, M.; Pitel, K.S.; Li, X.; Ke, H.Z.; Turner, R.T.; Iwaniec, U.T.; Hawse, J.R. Sclerostin antibody treatment rescues the osteopenic bone phenotype of TGFbeta inducible early gene-1 knockout female mice. J. Cell Physiol. 2020, 235, 5679–5688. [Google Scholar] [CrossRef]
- Yerges, L.M.; Klei, L.; Cauley, J.A.; Roeder, K.; Kammerer, C.M.; Ensrud, K.E.; Nestlerode, C.S.; Lewis, C.; Lang, T.F.; Barrett-Connor, E.; et al. Candidate gene analysis of femoral neck trabecular and cortical volumetric bone mineral density in older men. J. Bone Miner. Res. 2010, 25, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Hawse, J.R.; Cicek, M.; Grygo, S.B.; Bruinsma, E.S.; Rajamannan, N.M.; van Wijnen, A.J.; Lian, J.B.; Stein, G.S.; Oursler, M.J.; Subramaniam, M.; et al. TIEG1/KLF10 modulates Runx2 expression and activity in osteoblasts. PLoS ONE 2011, 6, e19429. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Pitel, K.S.; Withers, S.G.; Drissi, H.; Hawse, J.R. TIEG1 enhances Osterix expression and mediates its induction by TGFbeta and BMP2 in osteoblasts. Biochem. Biophys. Res. Commun. 2016, 470, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Cicek, M.; Vrabel, A.; Sturchio, C.; Pederson, L.; Hawse, J.R.; Subramaniam, M.; Spelsberg, T.C.; Oursler, M.J. TGF-beta inducible early gene 1 regulates osteoclast differentiation and survival by mediating the NFATc1, AKT, and MEK/ERK signaling pathways. PLoS ONE 2011, 6, e17522. [Google Scholar] [CrossRef]
- Xiong, Y.; Svingen, P.A.; Sarmento, O.O.; Smyrk, T.C.; Dave, M.; Khanna, S.; Lomberk, G.A.; Urrutia, R.A.; Faubion, W.A., Jr. Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R608–R620. [Google Scholar] [CrossRef]
- Lin, C.H.; Lin, S.Y.; Chang, H.W.; Ko, L.J.; Tseng, Y.S.; Chang, V.H.; Yu, W.C. CDK2 phosphorylation regulates the protein stability of KLF10 by interfering with binding of the E3 ligase SIAH1. Biochim. Et Biophys. Acta 2015, 1853, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.; Gebelein, B.; Mesa, K.; Mladek, A.; Urrutia, R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 1998, 273, 25929–25936. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Mahner, S.; Jeschke, U.; Hester, A. The Distinct Roles of Transcriptional Factor KLF11 in Normal Cell Growth Regulation and Cancer as a Mediator of TGF-beta Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 2928. [Google Scholar] [CrossRef] [PubMed]
- Song, C.Z.; Gavriilidis, G.; Asano, H.; Stamatoyannopoulos, G. Functional study of transcription factor KLF11 by targeted gene inactivation. Blood Cells Mol. Dis. 2005, 34, 53–59. [Google Scholar] [CrossRef]
- Zhao, G.; Luo, W.D.; Yuan, Y.; Lin, F.; Guo, L.M.; Ma, J.J.; Chen, H.B.; Tang, H.; Shu, J. LINC02381, a sponge of miR-21, weakens osteogenic differentiation of hUC-MSCs through KLF12-mediated Wnt4 transcriptional repression. J. Bone Miner. Metab. 2022, 40, 66–80. [Google Scholar] [CrossRef]
- Yu, B.; Chang, J.; Liu, Y.; Li, J.; Kevork, K.; Al-Hezaimi, K.; Graves, D.T.; Park, N.H.; Wang, C.Y. Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-kappaB. Nat. Med. 2014, 20, 1009–1017. [Google Scholar] [CrossRef]
- Song, A.; Chen, Y.F.; Thamatrakoln, K.; Storm, T.A.; Krensky, A.M. RFLAT-1: A new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity 1999, 10, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Lavallee, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; Horb, M.E.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213. [Google Scholar] [CrossRef]
- Leclerc, N.; Luppen, C.A.; Ho, V.V.; Nagpal, S.; Hacia, J.G.; Smith, E.; Frenkel, B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J. Mol. Endocrinol. 2004, 33, 175–193. [Google Scholar] [CrossRef]
- Parker-Katiraee, L.; Carson, A.R.; Yamada, T.; Arnaud, P.; Feil, R.; Abu-Amero, S.N.; Moore, G.E.; Kaneda, M.; Perry, G.H.; Stone, A.C.; et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007, 3, e65. [Google Scholar] [CrossRef] [PubMed]
- Small, K.S.; Hedman, A.K.; Grundberg, E.; Nica, A.C.; Thorleifsson, G.; Kong, A.; Thorsteindottir, U.; Shin, S.Y.; Richards, H.B.; Consortium, G.; et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011, 43, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Wu, J.; Chen, W.; Fan, H.; Liu, H. KLF14 inhibits osteogenic differentiation of human bone marrow mesenchymal stem cells by downregulating WNT3A. Am. J. Transl. Res. 2020, 12, 4445–4455. [Google Scholar] [PubMed]
- Compston, J. Management of glucocorticoid-induced osteoporosis: What is new? Int. J. Rheum. Dis. 2019, 22, 1595–1597. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Su, Y.; Wang, D.; Chen, Y.; Liu, Y.; Luo, S.; Wu, T.; Cui, L. Tanshinol Rescues the Impaired Bone Formation Elicited by Glucocorticoid Involved in KLF15 Pathway. Oxidative Med. Cell Longev. 2016, 2016, 1092746. [Google Scholar] [CrossRef]
- Mori, T.; Sakaue, H.; Iguchi, H.; Gomi, H.; Okada, Y.; Takashima, Y.; Nakamura, K.; Nakamura, T.; Yamauchi, T.; Kubota, N.; et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 2005, 280, 12867–12875. [Google Scholar] [CrossRef]
- Yang, Y.J.; Zhu, Z.; Wang, D.T.; Zhang, X.L.; Liu, Y.Y.; Lai, W.X.; Mo, Y.L.; Li, J.; Liang, Y.L.; Hu, Z.Q.; et al. Tanshinol alleviates impaired bone formation by inhibiting adipogenesis via KLF15/PPARgamma2 signaling in GIO rats. Acta Pharmacol. Sin. 2018, 39, 633–641. [Google Scholar] [CrossRef]
- Song, Z.; Lian, X.; Wang, Y.; Xiang, Y.; Li, G. KLF15 regulates in vitro chondrogenic differentiation of human mesenchymal stem cells by targeting SOX9. Biochem. Biophys. Res. Commun. 2017, 493, 1082–1088. [Google Scholar] [CrossRef]
- Hwang, C.K.; D’Souza, U.M.; Eisch, A.J.; Yajima, S.; Lammers, C.H.; Yang, Y.; Lee, S.H.; Kim, Y.M.; Nestler, E.J.; Mouradian, M.M. Dopamine receptor regulating factor, DRRF: A zinc finger transcription factor. Proc. Natl. Acad. Sci. USA 2001, 98, 7558–7563. [Google Scholar] [CrossRef]
- Jang, M.K.; Lee, S.; Jung, M.H. RNA-Seq Analysis Reveals a Negative Role of KLF16 in Adipogenesis. PLoS ONE 2016, 11, e0162238. [Google Scholar] [CrossRef]
- van Vliet, J.; Crofts, L.A.; Quinlan, K.G.; Czolij, R.; Perkins, A.C.; Crossley, M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics 2006, 87, 474–482. [Google Scholar] [CrossRef]
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef] [PubMed]
- Fellows, C.R.; Matta, C.; Mobasheri, A. Applying Proteomics to Study Crosstalk at the Cartilage-Subchondral Bone Interface in Osteoarthritis: Current Status and Future Directions. EBioMedicine 2016, 11, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Tan, H.; Dai, P. Kruppel-Like Factor 2 Regulates Degradation of Type II Collagen by Suppressing the Expression of Matrix Metalloproteinase (MMP)-13. Cell Physiol. Biochem. 2017, 42, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- Fisch, K.M.; Gamini, R.; Alvarez-Garcia, O.; Akagi, R.; Saito, M.; Muramatsu, Y.; Sasho, T.; Koziol, J.A.; Su, A.I.; Lotz, M.K. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthr. Cartil. 2018, 26, 1531–1538. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, M.; Xiao, W. KLF15 Regulates the Expression of MMP-3 in Human Chondrocytes. J. Interferon Cytokine Res. 2018, 38, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Huang, H.; Pan, X.; Li, S.; Xie, Z.; Ma, Y.; Hu, B.; Wang, J.; Chen, Z.; Shi, P. EGR1 promotes the cartilage degeneration and hypertrophy by activating the Kruppel-like factor 5 and beta-catenin signaling. Biochim. Et Biophys. Acta Mol. Basis Dis. 2019, 1865, 2490–2503. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, D.; Yan, Z.; Zhan, J.; Xue, X.; Pan, X.; Yu, H. LncRNA MEG3 Protects Chondrocytes From IL-1beta-Induced Inflammation via Regulating miR-9-5p/KLF4 Axis. Front. Physiol. 2021, 12, 617654. [Google Scholar] [CrossRef]
- Li, J.; Jiang, M.; Xiong, C.; Pan, J.; Jia, S.; Zhang, Y.; Zhang, J.; Xu, N.; Zhou, X.; Huang, Y. KLF4, negatively regulated by miR-7, suppresses osteoarthritis development via activating TGF-beta1 signaling. Int. Immunopharmacol. 2022, 102, 108416. [Google Scholar] [CrossRef]
- Gao, X.; Jiang, S.; Du, Z.; Ke, A.; Liang, Q.; Li, X. KLF2 Protects against Osteoarthritis by Repressing Oxidative Response through Activation of Nrf2/ARE Signaling In Vitro and In Vivo. Oxidative Med. Cell Longev. 2019, 2019, 8564681. [Google Scholar] [CrossRef]
- Yu, S.M.; Kim, S.J. Kruppel-like factor 4 (KLF-4) plays a crucial role in simvastatin (SVT)-induced differentiation of rabbit articular chondrocytes. Biochem. Biophys. Res. Commun. 2018, 501, 814–819. [Google Scholar] [CrossRef]
- Kawata, M.; Teramura, T.; Ordoukhanian, P.; Head, S.R.; Natarajan, P.; Sundaresan, A.; Olmer, M.; Asahara, H.; Lotz, M.K. Kruppel-like factor-4 and Kruppel-like factor-2 are important regulators of joint tissue cells and protect against tissue destruction and inflammation in osteoarthritis. Ann. Rheum. Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, Y.; Zeng, G.; Yang, T.; Song, W. INSR mediated by transcription factor KLF4 and DNA methylation ameliorates osteoarthritis progression via inactivation of JAK2/STAT3 signaling pathway. Am. J. Transl. Res. 2020, 12, 7953–7967. [Google Scholar]
- Lai, X.; Song, Y.; Tian, J. CircCDK14 ameliorates interleukin-1beta-induced chondrocyte damage by the miR-1183/KLF5 pathway in osteoarthritis. Autoimmunity 2022, 55, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Lu, H.; Li, H.; Xu, X.; Wang, D. KLF10 is upregulated in osteoarthritis and inhibits chondrocyte proliferation and migration by upregulating Acvr1 and suppressing inhbb expression. Acta Histochem. 2020, 122, 151528. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Jiang, H.; Qu, W.; Rui, Y.J. KLF11 protects chondrocytes via inhibiting p38 MAPK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6505–6516. [Google Scholar] [CrossRef] [PubMed]
- Compston, J.E.; McClung, M.R.; Leslie, W.D. Osteoporosis. Lancet 2019, 393, 364–376. [Google Scholar] [CrossRef]
- Miller, P.D.; Barlas, S.; Brenneman, S.K.; Abbott, T.A.; Chen, Y.T.; Barrett-Connor, E.; Siris, E.S. An approach to identifying osteopenic women at increased short-term risk of fracture. Arch. Intern. Med. 2004, 164, 1113–1120. [Google Scholar] [CrossRef]
- Siris, E.S.; Chen, Y.T.; Abbott, T.A.; Barrett-Connor, E.; Miller, P.D.; Wehren, L.E.; Berger, M.L. Bone mineral density thresholds for pharmacological intervention to prevent fractures. Arch. Intern. Med. 2004, 164, 1108–1112. [Google Scholar] [CrossRef]
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef]
- Viprakasit, V.; Ekwattanakit, S.; Riolueang, S.; Chalaow, N.; Fisher, C.; Lower, K.; Kanno, H.; Tachavanich, K.; Bejrachandra, S.; Saipin, J.; et al. Mutations in Kruppel-like factor 1 cause transfusion-dependent hemolytic anemia and persistence of embryonic globin gene expression. Blood 2014, 123, 1586–1595. [Google Scholar] [CrossRef]
- Kulczynska-Figurny, K.; Bieker, J.J.; Siatecka, M. Severe anemia caused by dominant mutations in Kruppel-like factor 1 (KLF1). Mutat. Res. Rev. Mutat. Res. 2020, 786, 108336. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Lin, C.L.; Su, C.H.; Lin, K.C.; Leong, K.H.; Wang, Y.T.; Kuo, C.F.; Tsai, S.Y. The Risk of Developing Osteoporosis in Hemolytic Anemia-What Aggravates the Bone Loss? J. Clin. Med. 2021, 10, 3364. [Google Scholar] [CrossRef] [PubMed]
- Kristjansdottir, H.L.; Mellstrom, D.; Johansson, P.; Karlsson, M.; Vandenput, L.; Lorentzon, M.; Herlitz, H.; Ohlsson, C.; Lerner, U.H.; Lewerin, C. Anemia is associated with increased risk of non-vertebral osteoporotic fractures in elderly men: The MrOS Sweden cohort. Arch. Osteoporos. 2022, 17, 85. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.G.; Wei, Z.X.; Liu, J.B. Reduced local blood supply to the tibial metaphysis is associated with ovariectomy-induced osteoporosis in mice. Connect. Tissue Res. 2011, 52, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Portal-Nunez, S.; Lozano, D.; Esbrit, P. Role of angiogenesis on bone formation. Histol. Histopathol. 2012, 27, 559–566. [Google Scholar] [CrossRef]
- Tong, X.; Chen, X.; Zhang, S.; Huang, M.; Shen, X.; Xu, J.; Zou, J. The Effect of Exercise on the Prevention of Osteoporosis and Bone Angiogenesis. BioMed Res. Int. 2019, 2019, 8171897. [Google Scholar] [CrossRef] [PubMed]
- Rodda, S.J.; McMahon, A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 2006, 133, 3231–3244. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Rowland, B.D.; Peeper, D.S. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 2006, 6, 11–23. [Google Scholar] [CrossRef]
- Zhang, M.; Xuan, S.; Bouxsein, M.L.; von Stechow, D.; Akeno, N.; Faugere, M.C.; Malluche, H.; Zhao, G.; Rosen, C.J.; Efstratiadis, A.; et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 2002, 277, 44005–44012. [Google Scholar] [CrossRef]
- Lu, Y.; Xie, Y.; Zhang, S.; Dusevich, V.; Bonewald, L.F.; Feng, J.Q. DMP1-targeted Cre expression in odontoblasts and osteocytes. J. Dent. Res. 2007, 86, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Logan, M.; Martin, J.F.; Nagy, A.; Lobe, C.; Olson, E.N.; Tabin, C.J. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 2002, 33, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Sosic, D.; Richardson, J.A.; Yu, K.; Ornitz, D.M.; Olson, E.N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell 2003, 112, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Xu, J.; Liu, Z.; Sosic, D.; Shao, J.; Olson, E.N.; Towler, D.A.; Ornitz, D.M. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 2003, 130, 3063–3074. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, J.; Tomishima, M.; Liu, H.; Zhao, C.; Cai, X.; Marth, J.D.; Enquist, L.; Friedman, J.M. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 2001, 291, 2608–2613. [Google Scholar] [CrossRef]
- Hopwood, B.; Tsykin, A.; Findlay, D.M.; Fazzalari, N.L. Gene expression profile of the bone microenvironment in human fragility fracture bone. Bone 2009, 44, 87–101. [Google Scholar] [CrossRef]
- You, M.; Zhang, L.; Zhang, X.; Fu, Y.; Dong, X. MicroRNA-197-3p Inhibits the Osteogenic Differentiation in Osteoporosis by Down-Regulating KLF 10. Clin. Interv. Aging 2021, 16, 107–117. [Google Scholar] [CrossRef]
- Tau, K.R.; Hefferan, T.E.; Waters, K.M.; Robinson, J.A.; Subramaniam, M.; Riggs, B.L.; Spelsberg, T.C. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology 1998, 139, 1346–1353. [Google Scholar] [CrossRef]
- Asada, M.; Rauch, A.; Shimizu, H.; Maruyama, H.; Miyaki, S.; Shibamori, M.; Kawasome, H.; Ishiyama, H.; Tuckermann, J.; Asahara, H. DNA binding-dependent glucocorticoid receptor activity promotes adipogenesis via Kruppel-like factor 15 gene expression. Lab. Investig. A J. Tech. Methods Pathol. 2011, 91, 203–215. [Google Scholar] [CrossRef]
- Lam, F.F.; Yeung, J.H.; Chan, K.M.; Or, P.M. Relaxant effects of danshen aqueous extract and its constituent danshensu on rat coronary artery are mediated by inhibition of calcium channels. Vasc. Pharmacol. 2007, 46, 271–277. [Google Scholar] [CrossRef]
- Cui, L.; Liu, Y.Y.; Wu, T.; Ai, C.M.; Chen, H.Q. Osteogenic effects of D+beta-3,4-dihydroxyphenyl lactic acid (salvianic acid A, SAA) on osteoblasts and bone marrow stromal cells of intact and prednisone-treated rats. Acta Pharmacol. Sin. 2009, 30, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Yang, Y.; Chen, J.; Zhong, Z.; Huang, H.; Zhang, J.; Cui, L. Tanshinol stimulates bone formation and attenuates dexamethasone-induced inhibition of osteogenesis in larval zebrafish. J. Orthop. Transl. 2016, 4, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Tetreault, M.P.; Yang, Y.; Katz, J.P. Kruppel-like factors in cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Zapico, M.E.; Mladek, A.; Ellenrieder, V.; Folch-Puy, E.; Miller, L.; Urrutia, R. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 2003, 22, 4748–4758. [Google Scholar] [CrossRef]
- Rowland, B.D.; Bernards, R.; Peeper, D.S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005, 7, 1074–1082. [Google Scholar] [CrossRef]
- Guo, P.; Dong, X.Y.; Zhang, X.; Zhao, K.W.; Sun, X.; Li, Q.; Dong, J.T. Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification. J. Biol. Chem. 2009, 284, 6071–6078. [Google Scholar] [CrossRef]
- Yang, Y.; Nakagawa, H.; Tetreault, M.P.; Billig, J.; Victor, N.; Goyal, A.; Sepulveda, A.R.; Katz, J.P. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res. 2011, 71, 6475–6484. [Google Scholar] [CrossRef]
- Yang, Y.; Tarapore, R.S.; Jarmel, M.H.; Tetreault, M.P.; Katz, J.P. p53 mutation alters the effect of the esophageal tumor suppressor KLF5 on keratinocyte proliferation. Cell Cycle 2012, 11, 4033–4039. [Google Scholar] [CrossRef]
- Messerschmitt, P.J.; Garcia, R.M.; Abdul-Karim, F.W.; Greenfield, E.M.; Getty, P.J. Osteosarcoma. J. Am. Acad. Orthop. Surg. 2009, 17, 515–527. [Google Scholar] [CrossRef]
- Raymond, A.K.; Jaffe, N. Osteosarcoma multidisciplinary approach to the management from the pathologist’s perspective. Cancer Treat. Res. 2009, 152, 63–84. [Google Scholar] [CrossRef]
- Taniguchi, H.; Jacinto, F.V.; Villanueva, A.; Fernandez, A.F.; Yamamoto, H.; Carmona, F.J.; Puertas, S.; Marquez, V.E.; Shinomura, Y.; Imai, K.; et al. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene 2012, 31, 1988–1994. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Qian, Z.; Jiang, F.; Ge, D.; Tang, J.; Chen, H.; Yang, J.; Yao, Y.; Yan, J.; Zhao, L.; et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am. J. Transl. Res. 2019, 11, 4126–4138. [Google Scholar] [PubMed]
- Qi, X.T.; Li, Y.L.; Zhang, Y.Q.; Xu, T.; Lu, B.; Fang, L.; Gao, J.Q.; Yu, L.S.; Zhu, D.F.; Yang, B.; et al. KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol. Sin. 2019, 40, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xian, M.; Yang, B.; Ying, M.; He, Q. Inhibition of KLF4 by Statins Reverses Adriamycin-Induced Metastasis and Cancer Stemness in Osteosarcoma Cells. Stem Cell Rep. 2017, 8, 1617–1629. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, P.; Liu, Q.; Wang, J.; Yan, F.; Duan, L.; Lin, F. KLF8 promotes cancer stem cell-like phenotypes in osteosarcoma through miR-429-SOX2 signaling. Neoplasma 2020, 67, 519–527. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Chen, H.; Yang, Y.; Xiao, C.; Yi, X.; Shi, C.; Zhong, K.; He, H.; Li, Y.; et al. Genome-wide CRISPR-Cas9 screen identified KLF11 as a druggable suppressor for sarcoma cancer stem cells. Sci. Adv. 2021, 7, eabe3445. [Google Scholar] [CrossRef]
- Lin, F.; Shen, Z.; Tang, L.N.; Zheng, S.E.; Sun, Y.J.; Min, D.L.; Yao, Y. KLF8 knockdown suppresses proliferation and invasion in human osteosarcoma cells. Mol. Med. Rep. 2014, 9, 1613–1617. [Google Scholar] [CrossRef][Green Version]
- Wei, W.; Ji, L.; Duan, W.; Zhu, J. CircSAMD4A contributes to cell doxorubicin resistance in osteosarcoma by regulating the miR-218-5p/KLF8 axis. Open Life Sci. 2020, 15, 848–859. [Google Scholar] [CrossRef]
- Wang, B.; Yan, L.; Shi, W.; Xie, H.; Chen, R.; Shao, Y.; Liang, W. CircRNA PVT1 promotes proliferation and chemoresistance of osteosarcoma cells via the miR-24-3p/KLF8 axis. Int. J. Clin. Oncol. 2022, 27, 811–822. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Desouza, C.V.; Shivaswamy, V. Pioglitazone in the treatment of type 2 diabetes: Safety and efficacy review. Clin. Med. Insights. Endocrinol. Diabetes 2010, 3, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Liu, H.; Huang, C. KLF5-induced miR-487a augments the progression of osteosarcoma cells by targeting NKX3-1 in vitro. Oncol. Lett. 2022, 24, 258. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Han, Y.; Chen, Z.; Pan, X.; Yuan, P.; Zhao, X.; Zhu, H.; Wang, J.; Sun, X.; Shi, P. ML264 inhibits osteosarcoma growth and metastasis via inhibition of JAK2/STAT3 and WNT/beta-catenin signalling pathways. J. Cell Mol. Med. 2020, 24, 5652–5664. [Google Scholar] [CrossRef]
- Jianwei, Z.; Enzhong, B.; Fan, L.; Jian, L.; Ning, A. Effects of Kruppel-like factor 6 on osteosarcoma cell biological behavior. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2013, 34, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abe, M.; Saeki, N.; Ikeda, Y.; Ohba, S. Kruppel-like Factors in Skeletal Physiology and Pathologies. Int. J. Mol. Sci. 2022, 23, 15174. https://doi.org/10.3390/ijms232315174
Abe M, Saeki N, Ikeda Y, Ohba S. Kruppel-like Factors in Skeletal Physiology and Pathologies. International Journal of Molecular Sciences. 2022; 23(23):15174. https://doi.org/10.3390/ijms232315174
Chicago/Turabian StyleAbe, Makoto, Naoya Saeki, Yuki Ikeda, and Shinsuke Ohba. 2022. "Kruppel-like Factors in Skeletal Physiology and Pathologies" International Journal of Molecular Sciences 23, no. 23: 15174. https://doi.org/10.3390/ijms232315174
APA StyleAbe, M., Saeki, N., Ikeda, Y., & Ohba, S. (2022). Kruppel-like Factors in Skeletal Physiology and Pathologies. International Journal of Molecular Sciences, 23(23), 15174. https://doi.org/10.3390/ijms232315174