CD200 as a Potential New Player in Inflammation during Rotator Cuff Tendon Injury/Repair: An In Vitro Model
Abstract
:1. Introduction
2. Results
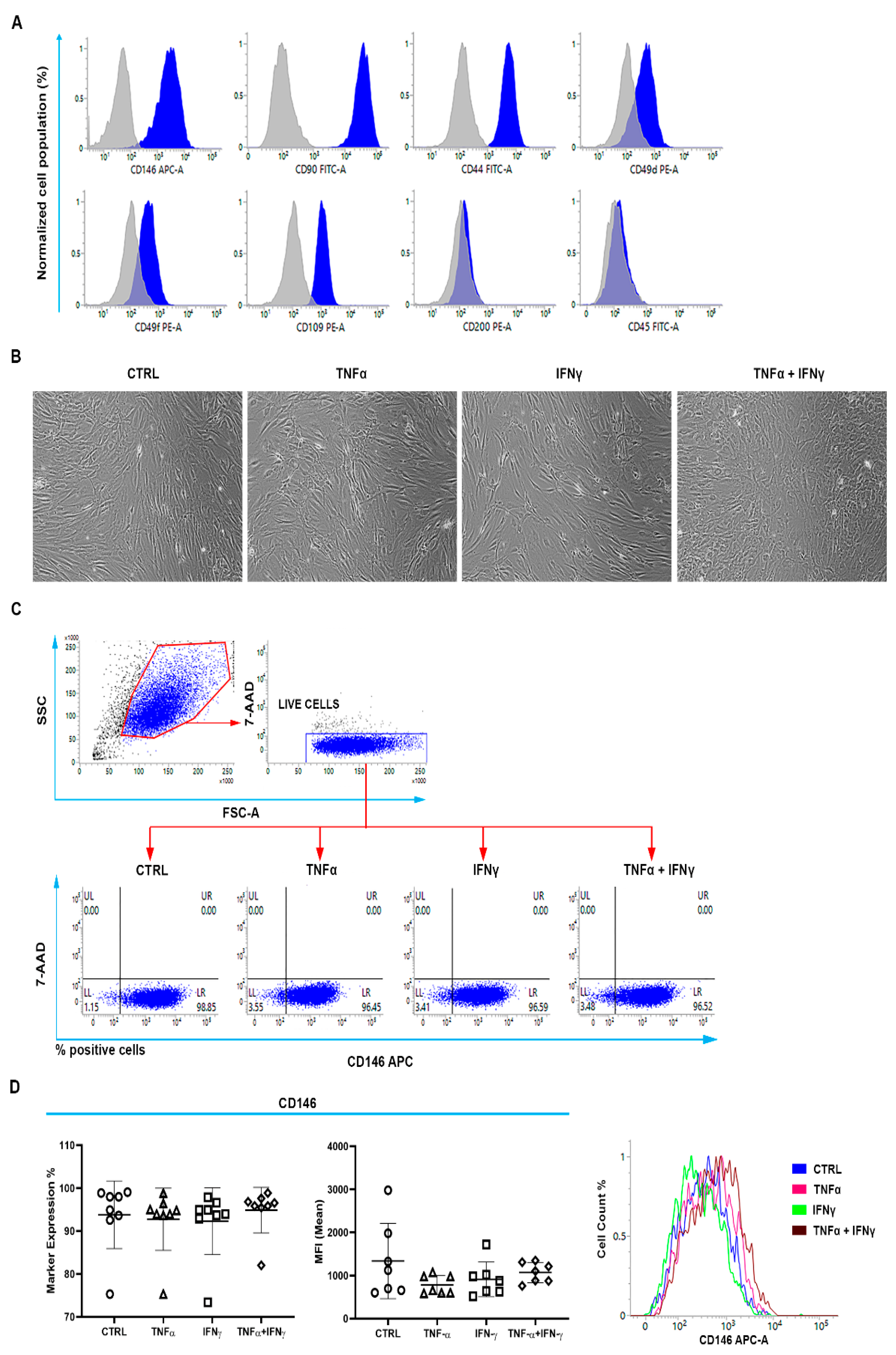
2.1. Identification of Cell Surface Markers Characterizing the TSPC Population
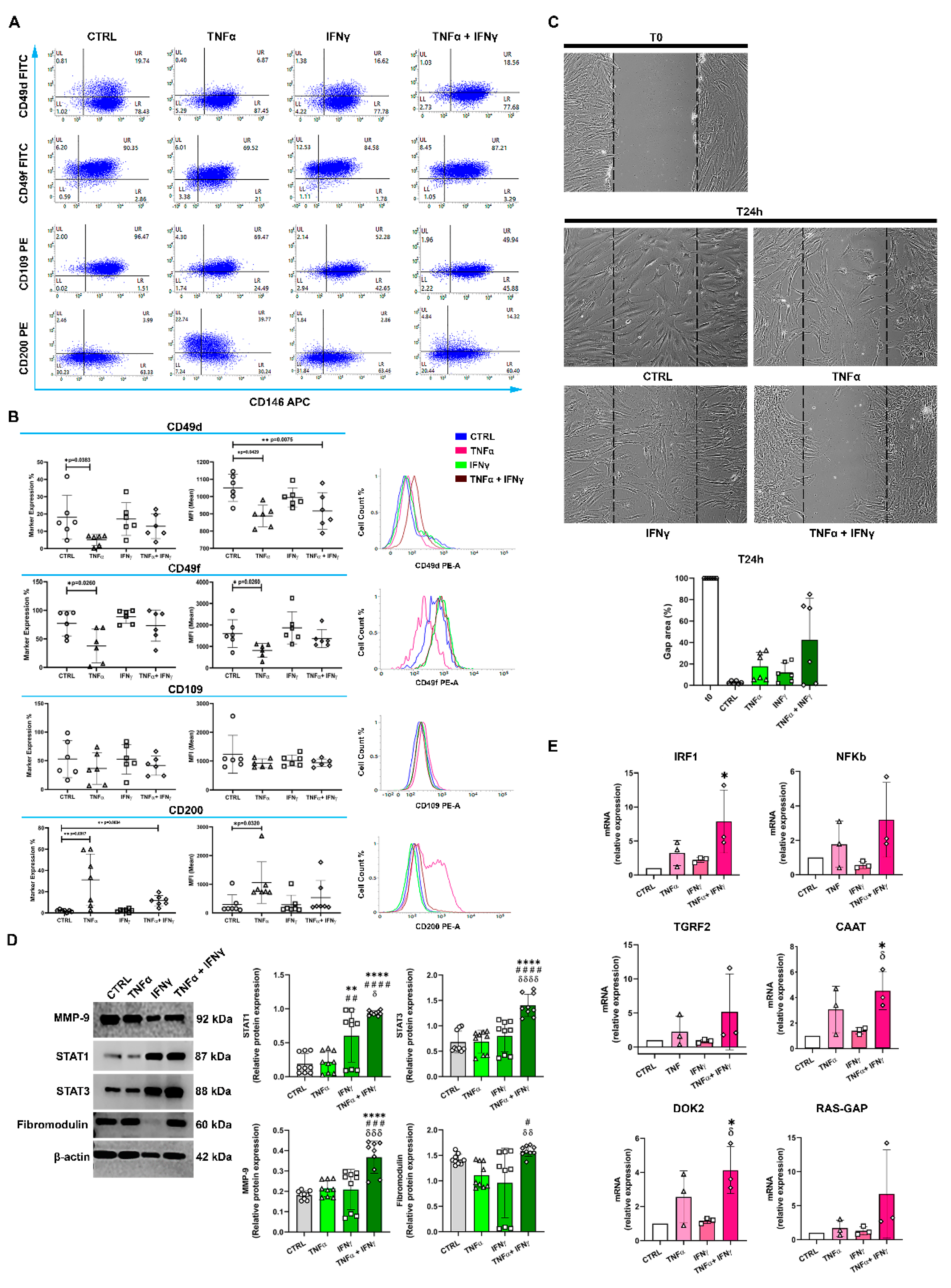
2.2. CD146+TSPCs Response to TNFα and IFNγ
2.3. TNFα and IFNγ Increased the Expression of CD146+CD200+ TSPCs
2.4. The In Vitro Gap Repair Assay
2.5. IFNγ+TNFα Increased the Expression of STAT1, STAT3, and MMP9 Proteins
2.6. IFNγ+TNFα Increased the Expression of IRF1, CAAT and DOK2 mRNA
3. Discussion
4. Materials and Methods
4.1. Rotator Cuff Tendon-Derived Cells Cultures
4.2. Flow Cytometry
4.3. In Vitro Gap Repair Assay
4.4. Immunoblotting
4.5. Gene Expression Analysis
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chisari, E.; Rehak, L.; Khan, W.S.; Maffulli, N. Tendon healing is adversely affected by low-grade inflammation. J. Orthop. Surg. Res. 2021, 16, 700. [Google Scholar] [CrossRef] [PubMed]
- Chisari, E.; Rehak, L.; Khan, W.S.; Maffulli, N. Tendon healing in presence of chronic low-level inflammation: A systematic review. Br. Med. Bull. 2019, 132, 97–116. [Google Scholar] [CrossRef]
- Cipollaro, L.; Sahemey, R.; Oliva, F.; Maffulli, N. Immunohistochemical features of rotator cuff tendinopathy. Br. Med. Bull. 2019, 130, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Riley, G.P. Gene expression and matrix turnover in overused and damaged tendons. Scand. J. Med. Sci. Sports 2005, 15, 241–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dakin, S.G.; Martinez, F.O.; Yapp, C.; Wells, G.; Oppermann, U.; Dean, B.J.; Smith, R.D.; Wheway, K.; Watkins, B.; Roche, L.; et al. Inflammation activation and resolution in human tendon disease. Sci. Transl. Med. 2015, 7, 311ra173. [Google Scholar] [CrossRef] [Green Version]
- Kendal, A.R.; Layton, T.; Al-Mossawi, H.; Appleton, L.; Dakin, S.; Brown, R.; Loizou, C.; Rogers, M.; Sharp, R.; Carr, A. Multi-omic single cell analysis resolves novel stromal cell populations in healthy and diseased human tendon. Sci. Rep. 2020, 10, 13939. [Google Scholar] [CrossRef]
- Russo, V.; El Khatib, M.; Prencipe, G.; Citeroni, M.R.; Faydaver, M.; Mauro, A.; Berardinelli, P.; Cerveró-Varona, A.; Haidar-Montes, A.A.; Turriani, M.; et al. Tendon Immune Regeneration: Insights on the Synergetic Role of Stem and Immune Cells during Tendon Regeneration. Cells 2022, 11, 434. [Google Scholar] [CrossRef]
- Bi, Y.; Ehirchiou, D.; Kilts, T.M.; Inkson, C.A.; Embree, M.C.; Sonoyama, W.; Li, L.; Leet, A.I.; Seo, B.M.; Zhang, L.; et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 2007, 13, 1219. [Google Scholar] [CrossRef]
- Tarafder, S.; Chen, E.; Jun, Y.; Kao, K.; Hee Sim, K.; Back, J.; Lee, F.Y.; Lee, C.H. Tendon stem/progenitor cells regulate inflammation in tendon healing via JNK and STAT3 signaling. FASEB J. 2017, 31, 3991–3998. [Google Scholar] [CrossRef] [Green Version]
- Vinhas, A.; Rodrigues, M.T.; Gomes, M.E. Exploring Stem Cells and Inflammation in Tendon Repair and Regeneration. Adv. Exp. Med. Biol. 2018, 1089, 37–46. [Google Scholar] [CrossRef]
- Hoek, R.M.; Ruuls, S.R.; Murphy, C.A.; Wright, G.J.; Goddard, R.; Zurawski, S.M.; Blom, B.; Homola, M.E.; Streit, W.J.; Brown, M.H.; et al. Downregulation of the macrophage lineage through interaction with OX2 (CD200). Science 2000, 290, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.J.; Puklavec, M.J.; Willis, A.C.; Hoek, R.M.; Sedgwick, J.D.; Brown, M.H.; Barclay, A.N. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity 2000, 13, 233–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorczynski, R.; Chen, Z.; Kai, Y.; Lee, L.; Wong, S.; Marsden, P.A. CD200 is a ligand for all members of the CD200R family of immunoregulatory molecules. J. Immunol. 2004, 172, 7744–7749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotwica-Mojzych, K.; Jodłowska-Jędrych, B.; Mojzych, M. CD200:CD200R Interactions and Their Importance in Immunoregulation. Int. J. Mol. Sci. 2021, 22, 1602. [Google Scholar] [CrossRef]
- Chen, Z.; Marsden, P.A.; Gorczynski, R.M. Role of distal enhancer in the transcriptional responsiveness of the human CD200 gene to interferon-y and tumor necrosis factor-α. Mol. Immunol. 2009, 46, 1951–1963. [Google Scholar] [CrossRef]
- Grol, M.W.; Haelterman, N.A.; Lim, J.; Munivez, E.M.; Archer, M.; Hudson, D.M.; Tufa, S.F.; Keene, D.R.; Lei, K.; Park, D.; et al. Tendon and motor phenotypes in the Crtap−/− mouse model of recessive osteogenesis imperfecta. eLife 2021, 26, e63488. [Google Scholar] [CrossRef]
- Alshoubaki, Y.K.; Nayer, B.; Das, S.; Martino, M.M. Modulation of the Activity of Stem and Progenitor Cells by Immune Cells. Stem Cells Transl. Med. 2022, 11, 248–258. [Google Scholar] [CrossRef]
- Baiula, M.; Spampinato, S.; Gentilucci, L.; Tolomelli, A. Novel Ligands Targeting α4β1 Integrin: Therapeutic Applications and Perspectives. Front. Chem. 2019, 7, 489. [Google Scholar] [CrossRef]
- Krebsbach, P.H.; Villa-Diaz, L.G. The Role of Integrin α6 (CD49f) in Stem Cells: More than a Conserved Biomarker. Stem Cells Dev. 2017, 26, 1090–1099. [Google Scholar] [CrossRef]
- Kumar, S.; Ponnazhagan, S. Bone homing of mesenchymal stem cells by ectopic alpha 4 integrin expression. FASEB J. 2007, 21, 3917–3927. [Google Scholar] [CrossRef]
- Mii, S.; Hoshino, A.; Enomoto, A.; Murakumo, Y.; Ito, M.; Yamaguchi, A.; Takahashi, M. CD109 deficiency induces osteopenia with an osteoporosis-like phenotype in vivo. Genes Cells 2018, 23, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Pontikoglou, C.; Langonné, A.; Ba, M.A.; Varin, A.; Rosset, P.; Charbord, P.; Sensébé, L.; Deschaseaux, F. CD200 expression in human cultured bone marrow mesenchymal stem cells is induced by pro-osteogenic and pro-inflammatory cues. J. Cell. Mol. Med. 2016, 20, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Butturini, E.; Carcereri de Prati, A.; Mariotto, S. Redox Regulation of STAT1 and STAT3 Signaling. Int. J. Mol. Sci. 2020, 21, 7034. [Google Scholar] [CrossRef] [PubMed]
- Wesemann, D.R.; Benveniste, E.N. STAT-1 alpha and IFN-gamma as modulators of TNF-alpha signaling in macrophages: Regulation and functional implications of the TNF receptor 1-STAT-1 alpha complex. J. Immunol. 2003, 171, 5313–5319. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.S.; Han, X.; Liu, X.H. STAT3: A Potential Drug Target for Tumor and Inflammation. Curr. Top. Med. Chem. 2019, 19, 1305–1317. [Google Scholar] [CrossRef]
- Del Buono, A.; Oliva, F.; Osti, L.; Maffulli, N. Metalloproteases and tendinopathy. Muscles Ligaments Tendons J. 2013, 3, 51–57. [Google Scholar] [CrossRef]
- Delalande, A.; Gosselin, M.P.; Suwalski, A.; Guilmain, W.; Leduc, C.; Berchel, M.; Jaffrès, P.A.; Baril, P.; Midoux, P.; Pichon, C. Enhanced Achilles tendon healing by fibromodulin gene transfer. Nanomedicine 2015, 11, 1735–1744. [Google Scholar] [CrossRef]
- Vila-del Sol, V.; Punzón, C.; Fresno, M. IFN-gamma-induced TNF-alpha expression is regulated by interferon regulatory factors 1 and 8 in mouse macrophages. J. Immunol. 2008, 181, 4461–4470. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Zhang, Y.B.; Gui, J.F.; Lemon, S.M.; Yamane, D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021, 17, 1009220. [Google Scholar] [CrossRef]
- Michalska, A.; Blaszczyk, K.; Wesoly, J.; Bluyssen, H.A.R. A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses. Front. Immunol. 2018, 9, 1135. [Google Scholar] [CrossRef]
- Bonelli, M.; Dalwigk, K.; Platzer, A.; Olmos Calvo, I.; Hayer, S.; Niederreiter, B.; Holinka, J.; Sevelda, F.; Pap, T.; Steiner, G.; et al. IRF1 is critical for the TNF-driven interferon response in rheumatoid fibroblast-like synoviocytes: JAKinibs suppress the interferon response in RA-FLSs. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dakin, S.G.; Newton, J.; Martinez, F.O.; Hedley, R.; Gwilym, S.; Jones, N.; Reid, H.A.B.; Wood, S.; Wells, G.; Appleton, L.; et al. Chronic inflammation is a feature of Achilles tendinopathy and rupture. Br. J. Sports Med. 2018, 52, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Courtois, G.; Gilmore, T.D. Mutations in the NF-KappaB signaling pathways: Implications for the human disease. Oncogene 2006, 25, 6831–6843. [Google Scholar] [CrossRef] [Green Version]
- Rius, J.; Guma, M.; Schachtrup, C.; Akassoglou, K.; Zinkernagel, A.S.; Nizet, V.; Johnson, R.S.; Haddad, G.G.; Karin, M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature 2008, 453, 807–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, A.C.; Shah, S.A.; Golman, M.; Song, L.; Li, X.; Kurtaliaj, I.; Akbar, M.; Millar, N.L.; Abu-Amer, Y.; Galatz, L.M.; et al. Targeting the NF-κB signaling pathway in chronic tendon disease. Sci. Transl. Med. 2019, 11, eaav4319. [Google Scholar] [CrossRef]
- Xu, K.; Lin, C.; Ma, D.; Chen, M.; Zhou, X.; He, Y.; Moqbel, S.A.A.; Ma, C.; Wu, L. Spironolactone Ameliorates Senescence and Calcification by Modulating Autophagy in Rat Tendon-Derived Stem Cells via the NF-κB/MAPK Pathway. Oxidative Med. Cell. Longev. 2021, 2021, 5519587. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Yu, T.; An, S.; Deng, R.; Tan, X.; Crane, J.; Zhang, W.; Pan, D.; Wan, M.; et al. Inhibition of Integrin αvβ6 Activation of TGF-β Attenuates Tendinopathy. Adv. Sci. 2022, 9, e2104469. [Google Scholar] [CrossRef]
- Ko, C.Y.; Chang, W.C.; Wang, J.M. Biological roles of CCAAT/Enhancer-binding protein delta during inflammation. J. Biomed. Sci. 2015, 22, 6. [Google Scholar] [CrossRef] [Green Version]
- Suzu, S.; Tanaka-Douzono, M.; Nomaguchi, K.; Yamada, M.; Hayasawa, H.; Kimura, F.; Motoyoshi, K. p56(dok-2) as a cytokine-inducible inhibitor of cell proliferation and signal transduction. EMBO J. 2000, 19, 5114–5122. [Google Scholar] [CrossRef] [Green Version]
- Oliva, F.; Berardi, A.C.; Misiti, S.; Verga Falzacappa, C.; Iacone, A.; Maffulli, N. Thyroid hormones enhance growth and counteract apoptosis in human tenocytes isolated from rotator cuff tendons. Cell Death Dis. 2013, 4, e705. [Google Scholar] [CrossRef]
- Osti, L.; Berardocco, M.; di Giacomo, V.; Di Bernardo, G.; Oliva, F.; Berardi, A.C. Hyaluronic acid increases tendon derived cell viability and collagen type I expression in vitro: Comparative study of four different Hyaluronic acid preparations by molecular weight. BMC Musculoskelet. Disord. 2015, 16, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallorini, M.; Berardi, A.C.; Gissi, C.; Cataldi, A.; Osti, L. Nrf2-mediated cytoprotective effect of four different hyaluronic acids by molecular weight in human tenocytes. J. Drug Target. 2020, 28, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Berardi, A.C.; Oliva, F.; Berardocco, M.; la Rovere, M.; Accorsi, P.; Maffulli, N. Thyroid hormones increase collagen I and cartilage oligomeric matrix protein (COMP) expression in vitro human tenocytes. Muscles Ligaments Tendons J. 2014, 4, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Bestwick, C.S.; Bestwick, L.A.; Maffulli, N.; Aspden, R.M. Phenotypic drift in human tenocyte culture. Tissue Eng. 2006, 12, 1843–1849. [Google Scholar] [CrossRef]
- Stolk, M.; Klatte-Schulz, F.; Schmock, A.; Minkwitz, S.; Wildemann, B.; Seifert, M. New insights into tenocyte-immune cell interplay in an in vitro model of inflammation. Sci. Rep. 2017, 7, 9801. [Google Scholar] [CrossRef]
- Gissi, C.; Radeghieri, A.; Antonetti Lamorgese Passeri, C.; Gallorini, M.; Calciano, L.; Oliva, F.; Veronesi, F.; Zendrini, A.; Cataldi, A.; Bergese, P.; et al. Extracellular vesicles from rat-bone-marrow mesenchymal stromal/stem cells improve tendon repair in rat Achilles tendon injury model in dose-dependent manner: A pilot study. PLoS ONE 2020, 15, e0229914. [Google Scholar] [CrossRef]
- Colosimo, A.; Di Rocco, G.; Curini, V.; Russo, V.; Capacchietti, G.; Berardinelli, P.; Mattioli, M.; Barboni, B. Characterization of the methylation status of five imprinted genes in sheep gametes. Anim. Genet. 2009, 40, 900–908. [Google Scholar]
- Cimini, C.; Moussa, F.; Taraschi, A.; Ramal-Sanchez, M.; Colosimo, A.; Giulia Capacchietti, G.; Mokh, S.; Valbonetti, L.; Tagaram, I.; Bernabò, N.; et al. Pre-treatment of swine oviductal epithelial cells with progesterone increases the sperm fertilizing ability in an IVF model. Animals 2022, 12, 1191. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Ragni, E.; Perucca Orfei, C.; Bowles, A.C.; de Girolamo, L.; Correa, D. Reliable Reference Genes for Gene Expression Assessment in Tendon-Derived Cells under Inflammatory and Pro-Fibrotic/Healing Stimuli. Cells 2019, 8, 1188. [Google Scholar] [CrossRef] [Green Version]
- Klatte-Schulz, F.; Pauly, S.; Scheibel, M.; Greiner, S.; Gerhardt, C.; Schmidmaier, G.; Wildemann, B. Influence of age on the cell biological characteristics and the stimulation potential of male human tenocyte-like cells. Eur. Cells Mater. 2012, 24, 74–89. [Google Scholar] [CrossRef]
- Hellebrekers, D.M.; Castermans, K.; Viré, E.; Dings, R.P.; Hoebers, N.T.; Mayo, K.H.; Oude Egbrink, M.G.; Molema, G.; Fuks, F.; van Engeland, M.; et al. Epigenetic regulation of tumor endothelial cell anergy: Silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res. 2006, 66, 10770–10777. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Mosig, R.; Moshier, E.; Pereira, E.; Rahaman, J.; Prasad-Hayes, M.; Halpert, R.; Billaud, J.N.; Dottino, P.; Martignetti, J.A. Interferon regulatory factor 1 is an independent predictor of platinum resistance and survival in high-grade serous ovarian carcinoma. Gynecol. Oncol. 2014, 134, 591–598. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giancola, R.; Oliva, F.; Gallorini, M.; Michetti, N.; Gissi, C.; Moussa, F.; Antonetti Lamorgese Passeri, C.; Colosimo, A.; Berardi, A.C. CD200 as a Potential New Player in Inflammation during Rotator Cuff Tendon Injury/Repair: An In Vitro Model. Int. J. Mol. Sci. 2022, 23, 15165. https://doi.org/10.3390/ijms232315165
Giancola R, Oliva F, Gallorini M, Michetti N, Gissi C, Moussa F, Antonetti Lamorgese Passeri C, Colosimo A, Berardi AC. CD200 as a Potential New Player in Inflammation during Rotator Cuff Tendon Injury/Repair: An In Vitro Model. International Journal of Molecular Sciences. 2022; 23(23):15165. https://doi.org/10.3390/ijms232315165
Chicago/Turabian StyleGiancola, Raffaella, Francesco Oliva, Marialucia Gallorini, Noemi Michetti, Clarissa Gissi, Fadl Moussa, Cristina Antonetti Lamorgese Passeri, Alessia Colosimo, and Anna Concetta Berardi. 2022. "CD200 as a Potential New Player in Inflammation during Rotator Cuff Tendon Injury/Repair: An In Vitro Model" International Journal of Molecular Sciences 23, no. 23: 15165. https://doi.org/10.3390/ijms232315165
APA StyleGiancola, R., Oliva, F., Gallorini, M., Michetti, N., Gissi, C., Moussa, F., Antonetti Lamorgese Passeri, C., Colosimo, A., & Berardi, A. C. (2022). CD200 as a Potential New Player in Inflammation during Rotator Cuff Tendon Injury/Repair: An In Vitro Model. International Journal of Molecular Sciences, 23(23), 15165. https://doi.org/10.3390/ijms232315165








