Functional Analyses of a Small Secreted Cysteine-Rich Protein ThSCSP_14 in Tilletia horrida
Abstract
:1. Introduction
2. Results
2.1. ThSCSP_14 in T. horrida Induces Cell Death in N. benthamiana
2.2. ThSCSP_14 Is Unique to Tilletia fungi
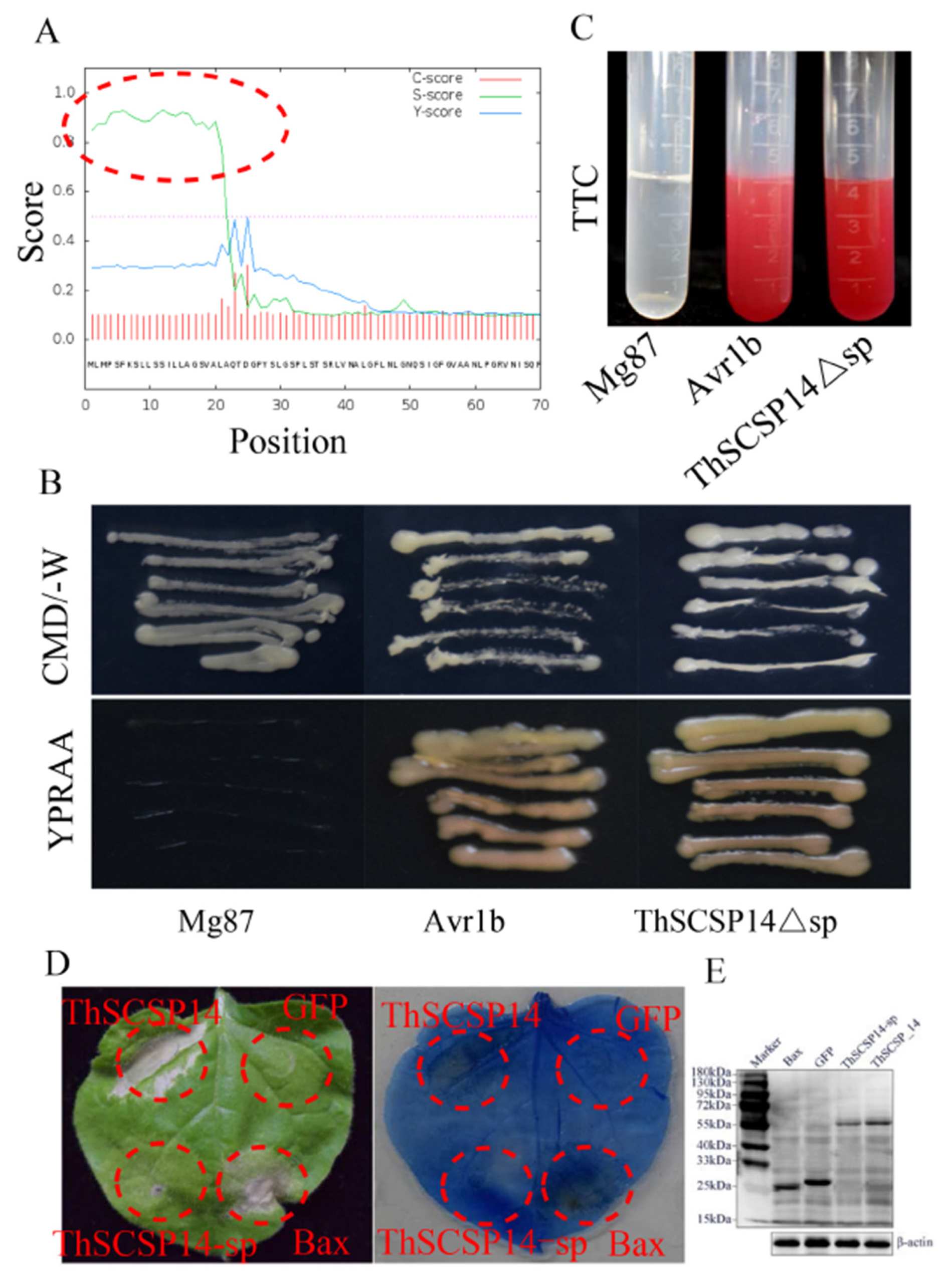
2.3. Functional Validation of the Predicted SP of ThSCSP_14
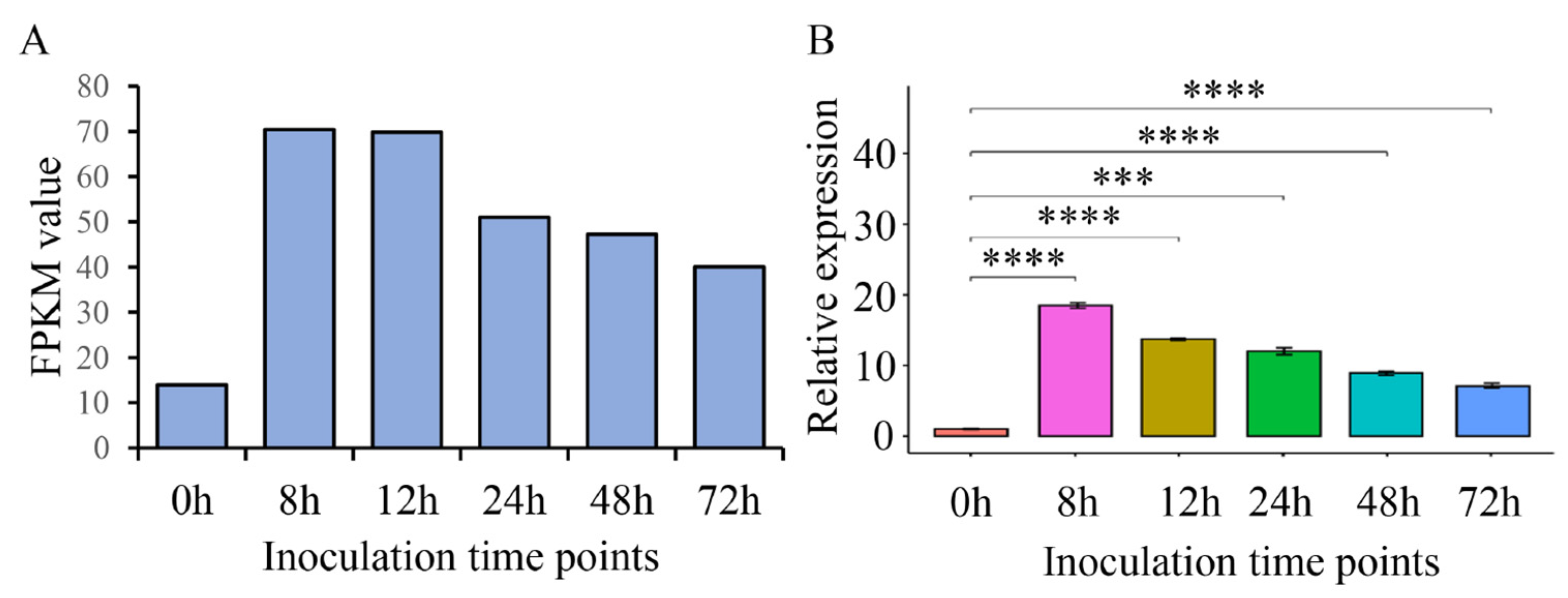
2.4. ThSCSP_14 Expression during T. horrida Infection
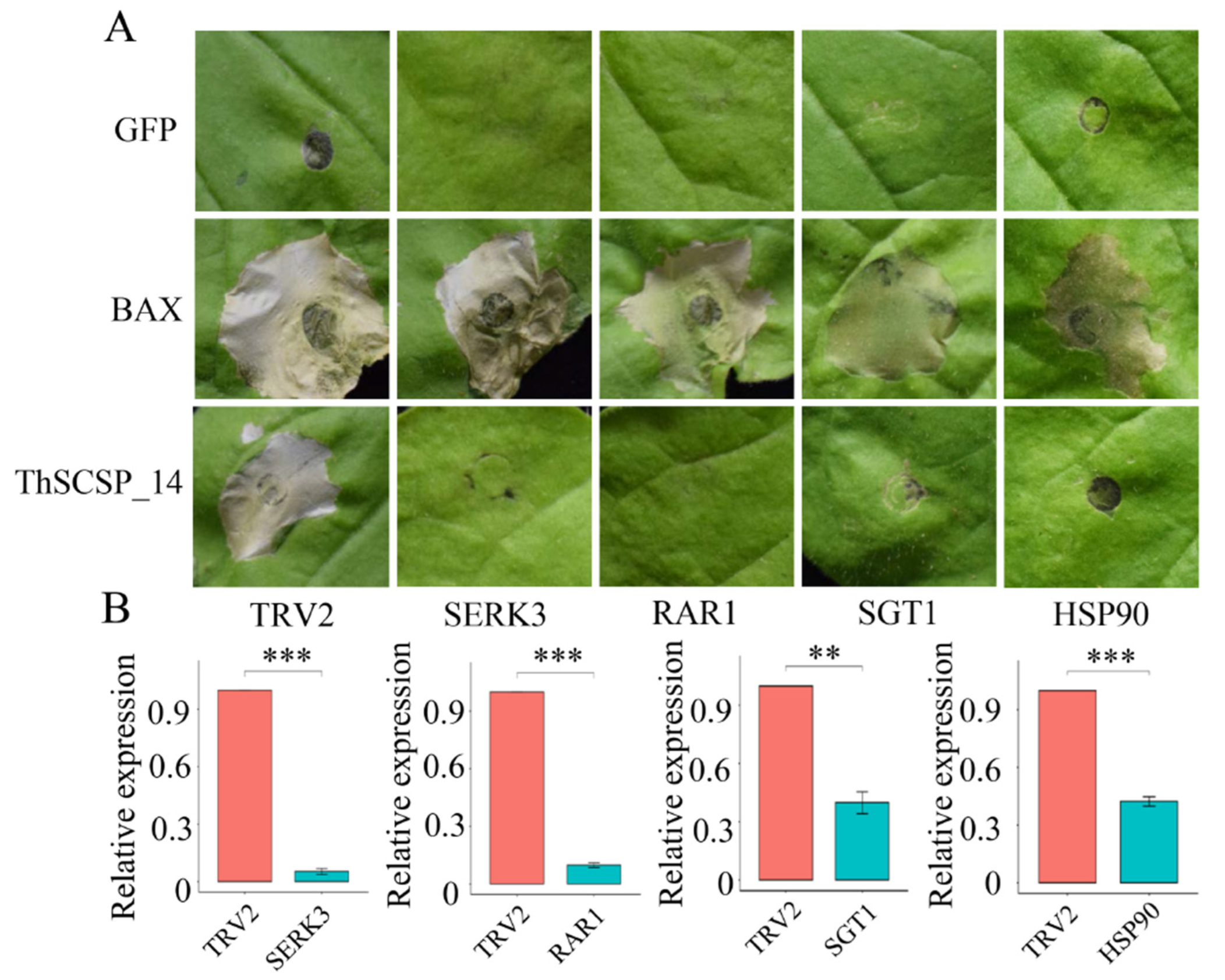
2.5. ThSCSP_14-triggered Cell Death in N. benthamiana Depends on SGT1, RAR1, HSP90, and SERK3/Bak1
2.6. Subcellular Localization of ThSCSP_14
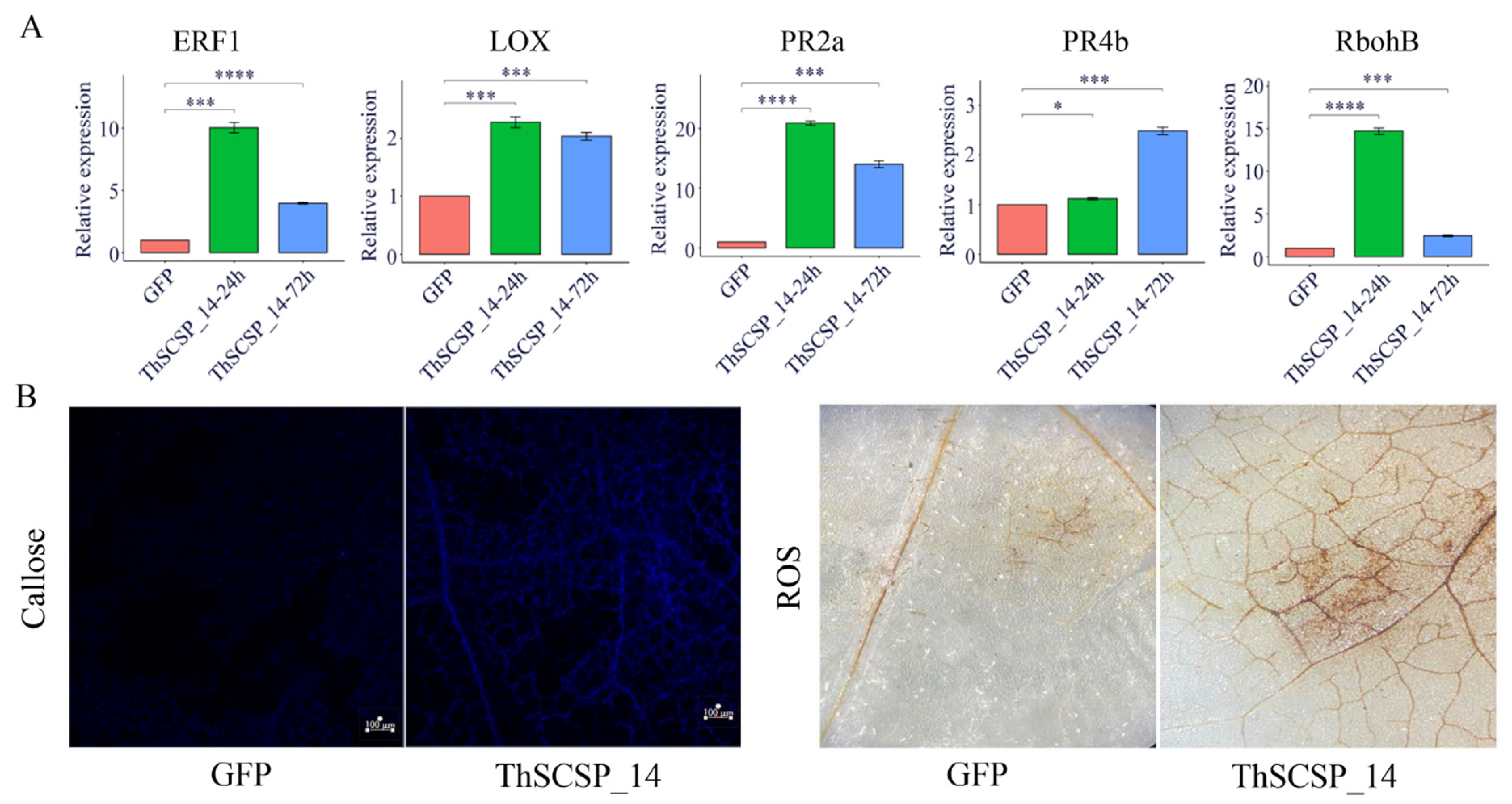
2.7. ThSCSP_14 Triggers Plant Immunity Responses
3. Discussion
4. Materials and Methods
4.1. Strains, Plant Materials, and Growth Conditions
4.2. Candidate Effectors
4.3. Plasmid Construction of Candidate Effectors
4.4. Transient Expression Assays
4.5. Secretory Function Assays of ThSCSP_14′s SP
4.6. RNA Extraction and Gene Expression Analysis
4.7. Western Blot Analysis
4.8. Trypan Blue Staining, H2O2 Activity, and Callose Deposition Observation
4.9. Virus-Induced Gene Silencing in N. benthamiana
4.10. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Yang, X.; Yao, J.; Kyaw, E.P.; Zhang, A.F.; Li, Y.F.; Gu, C.Y.; Zang, H.Y.; Gao, T.C. Simple and rapid detection of Tilletia horrida causing rice kernel smut in rice seeds. Sci. Rep. 2016, 6, 33258. [Google Scholar]
- Webster, P.K.; Gunnell, P.S. Compendium of Rice Diseases; American Phytopathological Society Press: St. Paul, MN, USA, 1992; p. 110. [Google Scholar]
- Wang, N.; Ai, P.; Tang, Y.F.; Zhang, J.F.; Dai, X.J.; Li, P.; Zheng, A.P. Draft Genome Sequence of the Rice Kernel Smut Tilletia horrida Strain QB-1. Genome Announc. 2016, 3, e00621-15. [Google Scholar]
- Rapicavoli, J.; Ingel, B.; Blanco-Ulate, B.; Cantu, D.; Roper, C. Xylella fastidiosa: An examination of a re-emerging plant pathogen. Mol. Plant Pathol. 2018, 19, 786–800. [Google Scholar]
- Dou, D.; Zhou, J.M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 2012, 12, 484–495. [Google Scholar]
- Howden, A.J.; Huitema, E. Effector-triggered post-translational modifications and their role in suppression of plant immunity. Front. Plant Sci. 2012, 3, 160. [Google Scholar]
- Quentin, M.; Abad, P.; Favery, B. Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 2013, 4, 53. [Google Scholar]
- Fang, A.F.; Han, Y.Q.; Zhang, N.; Zhang, M.; Liu, L.J.; Li, S.; Lu, F.; Sun, W. Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens. Mol. Plant Microbe Interact. 2016, 29, 405–416. [Google Scholar]
- Xu, Q.; Wang, J.F.; Zhao, J.R.; Xu, J.H.; Sun, S.T.; Zhang, H.F.; Wu, J.J.; Tang, C.L.; Kang, Z.S.; Wang, X.J. A polysaccharide deacetylase from Puccinia striiformis f. sp. tritici is an important pathogenicity gene that suppresses plant immunity. Plant Biotechnol. J. 2020, 18, 1830–1842. [Google Scholar]
- Mueller, A.N.; Ziemann, S.; Treitschke, S.; Aßmann, D.; Doehlemann, G. Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 2013, 9, e1003177. [Google Scholar]
- Redkar, A.; Villajuana-Bonequi, M.; Doehlemann, G. Conservation of the Ustilago maydis effector See1 in related smuts. Plant Signal. Behav. 2015, 10, e1086855. [Google Scholar]
- Hemetsberger, C.; Herrberger, C.; Zechmann, B.; Hillmer, M.; Doehlemann, G. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 2012, 8, e1002684. [Google Scholar]
- Djamei, A.; Schipper, K.; Rabe, F.; Ghosh, A.; Vincon, V.; Kahnt, J.; Osorio, S.; Tohge, T.; Fernie, A.R.; Feussner, I.; et al. Metabolic priming by a secreted fungal effector. Nature 2011, 478, 395–398. [Google Scholar]
- Tanaka, S.; Brefort, T.; Neidig, N.; Djamei, A.; Kahnt, J.; Vermerris, W.; Koenig, S.; Feussner, K.; Feussner, I.; Kahmann, R. A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife 2014, 3, e01355. [Google Scholar]
- Wang, A.J.; Pan, L.X.; Niu, X.Y.; Shu, X.Y.; Yi, X.Q.; Yamamoto, N.; Li, S.C.; Deng, Q.M.; Zhu, J.; Liang, Y.Y.; et al. Comparative secretome analysis of different smut fungi and identification of plant cell death-inducing secreted proteins from Tilletia horrida. BMC Plant Biol. 2019, 19, 360. [Google Scholar]
- Lyu, X.L.; Shen, C.C.; Fu, Y.P.; Xie, J.T.; Jiang, D.H.; Li, G.Q.; Cheng, J.S. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 2016, 12, e1005435. [Google Scholar]
- Xu, Q.; Tang, C.L.; Wang, X.D.; Sun, S.T.; Zhao, J.R.; Kang, Z.S.; Wang, X.J. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 2019, 10, 5571. [Google Scholar]
- Van der Hoorn, R.A.; Laurent, F.; Roth, R.; De Wit, P.J. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 2000, 13, 439–446. [Google Scholar]
- Luderer, R.; Takken, F.L.; de Wit, P.J.; Joosten, M.H. Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 2002, 45, 875–884. [Google Scholar]
- Wang, A.J.; Pang, L.X.; Wang, N.; Ai, P.; Yin, D.S.; Li, S.C.; Deng, Q.M.; Zhu, J.; Liang, Y.; Zhu, J.; et al. The pathogenic mechanisms of Tilletia horrida as revealed by comparative and functional genomics. Sci. Rep. 2018, 8, 15413. [Google Scholar]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar]
- Lee, S.J.; Rose, J.K. A yeast secretion trap assay for identification of secreted proteins from eukaryotic phytopathogens and their plant hosts. Methods Mol. Biol. 2012, 835, 519–530. [Google Scholar]
- Gu, B.; Kale, S.D.; Wang, Q.H.; Wang, D.H.; Pan, Q.N.; Cao, H.; Meng, Y.L.; Kang, Z.S.; Tyler, B.M.; Shan, W. Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLoS ONE. 2011, 6, e27217. [Google Scholar]
- Saitoh, H.; Fujisawa, S.; Mitsuoka, C.; Ito, A.; Hirabuchi, A.; Ikeda, K.; Irieda, H.; Yoshino, K.; Yoshida, K.; Matsumura, H.; et al. Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathog. 2012, 8, e1002711. [Google Scholar]
- Stergiopoulos, I.; de Wit, P.J. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar]
- Shirasu, K.; Schulze-Lefert, P. Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci. 2003, 8, 252–258. [Google Scholar]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its history and future as a model for plant–pathogen interactions. MPMI. 2008, 21, 1015–1026. [Google Scholar]
- Wang, C.; Zien, C.A.; Afitlhile, M.; Welti, R.; Hildebrand, D.F.; Wang, X. Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in arabidopsis. Plant Cell 2000, 12, 2237–2246. [Google Scholar]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar]
- Lee, S.; Ishiga, Y.; Clermont, K.; Mysore, K.S. Coronatine inhibits stomatal closure and delays hypersensitive response cell death induced by nonhost bacterial pathogens. PeerJ 2013, 1, e34. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar]
- Cantu, D.; Segovia, V.; MacLean, D.; Bayles, R.; Chen, X.; Kamoun, S.; Dubcovsky, J.; Saunders, D.G.; Uauy, C. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genom. 2013, 14, 270. [Google Scholar]
- Hane, J.K.; Anderson, J.P.; Williams, A.H.; Sperschneider, J.; Singh, K.B. Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. 2014, 10, e1004281. [Google Scholar]
- Wei, M.M.; Wang, A.J.; Liu, Y.; Ma, L.; Niu, X.Y.; Zheng, A.P. Identification of the novel effector RsIA_NP8 in Rhizoctonia solani AG1 IA that induces cell death and triggers defense responses in non-host plants. Front. Microbiol. 2020, 11, 1115. [Google Scholar]
- Song, T.Q.; Zhang, Y.; Zhang, Q.; Zhang, X.; Shen, D.Y.; Yu, J.J.; Yu, M.N.; Pan, X.Y.; Cao, H.J.; Yong, M.L.; et al. The N-terminus of an Ustilaginoidea virens Ser-Thr-rich glycosylphosphatidylinositol-anchored protein elicits plant immunity as a MAMP. Nat. Commun. 2021, 12, 2451. [Google Scholar]
- Morgan, W.; Kamoun, S. RXLR effectors of plant pathogenic oomycetes. Curr. Opin. Microbiol. 2007, 10, 332–338. [Google Scholar]
- Nie, J.J.; Yin, Z.Y.; Li, Z.P.; Wu, Y.X.; Huang, L.L. A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 2019, 222, 995–1011. [Google Scholar]
- Xiang, J.; Li, X.J.; Yin, L.; Liu, Y.X.; Zhang, Y.L.; Qu, J.J.; Lu, J. A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana. BMC Plant Biol. 2017, 17, 75. [Google Scholar]
- Dölfors, F.; Holmquist, L.; Dixelius, C.; Tzelepis, G. A LysM effector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol. Genet. Genom. 2019, 294, 1211–1218. [Google Scholar]
- Zhang, Y.; Li, M.Y.; Luo, F. Advances in studies of the type III secretion system in Ralstonia solanacearum—A review. Acta Microbiol. Sin. 2015, 55, 675–682. [Google Scholar]
- Taylor, K.W.; Kim, J.G.; Su, X.B.; Aakre, C.D.; Roden, J.A.; Adams, C.M.; Mudgett, M.B. Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog. 2012, 8, e1002768. [Google Scholar]
- Azevedo, C.; Sadanandom, A.; Kitagawa, K.; Freialdenhoven, A.; Shirasu, K.; Schulze-Lefert, P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 2002, 295, 2073–2076. [Google Scholar]
- Huitema, E.; Vleeshouwers, V.G.; Cakir, C.; Kamoun, S.; Govers, F. Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INN, INF2A, and INF2B of Phytophthora infestans. Mol. Plant Microbe Interact. 2005, 18, 183–193. [Google Scholar]
- Botër, M.; Amigues, B.; Peart, J.; Breuer, C.; Kadota, Y.; Casais, C.; Moore, G.; Kleanthous, C.; Ochsenbein, F.; Shirasu, K. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 2007, 19, 3791–3804. [Google Scholar]
- Mehdy, M.C. Active oxygen species in plant defense against pathogens. Plant Physiol. 1994, 105, 467472. [Google Scholar]
- Wu, G.S.; Shortt, B.J.; Lawrence, E.B.; Leon, J.; Fitzsimmons, K.C.; Levine, E.B.; Raskin, I.; Shah, D.M. Activation of host defense mechanisms by elevated production of H2O2 in transgenic plants. Plant Physiol. 1997, 115, 427–435. [Google Scholar]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar]
- Martin-Hernandez, A.M.; Dufresne, M.; Hugouvieux, V.; Melton, R.; Osbourn, A. Effects of targeted replacement of the tomatinase gene on the interaction of Septoria lycopersici with tomato plants. Mol. Plant Microbe Interact. 2000, 13, 1301–1311. [Google Scholar]
- Hans, T.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—Powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, X.; Xu, D.; Jiang, Y.; Liang, J.; Xiang, T.; Wang, Y.; Zhang, W.; Han, X.; Jiao, C.; Zheng, A.; et al. Functional Analyses of a Small Secreted Cysteine-Rich Protein ThSCSP_14 in Tilletia horrida. Int. J. Mol. Sci. 2022, 23, 15042. https://doi.org/10.3390/ijms232315042
Shu X, Xu D, Jiang Y, Liang J, Xiang T, Wang Y, Zhang W, Han X, Jiao C, Zheng A, et al. Functional Analyses of a Small Secreted Cysteine-Rich Protein ThSCSP_14 in Tilletia horrida. International Journal of Molecular Sciences. 2022; 23(23):15042. https://doi.org/10.3390/ijms232315042
Chicago/Turabian StyleShu, Xinyue, Deze Xu, Yuqi Jiang, Juan Liang, Ting Xiang, Yuxuan Wang, Weike Zhang, Xue Han, Chunhai Jiao, Aiping Zheng, and et al. 2022. "Functional Analyses of a Small Secreted Cysteine-Rich Protein ThSCSP_14 in Tilletia horrida" International Journal of Molecular Sciences 23, no. 23: 15042. https://doi.org/10.3390/ijms232315042
APA StyleShu, X., Xu, D., Jiang, Y., Liang, J., Xiang, T., Wang, Y., Zhang, W., Han, X., Jiao, C., Zheng, A., Li, P., Yin, D., & Wang, A. (2022). Functional Analyses of a Small Secreted Cysteine-Rich Protein ThSCSP_14 in Tilletia horrida. International Journal of Molecular Sciences, 23(23), 15042. https://doi.org/10.3390/ijms232315042






