Estimating Metastatic Risk of Pancreatic Ductal Adenocarcinoma at Single-Cell Resolution
Abstract
1. Introduction
2. Results
2.1. Major Cell Types in PDAC Primary and Metastatic Patients
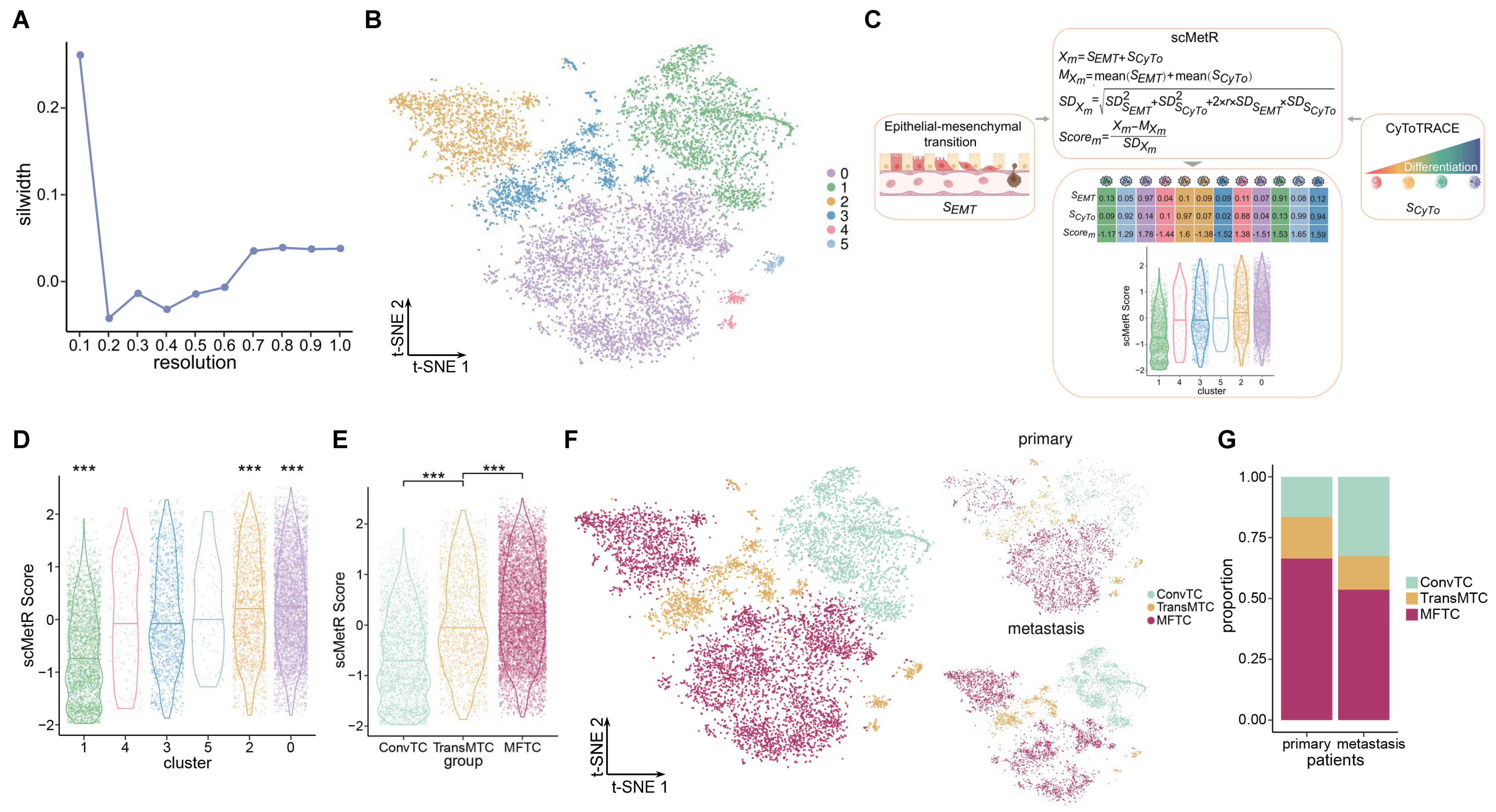
2.2. Heterogeneity in Tumor Cells
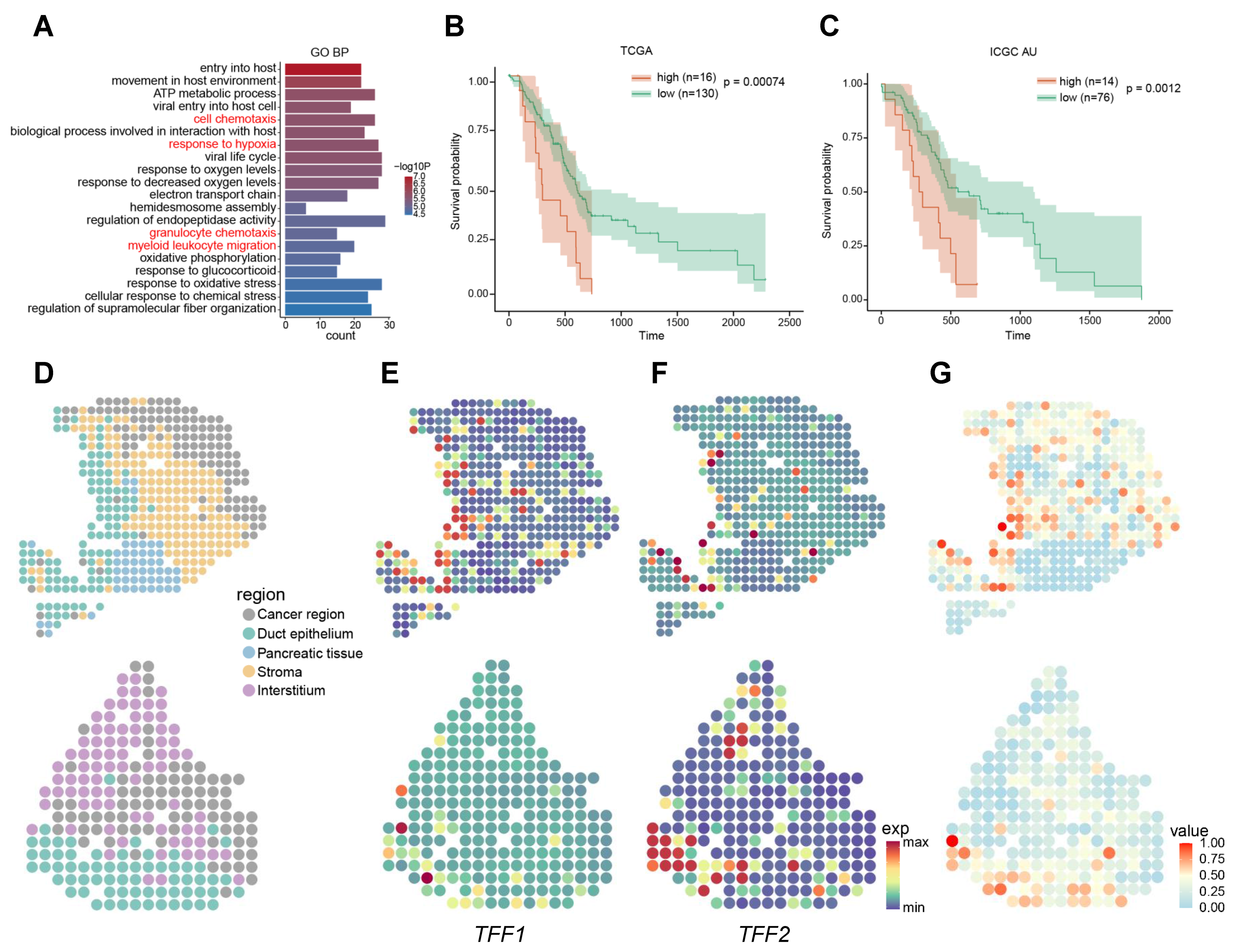
2.3. Signatures Associated with Metastasis
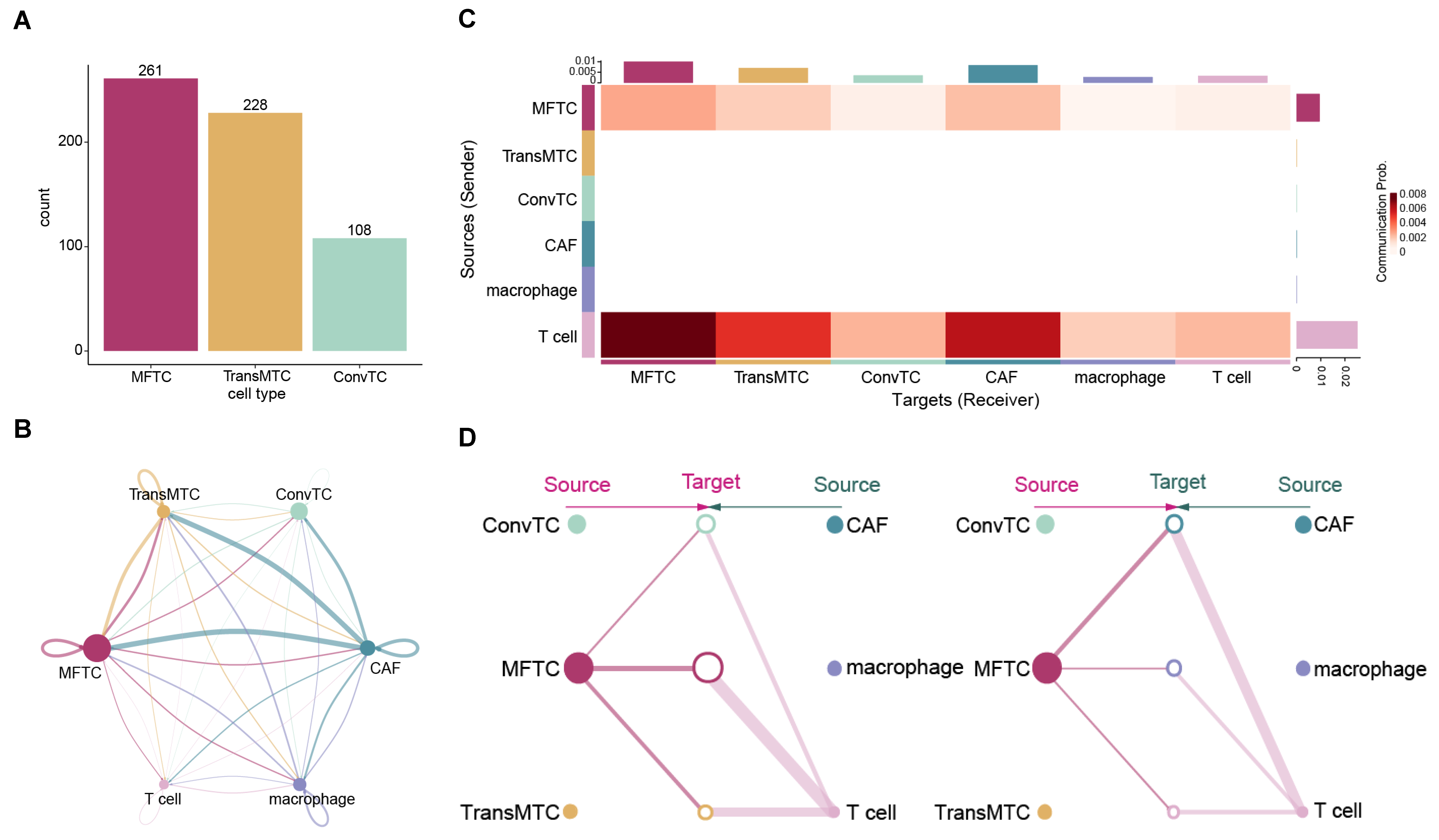
2.4. Cell–Cell Communications in PDAC
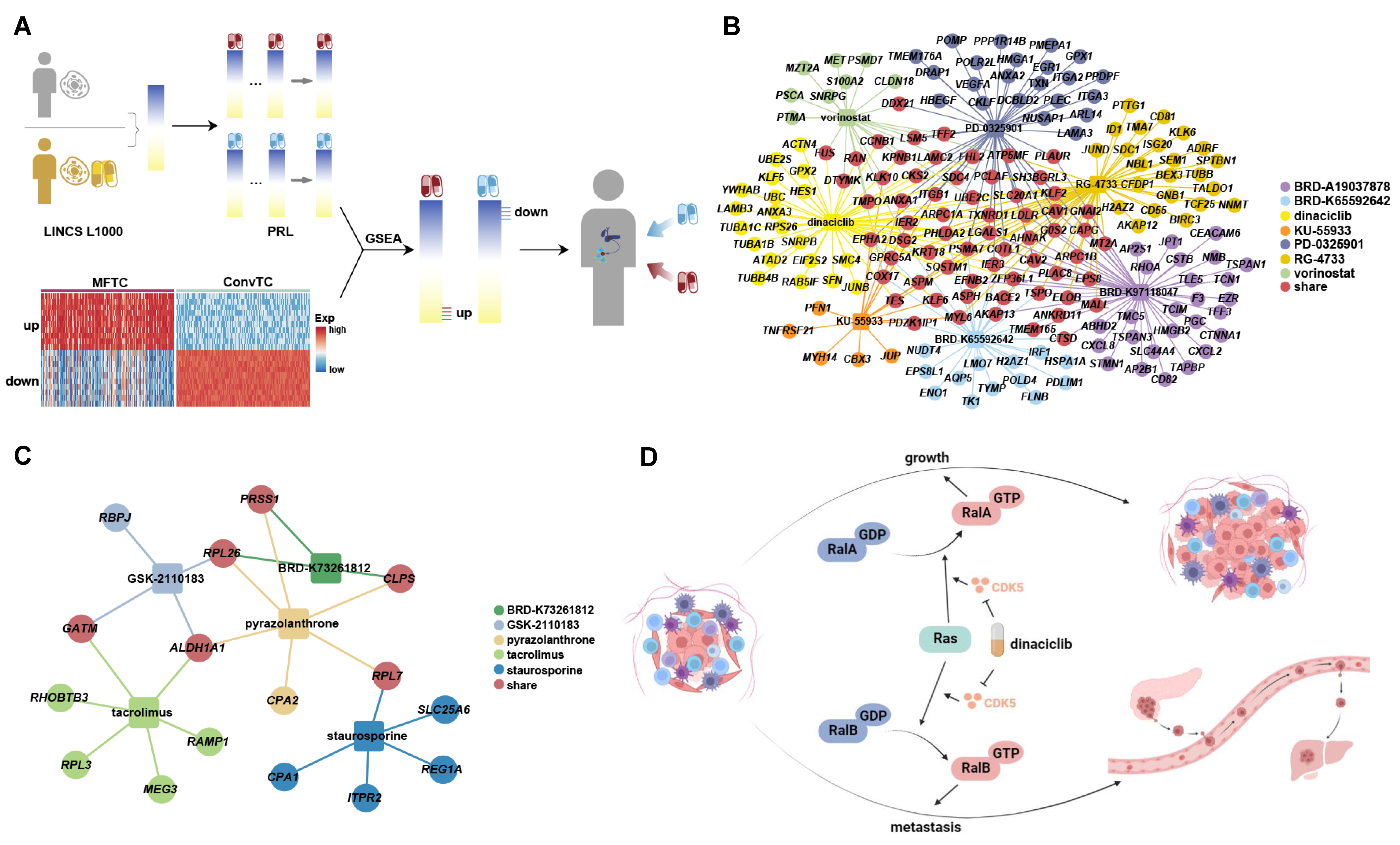
2.5. Potential Inhibitors Targeting the Signatures Associated with Metastasis
3. Discussion
4. Materials and Methods
4.1. Single-Cell RNA Sequencing Data Processing
4.2. Identifying Major Cell Types in PDAC
4.3. Re-Clustering of Tumor Cell to Explore Heterogeneity
4.4. Metastatic Risk Estimation of Tumor Cell Sub-Clusters
4.5. Functional and Survival Analysis Revealing Metastatic Risk of MFTC
4.6. Analysis of Spatial Transcriptomics Data to Discover Spatial Characteristic
4.7. Uncovering Cell–cell Communication Patterns Contributing to Metastasis
4.8. Prediction of Candidate Drugs for Reversing the Expression of MSGs
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, P.; Li, B.; Liu, F.; Zhang, M.; Wang, Q.; Liu, Y.; Yao, Y.; Li, D. The epithelial to mesenchymal transition (EMT) and cancer stem cells: Implication for treatment resistance in pancreatic cancer. Mol. Cancer 2017, 16, 52. [Google Scholar] [CrossRef]
- Ren, X.; Zhou, C.; Lu, Y.; Ma, F.; Fan, Y.; Wang, C. Single-cell RNA-seq reveals invasive trajectory and determines cancer stem cell-related prognostic genes in pancreatic cancer. Bioengineered 2021, 12, 5056–5068. [Google Scholar] [CrossRef]
- Miquel, M.; Zhang, S.; Pilarsky, C. Pre-clinical Models of Metastasis in Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 748631. [Google Scholar] [CrossRef]
- Beuran, M.; Negoi, I.; Paun, S.; Ion, A.D.; Bleotu, C.; Negoi, R.I.; Hostiuc, S. The epithelial to mesenchymal transition in pancreatic cancer: A systematic review. Pancreatology 2015, 15, 217–225. [Google Scholar] [CrossRef]
- Thomas, S.K.; Lee, J.; Beatty, G.L. Paracrine and cell autonomous signalling in pancreatic cancer progression and metastasis. EBioMedicine 2020, 53, 102662. [Google Scholar] [CrossRef]
- Yang, J.; Lin, P.; Yang, M.; Liu, W.; Fu, X.; Liu, D.; Tao, L.; Huo, Y.; Zhang, J.; Hua, R.; et al. Integrated genomic and transcriptomic analysis reveals unique characteristics of hepatic metastases and pro-metastatic role of complement C1q in pancreatic ductal adenocarcinoma. Genome Biol. 2021, 22, 4. [Google Scholar] [CrossRef]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Le Large, T.Y.S.; Bijlsma, M.F.; Kazemier, G.; van Laarhoven, H.W.M.; Giovannetti, E.; Jimenez, C.R. Key biological processes driving metastatic spread of pancreatic cancer as identified by multi-omics studies. Semin. Cancer Biol. 2017, 44, 153–169. [Google Scholar] [CrossRef]
- Jogi, A.; Vaapil, M.; Johansson, M.; Pahlman, S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups. J. Med. Sci. 2012, 117, 217–224. [Google Scholar] [CrossRef]
- Hart, I.R.; Easty, D. Tumor cell progression and differentiation in metastasis. Semin. Cancer Biol. 1991, 2, 87–95. [Google Scholar]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstrahle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 497–514.e22. [Google Scholar] [CrossRef]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Ravirala, D.; Pei, G.; Zhao, Z.; Zhang, X. Comprehensive characterization of tumor immune landscape following oncolytic virotherapy by single-cell RNA sequencing. Cancer Immunol Immunother. 2022, 71, 1479–1495. [Google Scholar] [CrossRef]
- Jiang, B.; Mu, Q.; Qiu, F.; Li, X.; Xu, W.; Yu, J.; Fu, W.; Cao, Y.; Wang, J. Machine learning of genomic features in organotropic metastases stratifies progression risk of primary tumors. Nat. Commun. 2021, 12, 6692. [Google Scholar] [CrossRef]
- Nguyen, B.; Fong, C.; Luthra, A.; Smith, S.A.; DiNatale, R.G.; Nandakumar, S.; Walch, H.; Chatila, W.K.; Madupuri, R.; Kundra, R.; et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 2022, 185, 563–575.e511. [Google Scholar] [CrossRef]
- Lin, W.; Noel, P.; Borazanci, E.H.; Lee, J.; Amini, A.; Han, I.W.; Heo, J.S.; Jameson, G.S.; Fraser, C.; Steinbach, M.; et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020, 12, 80. [Google Scholar] [CrossRef]
- Pan, H.; Diao, H.; Zhong, W.; Wang, T.; Wen, P.; Wu, C. A Cancer Cell Cluster Marked by LincRNA MEG3 Leads Pancreatic Ductal Adenocarcinoma Metastasis. Front. Oncol. 2021, 11, 656564. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Lee, J.J.; Bernard, V.; Semaan, A.; Monberg, M.E.; Huang, J.; Stephens, B.M.; Lin, D.; Rajapakshe, K.I.; Weston, B.R.; Bhutani, M.S.; et al. Elucidation of Tumor-Stromal Heterogeneity and the Ligand-Receptor Interactome by Single-Cell Transcriptomics in Real-world Pancreatic Cancer Biopsies. Clin. Cancer Res. 2021, 27, 5912–5921. [Google Scholar] [CrossRef]
- Aissa, A.F.; Islam, A.; Ariss, M.M.; Go, C.C.; Rader, A.E.; Conrardy, R.D.; Gajda, A.M.; Rubio-Perez, C.; Valyi-Nagy, K.; Pasquinelli, M.; et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 2021, 12, 1628. [Google Scholar] [CrossRef]
- Yi, M.; Nissley, D.V.; McCormick, F.; Stephens, R.M. ssGSEA score-based Ras dependency indexes derived from gene expression data reveal potential Ras addiction mechanisms with possible clinical implications. Sci. Rep. 2020, 10, 10258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, Y.; Zheng, C.; Qu, H. dbEMT 2.0: An updated database for epithelial-mesenchymal transition genes with experimentally verified information and precalculated regulation information for cancer metastasis. J. Genet. Genom. 2019, 46, 595–597. [Google Scholar] [CrossRef] [PubMed]
- Gulati, G.S.; Sikandar, S.S.; Wesche, D.J.; Manjunath, A.; Bharadwaj, A.; Berger, M.J.; Ilagan, F.; Kuo, A.H.; Hsieh, R.W.; Cai, S.; et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020, 367, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Frede, J.; Anand, P.; Sotudeh, N.; Pinto, R.A.; Nair, M.S.; Stuart, H.; Yee, A.J.; Vijaykumar, T.; Waldschmidt, J.M.; Potdar, S.; et al. Dynamic transcriptional reprogramming leads to immunotherapeutic vulnerabilities in myeloma. Nat. Cell Biol. 2021, 23, 1199–1211. [Google Scholar] [CrossRef]
- Xie, Z.B.; Yao, L.; Jin, C.; Zhang, Y.F.; Fu, D.L. High cytoplasm HABP1 expression as a predictor of poor survival and late tumor stage in pancreatic ductal adenocarcinoma patients. Eur. J. Surg. Oncol. 2019, 45, 207–212. [Google Scholar] [CrossRef]
- Cao, L.; Huang, C.; Cui Zhou, D.; Hu, Y.; Lih, T.M.; Savage, S.R.; Krug, K.; Clark, D.J.; Schnaubelt, M.; Chen, L.; et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 2021, 184, 5031–5052.e26. [Google Scholar] [CrossRef]
- Yuen, A.; Diaz, B. The impact of hypoxia in pancreatic cancer invasion and metastasis. Hypoxia 2014, 2, 91–106. [Google Scholar] [CrossRef]
- Bausch, D.; Pausch, T.; Krauss, T.; Hopt, U.T.; Fernandez-del-Castillo, C.; Warshaw, A.L.; Thayer, S.P.; Keck, T. Neutrophil granulocyte derived MMP-9 is a VEGF independent functional component of the angiogenic switch in pancreatic ductal adenocarcinoma. Angiogenesis 2011, 14, 235–243. [Google Scholar] [CrossRef]
- Kim, J.; Bae, J.S. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediat. Inflamm. 2016, 2016, 6058147. [Google Scholar] [CrossRef]
- Prinz, M.; Erny, D.; Hagemeyer, N. Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol. 2017, 18, 385–392. [Google Scholar] [CrossRef]
- Moncada, R.; Barkley, D.; Wagner, F.; Chiodin, M.; Devlin, J.C.; Baron, M.; Hajdu, C.H.; Simeone, D.M.; Yanai, I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020, 38, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Brandt, W.; Ramachandran, V.; Moore, T.T.; Wang, H.; May, F.E.; Westley, B.R.; Hwang, R.F.; Logsdon, C.D. Trefoil factor 1 stimulates both pancreatic cancer and stellate cells and increases metastasis. Pancreas 2011, 40, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Guppy, N.J.; El-Bahrawy, M.E.; Kocher, H.M.; Fritsch, K.; Qureshi, Y.A.; Poulsom, R.; Jeffery, R.E.; Wright, N.A.; Otto, W.R.; Alison, M.R. Trefoil factor family peptides in normal and diseased human pancreas. Pancreas 2012, 41, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Kajioka, H.; Kagawa, S.; Ito, A.; Yoshimoto, M.; Sakamoto, S.; Kikuchi, S.; Kuroda, S.; Yoshida, R.; Umeda, Y.; Noma, K.; et al. Targeting neutrophil extracellular traps with thrombomodulin prevents pancreatic cancer metastasis. Cancer Lett. 2021, 497, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Khalafalla, F.G.; Khan, M.W. Inflammation and Epithelial-Mesenchymal Transition in Pancreatic Ductal Adenocarcinoma: Fighting Against Multiple Opponents. Cancer Growth Metastasis 2017, 10, 1179064417709287. [Google Scholar] [CrossRef]
- Steele, C.W.; Jamieson, N.B.; Evans, T.R.; McKay, C.J.; Sansom, O.J.; Morton, J.P.; Carter, C.R. Exploiting inflammation for therapeutic gain in pancreatic cancer. Br. J. Cancer 2013, 108, 997–1003. [Google Scholar] [CrossRef]
- Padoan, A.; Plebani, M.; Basso, D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int. J. Mol. Sci. 2019, 20, 676. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- He, Z.; Wu, H.; Jiao, Y.; Zheng, J. Expression and prognostic value of CD97 and its ligand CD55 in pancreatic cancer. Oncol. Lett. 2015, 9, 793–797. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Lim, K.H.; Baines, A.T.; Fiordalisi, J.J.; Shipitsin, M.; Feig, L.A.; Cox, A.D.; Der, C.J.; Counter, C.M. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 2005, 7, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; O’Hayer, K.; Adam, S.J.; Kendall, S.D.; Campbell, P.M.; Der, C.J.; Counter, C.M. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 2006, 16, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, G.; Mishra, A.; Hong, S.M.; Bisht, S.; Strock, C.J.; Ball, D.W.; Goggins, M.; Maitra, A.; Nelkin, B.D. Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras-Ral signaling. Cancer Res. 2010, 70, 4460–4469. [Google Scholar] [CrossRef] [PubMed]
- Riess, C.; Irmscher, N.; Salewski, I.; Struder, D.; Classen, C.F.; Grosse-Thie, C.; Junghanss, C.; Maletzki, C. Cyclin-dependent kinase inhibitors in head and neck cancer and glioblastoma-backbone or add-on in immune-oncology? Cancer Metastasis Rev. 2021, 40, 153–171. [Google Scholar] [CrossRef]
- Hu, C.; Dadon, T.; Chenna, V.; Yabuuchi, S.; Bannerji, R.; Booher, R.; Strack, P.; Azad, N.; Nelkin, B.D.; Maitra, A. Combined Inhibition of Cyclin-Dependent Kinases (Dinaciclib) and AKT (MK-2206) Blocks Pancreatic Tumor Growth and Metastases in Patient-Derived Xenograft Models. Mol. Cancer 2015, 14, 1532–1539. [Google Scholar] [CrossRef]
- Charles Jacob, H.K.; Charles Richard, J.L.; Signorelli, R.; Kashuv, T.; Lavania, S.; Vaish, U.; Boopathy, R.; Middleton, A.; Boone, M.M.; Sundaram, R.; et al. Modulation of Early Neutrophil Granulation: The Circulating Tumor Cell-Extravesicular Connection in Pancreatic Ductal Adenocarcinoma. Cancers 2021, 13, 2727. [Google Scholar] [CrossRef]
- Wang, X.; Hu, L.P.; Qin, W.T.; Yang, Q.; Chen, D.Y.; Li, Q.; Zhou, K.X.; Huang, P.Q.; Xu, C.J.; Li, J.; et al. Identification of a subset of immunosuppressive P2RX1-negative neutrophils in pancreatic cancer liver metastasis. Nat. Commun. 2021, 12, 174. [Google Scholar] [CrossRef]
- Andrews, T.S.; Hemberg, M. False signals induced by single-cell imputation. F1000Research 2018, 7, 1740. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Wang, S.; Zeng, X.; Zhang, W. Are dropout imputation methods for scRNA-seq effective for scATAC-seq data? Brief. Bioinform. 2022, 23, bbab442. [Google Scholar] [CrossRef]
- van Dijk, D.; Sharma, R.; Nainys, J.; Yim, K.; Kathail, P.; Carr, A.J.; Burdziak, C.; Moon, K.R.; Chaffer, C.L.; Pattabiraman, D.; et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 2018, 174, 716–729. [Google Scholar] [CrossRef]
- Hou, W.; Ji, Z.; Ji, H.; Hicks, S.C. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol. 2020, 21, 218. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Sun, T.; Song, D.; Li, J.J. Statistics or biology: The zero-inflation controversy about scRNA-seq data. Genome Biol. 2022, 23, 31. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef] [PubMed]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Evans, L.D. A two-score composite program for combining standard scores. Behav. Res. Methods Instrum. Comput. 1996, 28, 209–213. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol. 2022, 40, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Bosotti, R.; Scacheri, E.; Belcastro, V.; Mithbaokar, P.; Ferriero, R.; Murino, L.; Tagliaferri, R.; Brunetti-Pierri, N.; Isacchi, A.; et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. USA 2010, 107, 14621–14626. [Google Scholar] [CrossRef]
- Li, F.; Cao, Y.; Han, L.; Cui, X.; Xie, D.; Wang, S.; Bo, X. GeneExpressionSignature: An R package for discovering functional connections using gene expression signatures. OMICS 2013, 17, 116–118. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Zhou, S.; Huang, Y.-e.; Yuan, M.; Lei, W.; Chen, J.; Lin, K.; Jiang, W. Estimating Metastatic Risk of Pancreatic Ductal Adenocarcinoma at Single-Cell Resolution. Int. J. Mol. Sci. 2022, 23, 15020. https://doi.org/10.3390/ijms232315020
Chen S, Zhou S, Huang Y-e, Yuan M, Lei W, Chen J, Lin K, Jiang W. Estimating Metastatic Risk of Pancreatic Ductal Adenocarcinoma at Single-Cell Resolution. International Journal of Molecular Sciences. 2022; 23(23):15020. https://doi.org/10.3390/ijms232315020
Chicago/Turabian StyleChen, Sina, Shunheng Zhou, Yu-e Huang, Mengqin Yuan, Wanyue Lei, Jiahao Chen, Kongxuan Lin, and Wei Jiang. 2022. "Estimating Metastatic Risk of Pancreatic Ductal Adenocarcinoma at Single-Cell Resolution" International Journal of Molecular Sciences 23, no. 23: 15020. https://doi.org/10.3390/ijms232315020
APA StyleChen, S., Zhou, S., Huang, Y.-e., Yuan, M., Lei, W., Chen, J., Lin, K., & Jiang, W. (2022). Estimating Metastatic Risk of Pancreatic Ductal Adenocarcinoma at Single-Cell Resolution. International Journal of Molecular Sciences, 23(23), 15020. https://doi.org/10.3390/ijms232315020




