Evidence for Multilevel Chemopreventive Activities of Natural Phenols from Functional Genomic Studies of Curcumin, Resveratrol, Genistein, Quercetin, and Luteolin
Abstract
:1. Introduction
1.1. Curcumin, Resveratrol, Genistein, Quercetin, and Luteolin Are Small Natural Phenols That Are Important for Chemoprevention
1.2. Chemical and Biochemical Activities of the Natural Phenols
1.3. Carcinogenesis
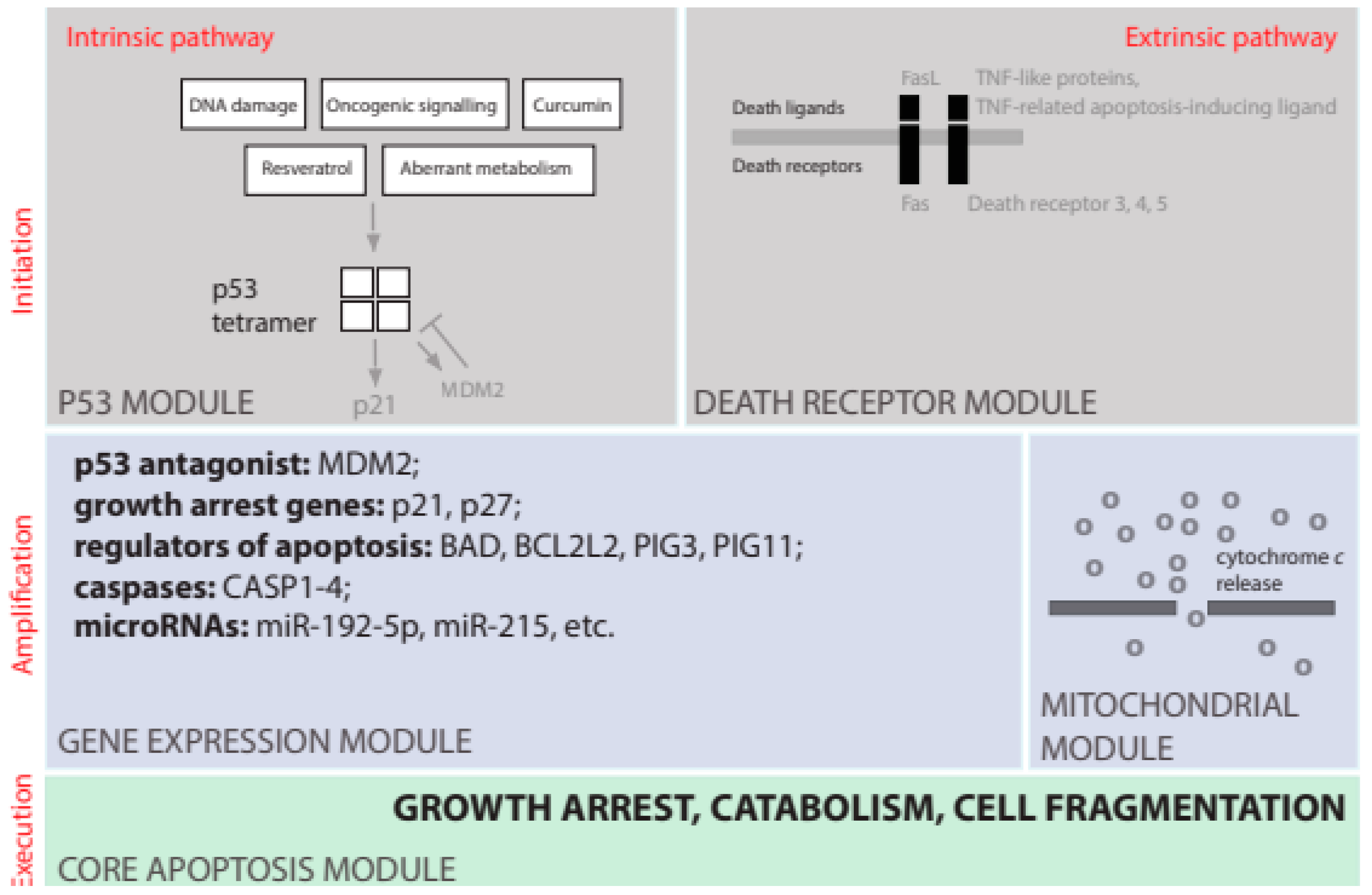
1.4. Apoptosis
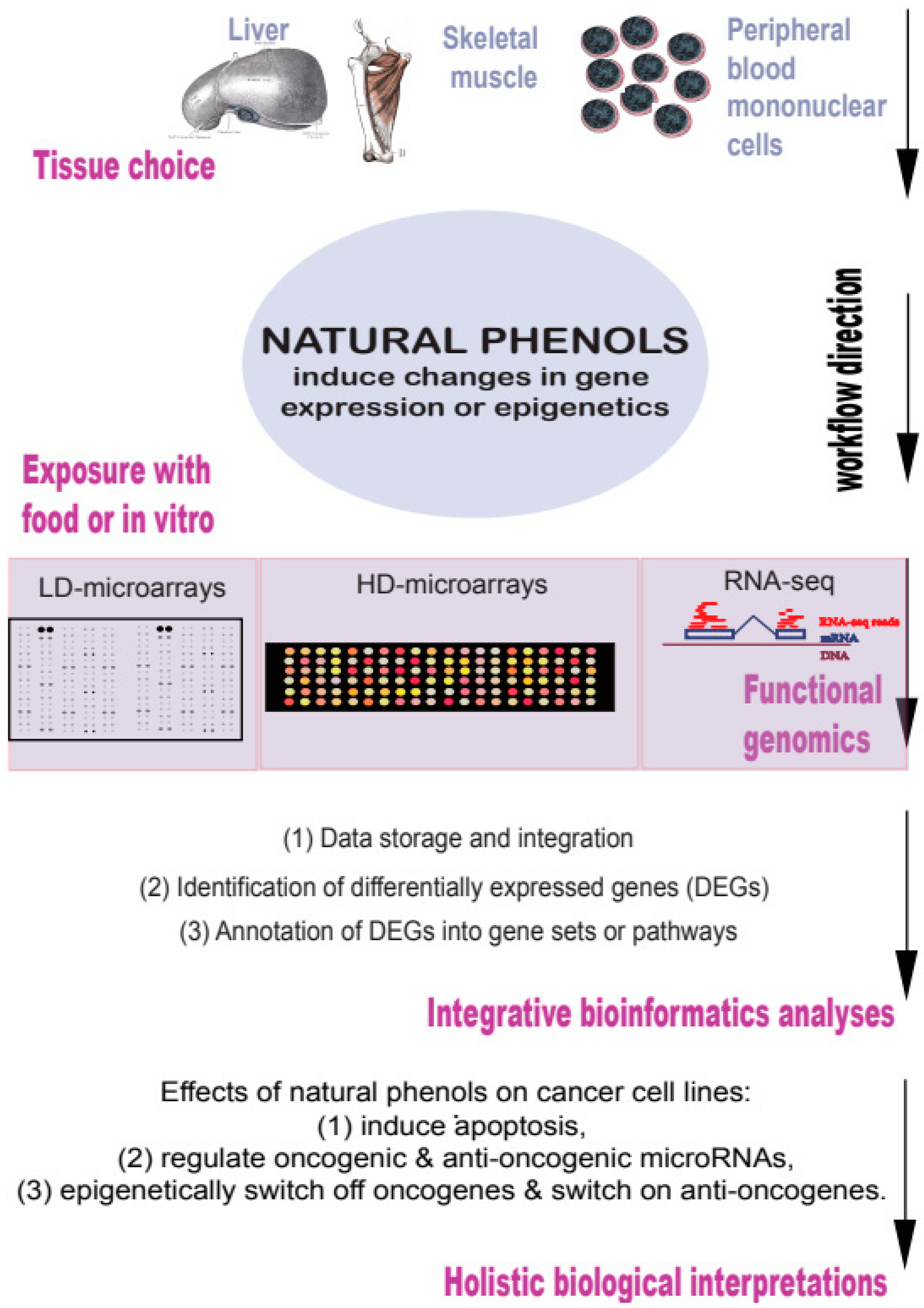
2. Results of Functional Genomic Studies of Natural Phenols
2.1. Natural Phenols Induce Apoptosis in Cancer Cell Lines in a P53-Dependent Manner
2.2. Natural Phenols as Potent Regulators of Expression of Oncogenic and Anti-Oncogenic microRNAs
2.3. Natural Phenols as Dietary Epidrugs
2.4. Epigenetic Changes Induced by Natural Phenols in Cancer Cells Switch off Oncogenes and Switch on Anti-Oncogenes
2.5. Similar Results Can Be Obtained for Other Natural Phenols
2.5.1. Genistein
2.5.2. Quercetin
2.5.3. Luteolin
3. Conclusions
Funding
Conflicts of Interest
References
- Cassidy, J.; Bissett, D.; Spence, R.; Payne, M. Oxford Handbook of Oncology; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Langner, E.; Rzeski, W. Dietary derived compounds in cancer chemoprevention. Contemp. Oncol. 2012, 16, 394–400. [Google Scholar] [CrossRef]
- Key, T.J. Fruit and vegetables and cancer risk. Br. J. Cancer 2011, 104, 6–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Diet, Nutrition and the Prevention of Chronic Diseases: Report of a Joint WHO/FAO expert Consultation, Geneva, 28 January–1 February 2002; World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Halliwell, B. Antioxidants in Human Health and Disease. Annu. Rev. Nutr. 1996, 16, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic diet: Impact on the epigenome and cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.A.; Gescher, A.J.; Steward, W.P. Curcumin: The story so far. Eur. J. Cancer 2005, 41, 1955–1968. [Google Scholar] [CrossRef]
- Priyadarsini, K.I. The Chemistry of Curcumin: From Extraction to Therapeutic Agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef] [Green Version]
- CID969516. PubChem Compound Summary for CID 969516, Curcumin. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/969516 (accessed on 28 June 2022).
- CID445154. PubChem Compound Summary for CID 445154, Resveratrol. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/445154 (accessed on 28 June 2022).
- CID5280961. PubChem Compound Summary for CID 5280961, Genistein. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5280961 (accessed on 28 June 2022).
- CID5280343. PubChem Compound Summary for CID 5280343, Quercetin. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5280343 (accessed on 28 June 2022).
- CID5280445. PubChem Compound Summary for CID 5280445, Luteolin. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5280445 (accessed on 28 June 2022).
- Iuga, C.R.; Alvarez-Idaboy, J.R.; Russo, N. Antioxidant Activity of trans-Resveratrol toward Hydroxyl and Hydroperoxyl Radicals: A Quantum Chemical and Computational Kinetics Study. J. Org. Chem. 2012, 77, 3868–3877. [Google Scholar] [CrossRef]
- Gülçin, I. Antioxidant properties of resveratrol: A structure–activity insight. Innov. Food Sci. Emerg. Technol. 2010, 11, 210–218. [Google Scholar] [CrossRef]
- Stojanović, S.; Sprinz, H.; Brede, O. Efficiency and Mechanism of the Antioxidant Action of trans-Resveratrol and Its Analogues in the Radical Liposome Oxidation. Arch. Biochem. Biophys. 2001, 391, 79–89. [Google Scholar] [CrossRef]
- Awasthi, S.; Pandya, U.; Singhal, S.S.; Lin, J.T.; Thiviyanathan, V.; Seifert, W.E.; Awasthi, Y.C.; Ansari, G. Curcumin‚ glutathione interactions and the role of human glutathione S-transferase P1-1. Chem.-Biol. Interact. 2000, 128, 19–38. [Google Scholar] [CrossRef]
- Magesh, S.; Chen, Y.; Hu, L. Small Molecule Modulators of Keap1-Nrf2-ARE Pathway as Potential Preventive and Therapeutic Agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef] [Green Version]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [Green Version]
- Athar, M.; Back, J.H.; Kopelovich, L.; Bickers, D.R.; Kim, A.L. Multiple molecular targets of resveratrol: Anti-carcinogenic mechanisms. Arch. Biochem. Biophys. 2009, 486, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Kasi, P.D.; Tamilselvam, R.; Skalicka-Woźniak, K.; Nabavi, S.F.; Daglia, M.; Bishayee, A.; Pazoki-Toroudi, H.; Nabavi, S.M. Molecular targets of curcumin for cancer therapy: An updated review. Tumour Biol. 2016, 37, 13017–13028. [Google Scholar] [CrossRef]
- Forman, H.J.; Davies, K.J.A.; Ursini, F. How Do Nutritional Antioxidants Really Work: Nucleophilic Tone and Para-Hormesis Versus Free Radical Scavenging in vivo. Free Radic. Biol. Med. 2014, 66, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Sirerol, J.A.; Rodríguez, M.L.; Mena, S.; Asensi, M.A.; Estrela, J.M.; Ortega, A.L. Role of Natural Stilbenes in the Prevention of Cancer. Oxidative Med. Cell. Longev. 2016, 2016, 3128951. [Google Scholar] [CrossRef] [Green Version]
- Shishodia, S. Molecular mechanisms of curcumin action: Gene expression. BioFactors 2013, 39, 37–55. [Google Scholar] [CrossRef]
- Huminiecki, L.; Horbanczuk, J. The functional genomic studies of resveratrol in respect to its anti-cancer effects. Biotechnol. Adv. 2018, 36, 1699–1708. [Google Scholar] [CrossRef]
- Huminiecki, L.; Atanasov, A.G.; Horbańczuk, J. The Functional Genomic Studies of Resveratrol in Animal Models in the Context of Atherosclerosis; Jastrzębiec, IGHZ-PAS: Bychawa, Poland, 2018. [Google Scholar]
- Huminiecki, L.; Horbańczuk, J.; Atanasov, A.G. The functional genomic studies of curcumin. Semin. Cancer Biol. 2017, 46, 107–118. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Biology of Cancer; Garland Science: New York, NY, USA, 2007. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Huminiecki, L. Telomerase as a therapeutic target. Acta Biochim. Pol. 1996, 43, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Plati, J.; Bucur, O.; Khosravi-Far, R. Dysregulation of Apoptotic Signaling in Cancer: Molecular Mechanisms and Therapeutic Opportunities. J. Cell. Biochem. 2008, 104, 1124–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danial, N.N.; Korsmeyer, S.J. Cell Death: Critical Control Points. Cell 2004, 116, 205–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, S.W.; Cepero, E.; Evan, G. Intrinsic tumour suppression. Nature 2004, 432, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. p53, the Cellular Gatekeeper for Growth and Division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Abbastabar, M.; Kheyrollah, M.; Azizian, K.; Bagherlou, N.; Tehrani, S.S.; Maniati, M.; Karimian, A. Multiple functions of p27 in cell cycle, apoptosis, epigenetic modification and transcriptional regulation for the control of cell growth: A double-edged sword protein. DNA Repair. 2018, 69, 63–72. [Google Scholar] [CrossRef]
- Wang, X.; Gorospe, M.; Huang, Y.; Holbrook, N.J. p27Kip1 overexpression causes apoptotic death of mammalian cells. Oncogene 1997, 15, 2991–2997. [Google Scholar] [CrossRef]
- Hui, L.; Zheng, Y.; Yan, Y.; Bargonetti, J.; A Foster, D. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene 2006, 25, 7305–7310. [Google Scholar] [CrossRef] [Green Version]
- Gartel, A.L.; Radhakrishnan, S.K. Lost in Transcription: p21 Repression, Mechanisms, and Consequences. Cancer Res. 2005, 65, 3980–3985. [Google Scholar] [CrossRef] [Green Version]
- Baehrecke, E.H. How death shapes life during development. Nat. Rev. Mol. Cell Biol. 2002, 3, 779–787. [Google Scholar] [CrossRef]
- Aravind, L.; Dixit, V.M.; Koonin, E.V. Apoptotic molecular machinery: Vastly increased complexity in vertebrates revealed by genome comparisons. Science 2001, 291, 1279–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huminiecki, L.; Heldin, C.H. 2R and remodeling of vertebrate signal transduction engine. BMC Biol. 2010, 8, 146. [Google Scholar] [CrossRef] [Green Version]
- Opferman, J.T.; Korsmeyer, S.J. Apoptosis in the development and maintenance of the immune system. Nat. Immunol. 2003, 4, 410–415. [Google Scholar] [CrossRef]
- Mortezaee, K.; Salehi, E.; Mirtavoos-Mahyari, H.; Motevaseli, E.; Najafi, M.; Farhood, B.; Rosengren, R.J.; Sahebkar, A. Mechanisms of apoptosis modulation by curcumin: Implications for cancer therapy. J. Cell. Physiol. 2019, 234, 12537–12550. [Google Scholar] [CrossRef]
- Karunagaran, D.; Rashmi, R.; Kumar, T.R.S. Induction of Apoptosis by Curcumin and Its Implications for Cancer Therapy. Curr. Cancer Drug Targets 2005, 5, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Taeb, S.; Haghi-Aminjan, H.; Afrashi, S.; Moloudi, K.; Musa, A.E.; Najafi, M.; Farhood, B. Resveratrol as an Enhancer of Apoptosis in Cancer: A Mechanistic Review. Anti-Cancer Agents Med. Chem. 2021, 21, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Dong, H.; Jia, S.; Yang, Z. Resveratrol as a modulatory of apoptosis and autophagy in cancer therapy. Clin. Transl. Oncol. 2022, 24, 1219–1230. [Google Scholar] [CrossRef]
- Jones, S.B.; DePrimo, S.E.; Whitfield, M.L.; Brooks, J.D. Resveratrol-induced gene expression profiles in human prostate cancer cells. Cancer Epidemiol. Biomark. Prev. 2005, 14, 596–604. [Google Scholar] [CrossRef] [Green Version]
- Whyte, L.; Huang, Y.-Y.; Torres, K.; Mehta, R.G. Molecular mechanisms of resveratrol action in lung cancer cells using dual protein and microarray analyses. Cancer Res. 2007, 67, 12007–12017. [Google Scholar] [CrossRef] [Green Version]
- Harati, K.; Slodnik, P.; Chromik, A.M.; Goertz, O.; Hirsch, T.; Kapalschinski, N.; Klein-Hitpass, L.; Kolbenschlag, J.; Uhl, W.; Lehnhardt, M.; et al. Resveratrol induces apoptosis and alters gene expression in human fibrosarcoma cells. Anticancer Res. 2015, 35, 767–774. [Google Scholar]
- Chin, Y.-T.; Hsieh, M.-T.; Yang, S.-H.; Tsai, P.-W.; Wang, S.-H.; Wang, C.-C.; Lee, Y.-S.; Cheng, G.-Y.; HuangFu, W.-C.; London, D.; et al. Anti-proliferative and gene expression actions of resveratrol in breast cancer cells in vitro. Oncotarget 2014, 5, 12891–12907. [Google Scholar] [CrossRef]
- Shi, T.; Liou, L.S.; Sadhukhan, P.; Duan, Z.-H.; Novick, A.C.; Hissong, J.G.; Almasan, A.; DiDonato, J.A. Effects of resveratrol on gene expression in renal cell carcinoma. Cancer Biol. Ther. 2004, 3, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, C.; Rodriguez, S.; Ramachandran, R.; Nair, P.K.R.; Fonseca, H.; Khatib, Z.; Escalon, E.; Melnick, S.J. Expression profiles of apoptotic genes induced by curcumin in human breast cancer and mammary epithelial cell lines. Anticancer Res. 2005, 25, 3293–3302. [Google Scholar]
- Marquardt, J.U.; Gomez-Quiroz, L.; Arreguin Camacho, L.O.; Pinna, F.; Lee, Y.H.; Kitade, M.; Domínguez, M.P.; Castven, D.; Breuhahn, K.; Conner, E.A.; et al. Curcumin effectively inhibits oncogenic NF-kappaB signaling and restrains stemness features in liver cancer. J. Hepatol. 2015, 63, 661–669. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.; Zhang, J.; Miao, Q.; Yao, L. Curcumin promotes apoptosis by activating the p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer. Cancer Lett. 2015, 357, 196–205. [Google Scholar] [CrossRef]
- Lin, H.Y.; Shih, A.I.; Davis, F.B.; Tang, H.Y.; Martino, L.J.; Bennett, J.A.; Davis, P.J. Resveratrol induced serine phosphorylation of p53 causes apoptosis in a mutant p53 prostate cancer cell line. J. Urol. 2002, 168, 748–755. [Google Scholar] [CrossRef]
- Nigro, J.M.; Baker, S.J.; Preisinger, A.C.; Jessup, J.M.; Hosteller, R.; Cleary, K.; Signer, S.H.; Davidson, N.; Baylin, S.; Devilee, P.; et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989, 342, 705–708. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Caldas, C.; Brenton, J.D. Sizing up miRNAs as cancer genes. Nat. Med. 2005, 11, 712–714. [Google Scholar] [CrossRef]
- Hermeking, H. p53 Enters the MicroRNA World. Cancer Cell 2007, 12, 414–418. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 2022, 38, 379–394. [Google Scholar] [CrossRef]
- Kang, H. MicroRNA-Mediated Health-Promoting Effects of Phytochemicals. Int. J. Mol. Sci. 2019, 20, 2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devi, K.P.; Rajavel, T.; Daglia, M.; Nabavi, M.S.; Bishayee, A.; Nabavi, S.M. Targeting miRNAs by polyphenols: Novel therapeutic strategy for cancer. Semin. Cancer Biol. 2017, 46, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Masoudifar, A.; Sahebkar, A.; Zare, N.; Nahand, J.S.; Rashidi, B.; Mehrabian, E.; Mohammadi, M.; Mirzaei, H.R.; Jaafari, M.R. MicroRNA: A novel target of curcumin in cancer therapy. J. Cell. Physiol. 2018, 233, 3004–3015. [Google Scholar] [CrossRef]
- Li, X.; Xie, W.; Xie, C.; Huang, C.; Zhu, J.; Liang, Z.; Deng, F.; Zhu, M.; Zhu, W.; Wu, R.; et al. Curcumin modulates miR-19/PTEN/AKT/p53 axis to suppress bisphenol A-induced MCF-7 breast cancer cell proliferation. Phytother. Res. 2014, 28, 1553–1560. [Google Scholar] [CrossRef]
- Gandhy, S.U.; Kim, K.; Larsen, L.; Rosengren, R.J.; Safe, S. Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (Sp) transcription factors by targeting microRNAs. BMC Cancer 2012, 12, 564. [Google Scholar] [CrossRef] [Green Version]
- Noratto, G.D.; Jutooru, I.; Safe, S.; Angel-Morales, G.; Mertens-Talcott, S.U. The drug resistance suppression induced by curcuminoids in colon cancer SW-480 cells is mediated by reactive oxygen species-induced disruption of the microRNA-27a-ZBTB10-Sp axis. Mol. Nutr. Food Res. 2013, 57, 1638–1648. [Google Scholar] [CrossRef]
- Akbari, A.; Sedaghat, M.; Heshmati, J.; Tabaeian, S.P.; Dehghani, S.; Pizarro, A.B.; Rostami, Z.; Agah, S. Molecular mechanisms underlying curcumin-mediated microRNA regulation in carcinogenesis; Focused on gastrointestinal cancers. Biomed. Pharmacother. 2021, 141, 111849. [Google Scholar] [CrossRef]
- Sun, M.; Estrov, Z.; Ji, Y.; Coombes, K.R.; Harris, D.H.; Kurzrock, R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol. Cancer Ther. 2008, 7, 464–473. [Google Scholar] [CrossRef] [Green Version]
- Deveraux, Q.L.; Reed, J.C. IAP family proteins—Suppressors of apoptosis. Genes Dev. 1999, 13, 239–252. [Google Scholar] [CrossRef]
- Bae, S.; Lee, E.-M.; Cha, H.J.; Kim, K.; Yoon, Y.; Lee, H.; Kim, J.; Kim, Y.-J.; Lee, H.G.; Jeung, H.-K.; et al. Resveratrol alters microRNA expression profiles in A549 human non-small cell lung cancer cells. Mol. Cells 2011, 32, 243–249. [Google Scholar] [CrossRef] [Green Version]
- Sheth, S.; Jajoo, S.; Kaur, T.; Mukherjea, D.; Sheehan, K.; Rybak, L.P.; Ramkumar, V. Resveratrol reduces prostate cancer growth and metastasis by inhibiting the Akt/MicroRNA-21 pathway. PLoS ONE 2012, 7, e51655. [Google Scholar] [CrossRef] [Green Version]
- Wach, S.; Nolte, E.; Szczyrba, J.; Stöhr, R.; Hartmann, A.; Ørntoft, T.; Dyrskjøt, L.; Eltze, E.; Wieland, W.; Keck, B.; et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int. J. Cancer 2012, 130, 611–621. [Google Scholar] [CrossRef]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef] [Green Version]
- Sayed, D.; He, M.; Hong, C.; Gao, S.; Rane, S.; Yang, Z.; Abdellatif, M. MicroRNA-21 Is a Downstream Effector of AKT That Mediates Its Antiapoptotic Effects via Suppression of Fas Ligand. J. Biol. Chem. 2010, 285, 20281–20290. [Google Scholar] [CrossRef] [Green Version]
- Link, A.B.F.; Goel, A. Cancer chemoprevention by dietary polyphenols: Promising role for epigenetics. Biochem. Pharmacol. 2010, 80, 1771–1792. [Google Scholar] [CrossRef] [Green Version]
- Link, A.; Balaguer, F.; Shen, Y.; Lozano, J.J.; Leung, H.-C.E.; Boland, C.R.; Goel, A. Curcumin modulates DNA methylation in colorectal cancer cells. PLoS ONE 2013, 8, e57709. [Google Scholar] [CrossRef] [Green Version]
- Gunderson, K.L. Whole-genome genotyping on bead arrays. Methods Mol. Biol. 2009, 529, 197–213. [Google Scholar]
- Lubecka, K.; Kurzava, L.; Flower, K.; Buvala, H.; Zhang, H.; Teegarden, D.; Camarillo, I.; Suderman, M.; Kuang, S.; Andrisani, O.; et al. Stilbenoids remodel the DNA methylation patterns in breast cancer cells and inhibit oncogenic NOTCH signaling through epigenetic regulation of MAML2 transcriptional activity. Carcinogenesis 2016, 37, 656–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina-Aguilar, R.; Pérez-Plasencia, C.; Marchat, L.A.; Gariglio, P.; García Mena, J.; Rodríguez Cuevas, S.; Ruíz-García, E.; Astudillo-de la Vega, H.; Hernández Juárez, J.; Flores-Pérez, A.; et al. Methylation Landscape of Human Breast Cancer Cells in Response to Dietary Compound Resveratrol. PLoS ONE 2016, 11, e0157866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beetch, M.; Lubecka, K.; Kristofzski, H.; Suderman, M.; Stefanska, B. Subtle Alterations in DNA Methylation Patterns in Normal Cells in Response to Dietary Stilbenoids. Mol. Nutr. Food Res. 2018, 62, 1800193. [Google Scholar] [CrossRef] [PubMed]
- Beetch, M.; Harandi-Zadeh, S.; Shen, K.; Lubecka, K.; Kitts, D.D.; O’Hagan, H.M.; Stefanska, B. Dietary antioxidants remodel DNA methylation patterns in chronic disease. Nat. Prod. Chemother. 2019, 177, 1382–1408. [Google Scholar] [CrossRef] [Green Version]
- Beetch, M.; Lubecka, K.; Shen, K.; Flower, K.; Harandi-Zadeh, S.; Suderman, M.; Flanagan, J.M.; Stefanska, B. Stilbenoid-Mediated Epigenetic Activation of Semaphorin 3A in Breast Cancer Cells Involves Changes in Dynamic Interactions of DNA with DNMT3A and NF1C Transcription Factor. Mol. Nutr. Food Res. 2019, 63, 1801386. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Chatterjee, B.; Ghosh, K.; Kanade, S.R. Curcumin-mediated demethylation of the proximal promoter CpG island enhances the KLF4 recruitment that leads to increased expression of p21Cip1 in vitro. J. Cell. Biochem. 2019, 120, 809–820. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Wu, R.; Gaspar, J.M.; Sargsyan, D.; Su, Z.Y.; Zhang, C.; Gao, L.; Cheng, D.; Li, W.; Wang, C.; et al. DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis 2018, 39, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Bilir, B.; Sharma, N.V.; Lee, J.; Hammarstrom, B.; Svindland, A.; Kucuk, O.; Moreno, C.S. Effects of genistein supplementation on genome-wide DNA methylation and gene expression in patients with localized prostate cancer. Int. J. Oncol. 2017, 51, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Simmen, F.A.; Xiao, R.; Simmen, R.C.M. Expression profiling of rat mammary epithelial cells reveals candidate signaling pathways in dietary protection from mammary tumors. Physiol. Genom. 2007, 30, 8–16. [Google Scholar] [CrossRef] [Green Version]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Yoshino, H.; Kinoshita, T.; Majid, S.; Saini, S.; Chang, I.; Tanaka, Y.; Enokida, H.; et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS ONE 2013, 8, e70372. [Google Scholar] [CrossRef]
- Soundararajan, R.; Wishart, A.D.; Rupasinghe, H.V.; Arcellana-Panlilio, M.; Nelson, C.M.; Mayne, M.; Robertson, G.S. Quercetin 2008, 3-glucoside protects neuroblastoma (SH-SY5Y) cells in vitro against oxidative damage by inducing sterol regulatory element-binding protein-2-mediated cholesterol biosynthesis. J. Biol. Chem. 2008, 283, 2231–2245. [Google Scholar] [CrossRef] [Green Version]
- Shike, M.; Doane, A.S.; Khanin, R.; Bromberg, J.; Gerald, W.L.; Norton, L. The effects of soy supplementation on gene expression in breast cancer: A randomized placebo-controlled study. Natl. Cancer Inst. 2014, 106, dju189. [Google Scholar] [CrossRef]
- Li, L.; Liu, J.; Wang, X.; Xiong, X.; Huang, S.; Wang, X. Microarray analysis of differentially expressed long non-coding RNAs in daidzein-treated lung cancer cells. Oncol. Lett. 2021, 22, 789. [Google Scholar] [CrossRef]
- Notas, G.; Nifli, A.P.; Kampa, M.; Pelekanou, V.; Alexaki, V.I.; Theodoropoulos, P.; Vercauteren, J.; Castanas, E. Quercetin accumulates in nuclear structures and triggers specific gene expression in epithelial cells. J. Nutr. Biochem. 2012, 23, 656–666. [Google Scholar] [CrossRef]
- Dihal, A.A.; van der Woude, H.; Hendriksen, P.J.M.; Charif, H.; Dekker, L.J.; Ijsselstijn, L.; de Boer, V.C.J.; Alink, G.M.; Burgers, P.C.; Rietjens, I.M.C.M.; et al. Transcriptome and proteome profiling of colon mucosa from quercetin fed F344 rats point to tumor preventive mechanisms, increased mitochondrial fatty acid degradation and decreased glycolysis. Proteomics 2008, 8, 45–61. [Google Scholar] [CrossRef]
- Sakurai, M.A.; Ozaki, Y.; Okuzaki, D.; Naito, Y.; Sasakura, T.; Okamoto, A.; Tabara, H.; Inoue, T.; Hagiyama, M.; Ito, A.; et al. Gefitinib and Luteolin Cause Growth Arrest of Human Prostate Cancer PC-3 Cells via Inhibition of Cyclin G-Associated Kinase and Induction of miR-630. PLoS ONE 2014, 9, e100124. [Google Scholar] [CrossRef] [Green Version]
- Selvi, R.B.; Swaminathan, A.; Chatterjee, S.; Shanmugam, M.K.; Li, F.; Ramakrishnan, G.B.; Siveen, K.S.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; et al. Inhibition of p300 lysine acetyltransferase activity by luteolin reduces tumor growth in head and neck squamous cell carcinoma (HNSCC) xenograft mouse model. Oncotarget 2015, 6, 43806–43818. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.S.; Prasad, S.K.; Shivamallu, C.; Prasad, K.S.; Syed, A.; Reddy, P.; Cull, C.A.; Amachawadi, R.G. Genistein: A Potent Anti-Breast Cancer Agent. Curr. Issues Mol. Biol. 2021, 43, 1502–1517. [Google Scholar] [CrossRef]
- Tuli, H.S.; Tuorkey, M.J.; Thakral, F.; Sak, K.; Kumar, M.; Sharma, A.K.; Sharma, U.; Jain, A.; Aggarwal, V.; Bishayee, A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front. Pharmacol. 2019, 6, 1336. [Google Scholar] [CrossRef] [Green Version]
- CID5281708. PubChem Compound Summary for CID 5281708, Daidzein. 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5281708 (accessed on 28 June 2022).
- Jose Merlin, J.P.; Rupasinghe, H.P.V.; Dellaire, G.; Murphy, K. Role of Dietary Antioxidants in p53-Mediated Cancer Chemoprevention and Tumor Suppression. Oxid. Med. Cell. Longev. 2021, 2021, 9924328. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F.; Gajate, C. Fas/CD95 death receptor and lipid rafts: New targets for apoptosis-directed cancer therapy. Drug Resist. Updates 2006, 9, 51–73. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Tang, H.Y.; Davis, F.B.; Davis, P.J. Resveratrol and apoptosis. Ann. N. Y. Acad. Sci. 2011, 1215, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Talib, W.H.; Al-Hadid, S.A.; Ali, M.B.W.; AL-Yasari, I.H.; Ali, M.R.A. Role of curcumin in regulating p53 in breast cancer: An overview of the mechanism of action. Breast Cancer 2018, 10, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Fulda, S.; Debatin, K.M. Sensitization for Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand-Induced Apoptosis by the Chemopreventive Agent Resveratrol. Cancer Res. 2004, 64, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Mader, I.; Wabitsch, M.; Debatin, K.M.; Fischer-Posovszky, P.; Fulda, S. Identification of a novel proapoptotic function of resveratrol in fat cells: SIRT1-independent sensitization to TRAIL-induced apoptosis. FASEB J. 2010, 24, 1997–2009. [Google Scholar] [CrossRef]
- Gogada, R.; Prabhu, V.; Amadori, M.; Scott, R.; Hashmi, S.; Chandra, D. Resveratrol induces p53-independent, X-linked inhibitor of apoptosis protein (XIAP)-mediated Bax protein oligomerization on mitochondria to initiate cytochrome c release and caspase activation. J. Biol. Chem. 2011, 286, 28749–28760. [Google Scholar] [CrossRef] [Green Version]
- Gosslau, A.; Pabbaraja, S.; Knapp, S.; Chen, K.Y. Trans- and cis-stilbene polyphenols induced rapid perinuclear mitochondrial clustering and p53-independent apoptosis in cancer cells but not normal cells. Eur. J. Pharmacol. 2008, 587, 25–34. [Google Scholar] [CrossRef]
- Watson, J.L.; Hill, R.; Yaffe, P.B.; Greenshields, A.; Walsh, M.; Lee, P.W.; Giacomantonio, C.A.; Hoskin, D.W. Curcumin causes superoxide anion production and p53-independent apoptosis in human colon cancer cells. Cancer Lett. 2010, 297, 1–8. [Google Scholar] [CrossRef]
- Li, F.; Chen, X.; Xu, B.; Zhou, H. Curcumin induces p53-independent necrosis in H1299 cells via a mitochondria-associated pathway. Mol. Med. Rep. 2015, 12, 7806–7814. [Google Scholar] [CrossRef] [Green Version]
- Watson, J.L.; Greenshields, A.; Hill, R.; Hilchie, A.; Lee, P.W.; Giacomantonio, C.A.; Hoskin, D.W. Curcumin-induced apoptosis in ovarian carcinoma cells is p53-independent and involves p38 mitogen-activated protein kinase activation and downregulation of Bcl-2 and survivin expression and Akt signaling. Mol. Carcinog. 2010, 49, 13–24. [Google Scholar] [CrossRef]
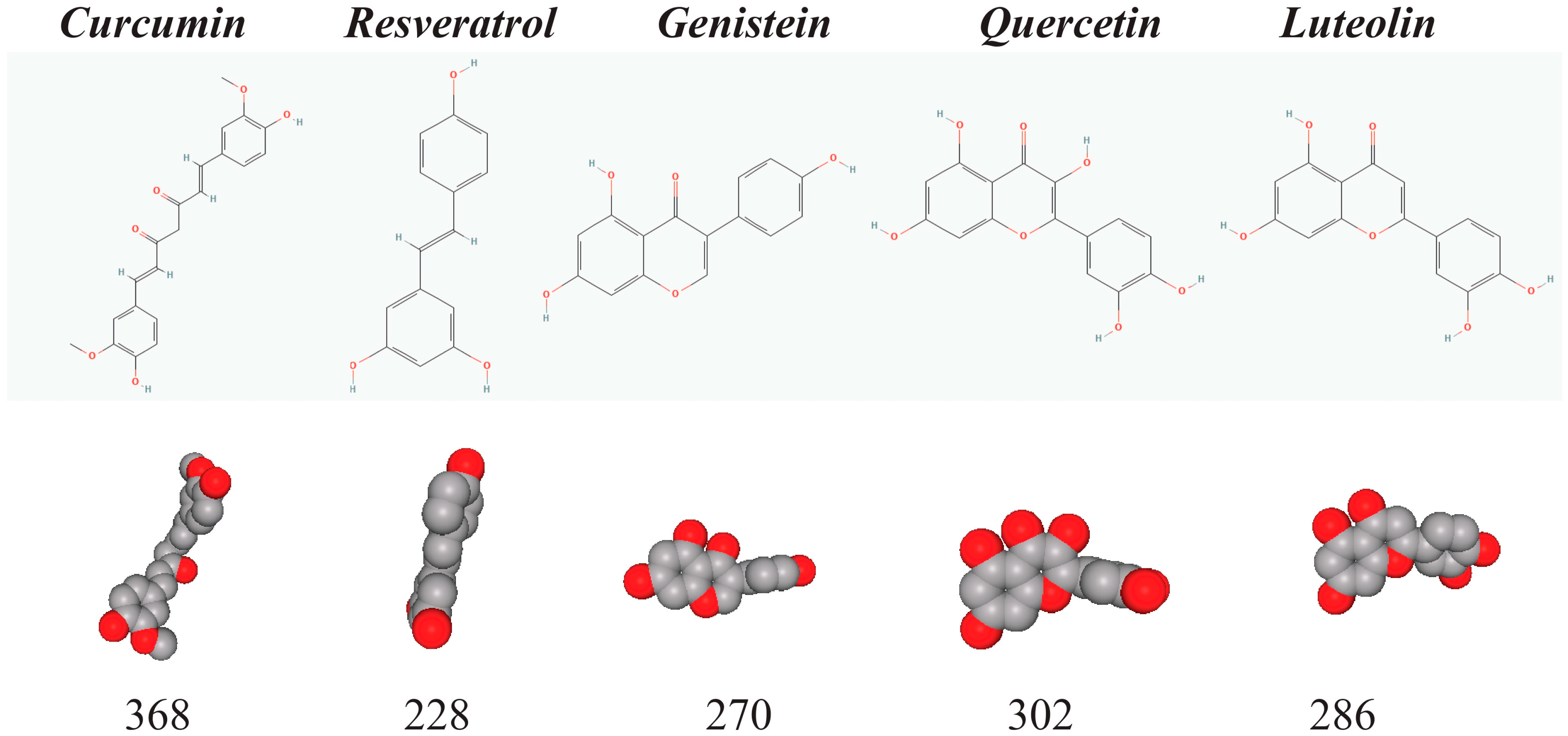
| Compound | Articles | Reviews | Clinical Trials | Supplements |
|---|---|---|---|---|
| Curcumin | 7157 | 1324 | 86 | 318 |
| Resveratrol | 4393 | 1067 | 22 | 214 |
| Genistein | 3474 | 489 | 33 | 15 |
| Quercetin | 4019 | 509 | 20 | 310 |
| Luteolin | 1048 | 102 | 3 | 100 |
| Compound | Synonyms | Reactive Group | PubChem * Reference | PubChem ID | Molecular Formula |
|---|---|---|---|---|---|
| Curcumin | Diferuloylmethane | Ether, ketone, phenol, unsaturated aliphatic hydrocarbon, hydroxyl | [9] | 969516 | C21H20O6 |
| Resveratrol | 3, 4′, 5-Trihydroxystilbene | Stilbene, phenol, hydroxyl | [10] | 445154 | C14H12O3 |
| Genistein | 4′, 5, 7-Trihydroxyisoflavone | Phenol, hydroxyl, ketone | [11] | 5280961 | C15H10O5 |
| Quercetin | 3, 3′, 4′, 5, 7-Pentahydroxyflavone | Phenol, hydroxyl, ketone | [12] | 5280343 | C15H10O7 |
| Luteolin | 3′, 4′, 5, 7-Tetrahydroxyflavone | Phenol, hydroxyl, ketone | [13] | 5280445 | C15H10O6 |
| Technological Platform | Cell Line and Treatment Conditions | Reported Mechanism | Other Conclusions | Reference |
|---|---|---|---|---|
| LD-microarray profiling expression of coding genes. | LNCaP human prostate adenocarcinoma cells (androgen-sensitive, p53 wt/wt). | Resveratrol induced the intrinsic apoptosis pathway (but only at the highest concentration tested). | At intermediate concentrations, resveratrol modulated cell cycle regulatory genes, and down-regulated markers of cellular proliferation. (At the lowest concentrations, resveratrol stimulated viability suggesting a hormetic response curve.) | [47] |
| The phytochemical was applied in cell media in increasing concentrations: 0.01, 0.1, 1, 10, 25, 40, and 100 μM. | The activation of p53-dependant apoptosis was evident by the transcriptional upregulation of p21 and MDM2. | Resveratrol also had a strong inhibitory effect on the androgen pathway (which included prostate-specific antigen—PSA). | ||
| HD-microarray profiling expression of coding genes. | The following human lung cancer cells were used: NCI H460 (p53 wt/wt), NCI H23 (with a homozygous missense mutation: methionine to isoleucine at codon 246), and A549 (p53 wt/wt). | Resveratrol induced the intrinsic apoptosis pathway in wild-type A549 cells. | Resveratrol also inhibited growth of the p53 mutated cancer cell line (NCI H23) suggesting that it can also induce p53-independent apoptosis or cell cycle arrest. | [48] |
| 25 μM phytochemical was applied in cell media (with the incubation time of 48 h). | The activation of p53-dependant apoptosis was evident by transcriptional upregulation of p21 and p27. | |||
| Human fibrosarcoma cells: HT1080 (p53 wt/wt). | Resveratrol induced the intrinsic apoptosis pathway. | Resveratrol also modulated the expression of genes associated with cell cycle, cytoskeleton, and cell-adhesion. | [49] | |
| The phytochemical was applied at 2195 ng/mL in media (with the incubation time of six hours). | The activation of p53-dependant apoptosis was evident by differential regulation of 13 genes in the KEGG’s p53 signaling pathway. | |||
| MDA-MB-231 human breast cancer cell line (estrogen receptor negative). | Resveratrol induced the intrinsic apoptosis pathway. | There was also evidence for cell cycle arrest (increased fraction of cells in the G1 phase and inhibition of the expression of cyclin B1). | [50] | |
| 10 μM phytochemical was applied in cell media (with the incubation time of six hours). | The activation of the p53-dependant apoptosis was evident by transcriptional upregulation of p21, PIG3, and BAD. | |||
| Human renal carcinoma cells of likely proximal tubule origin. (These cells are known to have a p53 wt/wt genotype, but p53 signaling is strongly repressed.) | Resveratrol induced the intrinsic apoptosis pathway. | Fifteen-fold induction of tumor necrosis factor α inducible protein 3 (TNFAIP3) also suggested the induction of the extrinsic apoptosis pathway. | [51] | |
| 50 μM phytochemical was applied in cell media (with the incubation time of 24 h). | The activation of the p53-dependant apoptosis was evident by sevenfold transcriptional upregulation of MDM2. |
| Technological Platform | Cell Line and Treatment Conditions | Reported Mechanism | Other Conclusions | Reference |
|---|---|---|---|---|
| LD-microarray profiling expression of coding genes. | MCF-7 human breast adenocarcinoma (p53 wt/wt). | Resveratrol induced the intrinsic apoptosis pathway. | Changes in expression of some apoptosis-related genes were dependent on the concentration of curcumin: shifting in opposite directions at the low versus the high concentration. (This might suggest hormetic effects.) | [52] |
| The activation of the p53-dependant apoptosis was evident by transcriptional up-regulation of CASP1, CASP2, CASP3, CASP4, BCL2L2, PIG3, and PIG11. | ||||
| Cells were incubated in media with 0, 25, or 50 μg/mL phytochemical (applied for 24 h). | ||||
| Observed changes also suggested indirect activation of apoptosis by down-regulation of pro-survival signals from growth factors and apoptosis inhibitors. | ||||
| HD-microarray profiling expression of coding genes. | Hepatocellular carcinoma cell lines: KMCH, WRL68, Huh7, PLC, and Pitts1. | Curcumin indirectly promoted apoptosis by silencing pro-survival oncogenic signaling, in particular the NF-κB pathway. | Curcumin also modulated MYC signaling, which is known to have mixed effects (partially mitogenic and partially pro-apoptotic). Curcumin also modulated genes involved in cytokine signaling, growth factor signaling, and the regulation of angiogenesis. | [53] |
| Cells were incubated with 25 μM phytochemical (applied for 72 h). | ||||
| A microarray profiling microRNAs (miRCURY). | Several human cell lines originating from non-small cell lung cancers. | Resveratrol induced the intrinsic apoptosis pathway. | Altogether, six microRNAs were up regulated by the curcumin treatment: miR-132-3p, miR-183-5p, miR-124-3p, miR-215, miR-192-5p, and miR-194-5p. Moreover, two microRNAs were down regulated (i.e., miR-602 and miR-223-3p). | [54] |
| The activation of the p53-dependant apoptosis was evident by up-regulation of pro-apoptotic miR-192-5p and miR-215. | ||||
| The microRNAs acts similarly to protein-coding anti-oncogenes inducing apoptosis in transformed cells. The induction of apoptosis involved the down-regulation of a known inhibitor of the intrinsic apoptosis pathway: X-linked inhibitor of apoptosis protein (XIAP—which normally binds and blocks initiator caspase 9). The resulting activation of the initiator caspase, in turn, led to the activation of effector caspases (in particular caspase-3) that rapidly effected cell death. | ||||
| Cells were incubated with 15 μM curcumin for 48 h. |
| Compound | Cancer Model | Chip | Data |
|---|---|---|---|
| Resveratrol | Prostate adenocarcinoma cell line [47] | LD-microarray with 30,000 UniGene clusters | GSE4399 |
| Non-small cell lung cancer [48] | Human Genome U133 Plus 2.0 Array (Affymetrix) | GDS2966 | |
| Fibrosarcoma cell line [49] | Human Genome U133 Plus 2.0 Array (Affymetrix) | GSE59704 | |
| Breast adenocarcinoma [50] | HD-microarray | n/a | |
| Kidney carcinoma [51] | LD-microarray | n/a | |
| Curcumin | Breast adenocarcinoma [52] | LD-microarray, 4069 I.M.A.G.E. clones spotted on microscopic slides | n/a |
| Liver hepatocellular carcinoma [53] | Human HT-12 V4.0 expression Beadchip (Illumina) | GSE59713 | |
| Non-small cell lung cancer [54] | MicroRNA microarray | n/a |
| Compound | Cancer Model | Chip | Data |
|---|---|---|---|
| Genistein | Prostate specimens from a clinical trial of genistein supplementation prior to prostatectomy [88] | HumanHT-12 v4 Expression BeadChip, HumanMethylation450 (Illumina) | GSE84748, GSE84749 |
| Rat mammary epithelial cells [89] | Rat 230A GeneChip (Affymetrix) | GSE6879 | |
| Prostate cancer cell line [90] | SurePrint G3 Human GE 8 × 60K Microarray (Agilent) | GSE29079 | |
| Human embryonic kidney cell line—HEK293, and breast cancer cell line—MCF-7 [91] | 14K microarray slides printed at the University of Calgary | GSE6199, GSE6200 | |
| Women with invasive breast adenocarcinoma [92] | Human U133 Plus 2.0 chip (Affymetrix) | GSE58792 | |
| Daidzein | H1299 lung cancer cells [93] | Capital Bio Technology human long non-coding RNA Array v4 | GSE181093 |
| Quercetin | HepG2 hepatocellular carcinoma cell line and breast adenocarcinoma T47D cells [94] | Human U133A Plus 2 chip (Affymetrix) | GSE15162 |
| Distal colon mucosa of rats [95] | Rat Genome 230 2.0 Array (Affymetrix) | GSE7479 | |
| Neuroblastoma cells in vitro [91] | GSE6200 | ||
| Luteolin | Prostate cancer cell line [96] | SurePrint Human v18.0 miRNA array, whole human genome oligonucleotide microarrays (Agilent) | GSE53180, GSE53178 |
| Mouse xenograft model of head and neck squamous cell carcinoma [97] | SurePrint G3 Human GE v2 8 × 60K array, human miRNA microarray (Agilent) | GSE75029 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huminiecki, L. Evidence for Multilevel Chemopreventive Activities of Natural Phenols from Functional Genomic Studies of Curcumin, Resveratrol, Genistein, Quercetin, and Luteolin. Int. J. Mol. Sci. 2022, 23, 14957. https://doi.org/10.3390/ijms232314957
Huminiecki L. Evidence for Multilevel Chemopreventive Activities of Natural Phenols from Functional Genomic Studies of Curcumin, Resveratrol, Genistein, Quercetin, and Luteolin. International Journal of Molecular Sciences. 2022; 23(23):14957. https://doi.org/10.3390/ijms232314957
Chicago/Turabian StyleHuminiecki, Lukasz. 2022. "Evidence for Multilevel Chemopreventive Activities of Natural Phenols from Functional Genomic Studies of Curcumin, Resveratrol, Genistein, Quercetin, and Luteolin" International Journal of Molecular Sciences 23, no. 23: 14957. https://doi.org/10.3390/ijms232314957




