From Nucleus to Organs: Insights of Aryl Hydrocarbon Receptor Molecular Mechanisms
Abstract
:1. Introduction
2. Epigenetics and Chromatin: AHR-Driven Barriers
2.1. Retrotransposons
2.2. Chromatin Structural Organization
3. AHR Signaling
3.1. The Wnt/β-Catenin Pathway
3.2. The PI3K/AKT Pathway
3.3. Interaction with TGF-β Signaling
3.4. NF-κβ and p65
3.5. Other Protein Interactions
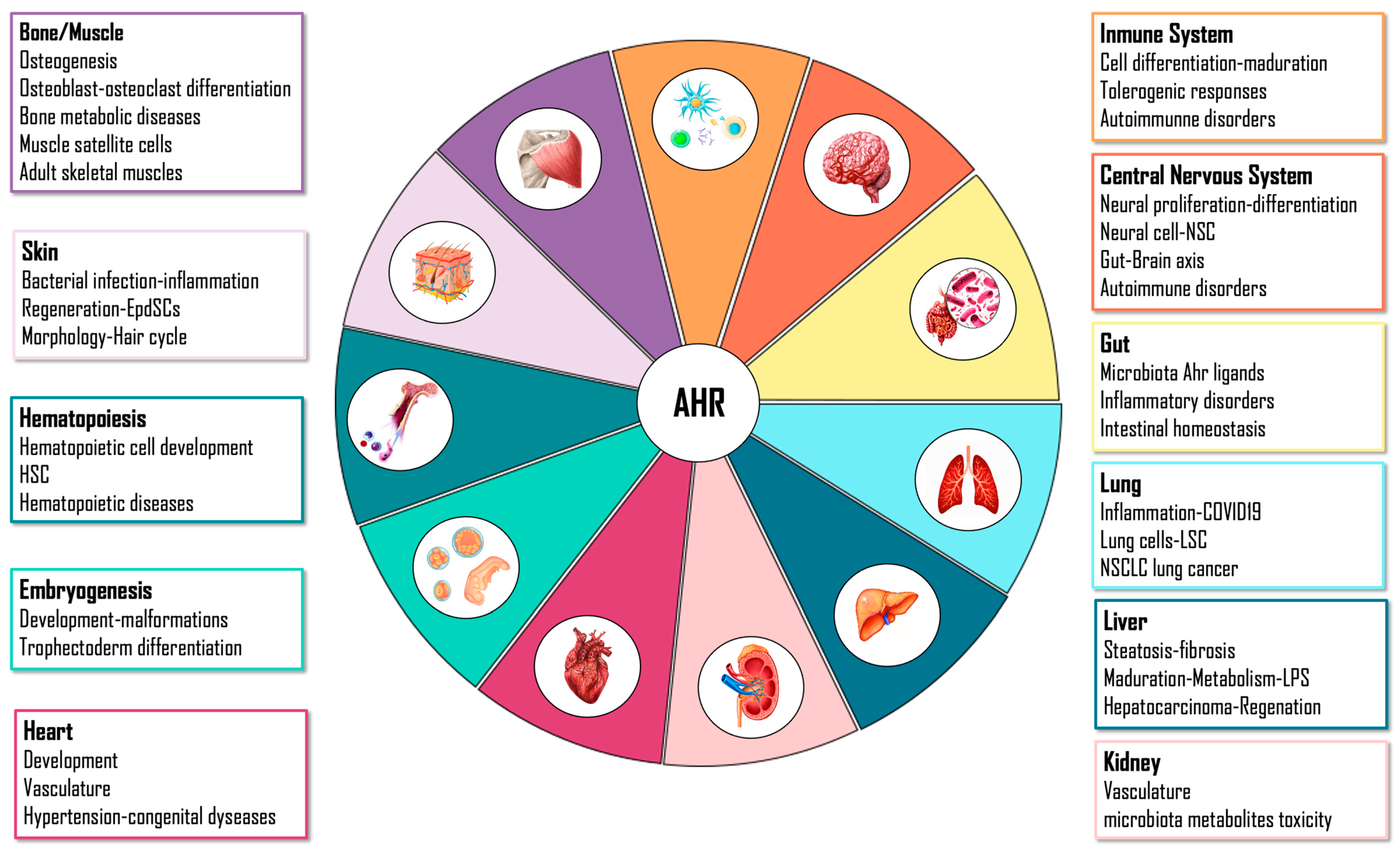
4. AHR Physiological Functions
4.1. Embryo
4.2. Liver
4.3. Gut
4.4. Immune System
4.5. Central Nervous System
4.6. Skin
4.7. Lung
5. Final Thoughts
Author Contributions
Funding
Conflicts of Interest
References
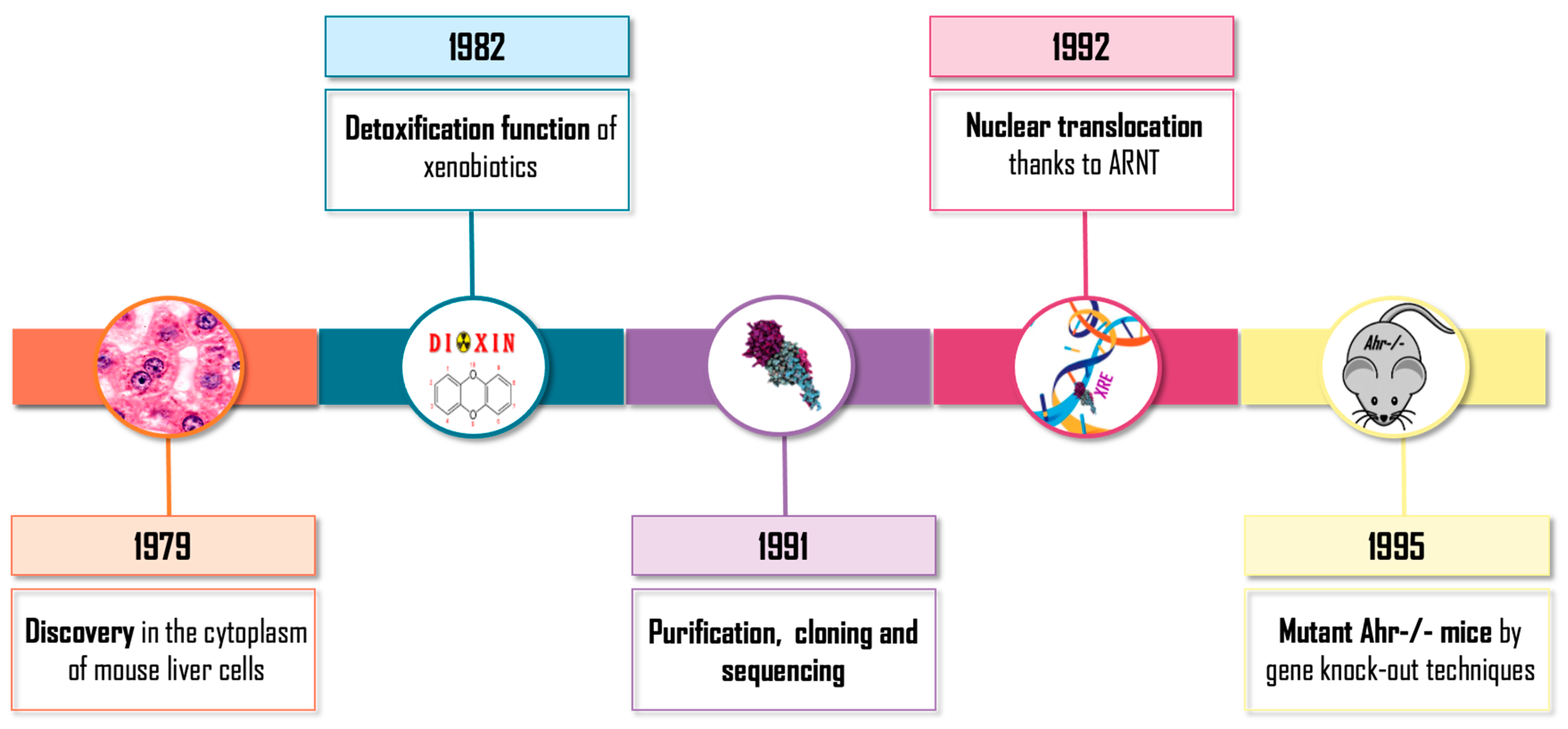
- Okey, A.B.; Bondy, G.P.; Mason, M.E.; Kahl, G.F.; Eisen, H.J.; Guenthner, T.M.; Nebert, D.W. Regulatory gene product of the Ah locus. Characterization of the cytosolic inducer-receptor complex and evidence for its nuclear translocation. J. Biol. Chem. 1979, 254, 11636–11648. [Google Scholar] [CrossRef]
- Poland, A.; Knutson, J.C. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982, 22, 517–554. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, W.F.; Poland, A. Nuclear uptake of 2,3,7,8-tetrachlorodibenzo-p-dioxin in C57BL/6J and DBA/2J mice. Role of the hepatic cytosol receptor protein. J. Biol. Chem. 1979, 254, 9814–9821. [Google Scholar] [CrossRef] [PubMed]
- Reyes, H.; Reisz-Porszasz, S.; Hankinson, O. Identification of the Ah Receptor Nuclear Translocator Protein (Arnt) as a Component of the DNA Binding Form of the Ah Receptor. Science 1992, 256, 1193–1195. [Google Scholar] [CrossRef] [PubMed]
- Bradfield, C.A.; Glover, E.; Poland, A. Purification and N-terminal amino acid sequence of the Ah receptor from the C57BL/6J mouse. Mol. Pharmacol. 1991, 39, 13–19. [Google Scholar] [PubMed]
- Burbach, K.M.; Poland, A.; Bradfield, C.A. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc. Natl. Acad. Sci. USA 1992, 89, 8185–8189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Salguero, P.M.; Hilbert, D.M.; Rudikoff, S.; Ward, J.M.; Gonzalez, F.J. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 1996, 140, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Mimura, J.; Yamashita, K.; Nakamura, K.; Morita, M.; Takagi, T.N.; Nakao, K.; Ema, M.; Sogawa, K.; Yasuda, M.; Katsuki, M.; et al. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 1997, 2, 645–654. [Google Scholar] [CrossRef]
- Shimizu, Y.; Nakatsuru, Y.; Ichinose, M.; Takahashi, Y.; Kume, H.; Mimura, J.; Fujii-Kuriyama, Y.; Ishikawa, T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2000, 97, 779–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marlowe, J.L.; Puga, A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell. Biochem. 2005, 96, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, A.; Poellinger, L.; Pongratz, I. Evidence That the Co-chaperone p23 Regulates Ligand Responsiveness of the Dioxin (Aryl Hydrocarbon) Receptor. J. Biol. Chem. 1999, 274, 13519–13524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazlauskas, A.; Sundström, S.; Poellinger, L.; Pongratz, I. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol. Cell. Biol. 2001, 21, 2594–2607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lees, M.J.; Whitelaw, M.L. Multiple Roles of Ligand in Transforming the Dioxin Receptor to an Active Basic Helix-Loop-Helix/PAS Transcription Factor Complex with the Nuclear Protein Arnt. Mol. Cell. Biol. 1999, 19, 5811–5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Duran, A.; Ballestar, E.; Carvajal-Gonzalez, J.M.; Marlowe, J.L.; Puga, A.; Esteller, M.; Fernandez-Salguero, P.M. Recruitment of CREB1 and Histone Deacetylase 2 (HDAC2) to the Mouse Ltbp-1 Promoter Regulates its Constitutive Expression in a Dioxin Receptor-dependent Manner. J. Mol. Biol. 2008, 380, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Niermann, T.; Schmutz, S.; Erne, P.; Resink, T. Aryl hydrocarbon receptor ligands repress T-cadherin expression in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2003, 300, 943–949. [Google Scholar] [CrossRef]
- Ule, J. Alu elements: At the crossroads between disease and evolution. Biochem. Soc. Trans. 2013, 41 Pt 6, 1532–1535. [Google Scholar] [CrossRef] [Green Version]
- Rejano-Gordillo, C.; Ordiales-Talavero, A.; Nacarino-Palma, A.; Merino, J.M.; González-Rico, F.J.; Fernández-Salguero, P.M. Aryl Hydrocarbon Receptor: From Homeostasis to Tumor Progression. Front. Cell Dev. Biol. 2022, 10, 884004. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.L.; Moran, J.V.; Batzer, M.A.; Kazazian, H.H. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003, 13, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Vasicek, T.J.; Zeng, L.; Guan, X.J.; Zhang, T.; Costantini, F.; Tilghman, S.M. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 1997, 147, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Kondo-Iida, E.; Kobayashi, K.; Watanabe, M.; Sasaki, J.; Kumagai, T.; Koide, H.; Saito, K.; Osawa, M.; Nakamura, Y.; Toda, T. Novel mutations and genotype-phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD). Hum. Mol. Genet. 1999, 8, 2303–2309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogvadze, E.; Buzdin, A. Retroelements and their impact on genome evolution and functioning. Cell. Mol. Life Sci. 2009, 66, 3727–3742. [Google Scholar] [CrossRef] [PubMed]
- Polak, P.; Domany, E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genom. 2006, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Kramerov, D.A.; Grigoryan, A.A.; Ryskov, A.P.; Georgiev, G.P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: Evidence from DNA cloning experiments. Nucleic Acids Res. 1979, 6, 697–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, C.M.; Houck, C.M.; Deininger, P.L.; Friedmann, T.; Schmid, C.W. Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature 1980, 284, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.L.; Jolly, D.J.; Rubin, C.M.; Friedmann, T.; Schmid, C.W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J. Mol. Biol. 1981, 151, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Roman, A.C.; Benitez, D.A.; Carvajal-Gonzalez, J.M.; Fernandez-Salguero, P.M. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 1632–1637. [Google Scholar] [CrossRef] [Green Version]
- Morales-Hernández, A.; González-Rico, F.J.; Román, A.C.; Rico-Leo, E.; Alvarez-Barrientos, A.; Sánchez, L.; Macia, Á.; Heras, S.R.; García-Pérez, J.L.; Merino, J.M.; et al. Alu retrotransposons promote differentiation of human carcinoma cells through the aryl hydrocarbon receptor. Nucleic Acids Res. 2016, 44, 4665–4683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Román, A.C.; González-Rico, F.J.; Moltó, E.; Hernando, H.; Neto, A.; Vicente-Garcia, C.; Ballestar, E.; Gómez-Skarmeta, J.L.; Vavrova-Anderson, J.; White, R.J.; et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011, 21, 422–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Rico, F.J.; Vicente-García, C.; Fernández, A.; Muñoz-Santos, D.; Montoliu, L.; Morales-Hernández, A.; Merino, J.M.; Román, A.-C.; Fernández-Salguero, P.M. Alu retrotransposons modulate Nanog expression through dynamic changes in regional chromatin conformation via aryl hydrocarbon receptor. Epigenetics Chromatin 2020, 13, 15. [Google Scholar] [CrossRef]
- Fraser, J.; Ferrai, C.; Chiariello, A.; Schueler, M.; Rito, T.; Laudanno, G.; Barbieri, M.; Moore, B.; Kraemer, D.; Aitken, S.; et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 2015, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- Narendra, V.; Bulajić, M.; Dekker, J.; Mazzoni, E.O.; Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016, 30, 2657–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonev, B.; Mendelson Cohen, N.; Szabo, Q.; Fritsch, L.; Papadopoulos, G.L.; Lubling, Y.; Xu, X.; Lv, X.; Hugnot, J.-P.; Tanay, A.; et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 2017, 171, 557–572.e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poterlowicz, K.; Yarker, J.L.; Malashchuk, I.; Lajoie, B.R.; Mardaryev, A.N.; Gdula, M.R.; Sharov, A.A.; Kohwi-Shigematsu, T.; Botchkarev, V.A.; Fessing, M.Y. 5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells. PLOS Genet. 2017, 13, e1006966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niskanen, H.; Tuszynska, I.; Zaborowski, R.; Heinäniemi, M.; Ylä-Herttuala, S.; Wilczynski, B.; Kaikkonen, M.U. Endothelial cell differentiation is encompassed by changes in long range interactions between inactive chromatin regions. Nucleic Acids Res. 2018, 46, 1724–1740. [Google Scholar] [CrossRef] [Green Version]
- Gialitakis, M.; Tolaini, M.; Li, Y.; Pardo, M.; Yu, L.; Toribio, A.; Choudhary, J.S.; Niakan, K.; Papayannopoulos, V.; Stockinger, B. Activation of the Aryl Hydrocarbon Receptor Interferes with Early Embryonic Development. Stem Cell Rep. 2017, 9, 1377–1386. [Google Scholar] [CrossRef] [Green Version]
- Mulero-Navarro, S.; Carvajal-Gonzalez, J.M.; Herranz, M.; Ballestar, E.; Fraga, M.F.; Ropero, S.; Esteller, M.; Fernandez-Salguero, P.M. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis 2006, 27, 1099–1104. [Google Scholar] [CrossRef]
- Nabirochkin, S.; Ossokina, M.; Heidmann, T. A Nuclear Matrix/Scaffold Attachment Region Co-localizes with the Gypsy Retrotransposon Insulator Sequence. J. Biol. Chem. 1998, 273, 2473–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glinsky, G.V. Contribution of transposable elements and distal enhancers to evolution of human-specific features of interphase chromatin architecture in embryonic stem cells. Chromosome Res. 2018, 26, 61–84. [Google Scholar] [CrossRef]
- Román, A.C.; González-Rico, F.J.; Fernández-Salguero, P.M. B1-SINE retrotransposons: Establishing genomic insulatory networks. Mob. Genet. Elem. 2011, 1, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Schwalie, P.C.; Wilson, M.D.; Ballester, B.; Gonçalves, A.; Kutter, C.; Brown, G.D.; Marshall, A.; Flicek, P.; Odom, D.T. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 2012, 148, 335–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef] [Green Version]
- Wendt, K.S.; Yoshida, K.; Itoh, T.; Bando, M.; Koch, B.; Schirghuber, E.; Tsutsumi, S.; Nagae, G.; Ishihara, K.; Mishiro, T.; et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008, 451, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Parelho, V.; Hadjur, S.; Spivakov, M.; Leleu, M.; Sauer, S.; Gregson, H.C.; Jarmuz, A.; Canzonetta, C.; Webster, Z.; Nesterova, T.; et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008, 132, 422–433. Available online: https://pubmed.ncbi.nlm.nih.gov/18237772/ (accessed on 7 January 2022). [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Sinha, M.; Peterson, C.L.; Weng, Z. The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome. PLoS Genet. 2008, 4, e1000138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larigot, L.; Juricek, L.; Dairou, J.; Coumoul, X. AhR signaling pathways and regulatory functions. Biochim. Open 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.S.; Soshilov, A.A.; He, G.; DeGroot, D.E.; Zhao, B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 2011, 124, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Celso, C.; Prowse, D.M.; Watt, F.M. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development 2004, 131, 1787–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faraldo, M.M.; Taddei-De La Hosseraye, I.; Teulière, J.; Deugnier, M.-A.; Moumen, M.; Thiery, J.-P.; Glukhova, M.A. Mammary gland development: Role of basal myoepithelial cells. J. Soc. Biol. 2006, 200, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chu, E.Y.; Watt, B.; Zhang, Y.; Gallant, N.M.; Andl, T.; Yang, S.H.; Lu, M.-M.; Piccolo, S.; Schmidt-Ullrich, R.; et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 2008, 313, 210–224. [Google Scholar] [CrossRef]
- van Amerongen, R.; Berns, A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006, 22, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, T.; Wend, P.; Klaus, A.; Birchmeier, W. Deciphering the function of canonical Wnt signals in development and disease: Conditional loss- and gain-of-function mutations of β-catenin in mice. Genes Dev. 2008, 22, 2308–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudloff, S.; Kemler, R. Differential requirements for β-catenin during mouse development. Dev. Camb. Engl. 2012, 139, 3711–3721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polakis, P. Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 2012, 4, a008052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, A.J.; Branam, A.M.; Peterson, R.E. Intersection of AHR and Wnt signaling in development, health, and disease. Int. J. Mol. Sci. 2014, 15, 17852–17885. [Google Scholar] [CrossRef] [Green Version]
- Procházková, J.; Kabátková, M.; Bryja, V.; Umannová, L.; Bernatík, O.; Kozubík, A.; Machala, M.; Vondrácek, J. The interplay of the aryl hydrocarbon receptor and β-catenin alters both AhR-dependent transcription and Wnt/β-catenin signaling in liver progenitors. Toxicol. Sci. 2011, 122, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Bjeldanes, L.F.; Kim, J.Y.; Grose, K.R.; Bartholomew, J.C.; Bradfield, C.A. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: Comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc. Natl. Acad. Sci. USA 1991, 88, 9543–9547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peter Guengerich, F.; Martin, M.V.; McCormick, W.A.; Nguyen, L.P.; Glover, E.; Bradfield, C.A. Aryl hydrocarbon receptor response to indigoids in vitro and in vivo. Arch. Biochem. Biophys. 2004, 423, 309–316. [Google Scholar] [CrossRef]
- Jeong, Y.-M.; Li, H.; Kim, S.Y.; Yun, H.-Y.; Baek, K.J.; Kwon, N.S.; Myung, S.C.; Kim, D.-S. Indole-3-carbinol inhibits prostate cancer cell migration via degradation of beta-catenin. Oncol. Res. 2011, 19, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Kawajiri, K.; Kobayashi, Y.; Ohtake, F.; Ikuta, T.; Matsushima, Y.; Mimura, J.; Pettersson, S.; Pollenz, R.S.; Sakaki, T.; Hirokawa, T.; et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc. Natl. Acad. Sci. USA 2009, 106, 13481–13486. [Google Scholar] [CrossRef]
- Moreno-Marín, N.; Merino, J.M.; Alvarez-Barrientos, A.; Patel, D.P.; Takahashi, S.; González-Sancho, J.M.; Gandolfo, P.; Rios, R.M.; Muñoz, A.; Gonzalez, F.J.; et al. Aryl Hydrocarbon Receptor Promotes Liver Polyploidization and Inhibits PI3K, ERK, and Wnt/β-Catenin Signaling. iScience 2018, 4, 44–63. [Google Scholar] [CrossRef] [PubMed]
- Branam, A.M.; Davis, N.M.; Moore, R.W.; Schneider, A.J.; Vezina, C.M.; Peterson, R.E. TCDD inhibition of canonical Wnt signaling disrupts prostatic bud formation in mouse urogenital sinus. Toxicol. Sci. 2013, 133, 42–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, L.K.; Sengupta, S.S.; Ladu, J.; Andreasen, E.A.; Tanguay, R.L. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008, 22, 3087–3096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelrahim, M.; Smith, R.; Safe, S. Aryl hydrocarbon receptor gene silencing with small inhibitory RNA differentially modulates Ah-responsiveness in MCF-7 and HepG2 cancer cells. Mol. Pharmacol. 2003, 63, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Pohjanvirta, R.; Miettinen, H.; Sankari, S.; Hegde, N.; Lindén, J. Unexpected gender difference in sensitivity to the acute toxicity of dioxin in mice. Toxicol. Appl. Pharmacol. 2012, 262, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Sun, L. Recent advances in the development of AHR antagonists in immuno-oncology. RSC Med. Chem. 2021, 12, 902–914. [Google Scholar] [CrossRef]
- Rodon, J.; Dienstmann, R.; Serra, V.; Tabernero, J. Development of PI3K inhibitors: Lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 2013, 10, 143–153. [Google Scholar] [CrossRef]
- Shi, X.; Wang, J.; Lei, Y.; Cong, C.; Tan, D.; Zhou, X. Research progress on the PI3K/AKT signaling pathway in gynecological cancer (Review). Mol. Med. Rep. 2019, 19, 4529–4535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.; Zhang, L.; Hoagland, M.S.; Swanson, H.I. Lack of the aryl hydrocarbon receptor leads to impaired activation of AKT/protein kinase B and enhanced sensitivity to apoptosis induced via the intrinsic pathway. J. Pharmacol. Exp. Ther. 2007, 320, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Buscà, R.; Pouysségur, J.; Lenormand, P. ERK1 and ERK2 Map Kinases: Specific Roles or Functional Redundancy? Front. Cell Dev. Biol. 2016, 4, 53. [Google Scholar] [CrossRef]
- Shi, F.; Aloufi, N.; Traboulsi, H.; Trempe, J.-F.; Eidelman, D.H.; Baglole, C.J. Endogenous regulation of the Akt pathway by the aryl hydrocarbon receptor (AhR) in lung fibroblasts. Sci. Rep. 2021, 11, 23189. [Google Scholar] [CrossRef] [PubMed]
- Koli, K.; Saharinen, J.; Hyytiäinen, M.; Penttinen, C.; Keski-Oja, J. Latency, activation, and binding proteins of TGF-beta. Microsc. Res. Tech. 2001, 52, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Rifkin, D.B. Latent transforming growth factor-beta (TGF-beta) binding proteins: Orchestrators of TGF-beta availability. J. Biol. Chem. 2005, 280, 7409–7412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, D.A. Latent-TGF-beta: An overview. Mol. Cell. Biochem. 2001, 219, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Duran, A.; Mulero-Navarro, S.; Chang, X.; Fernandez-Salguero, P.M. LTBP-1 blockade in dioxin receptor-null mouse embryo fibroblasts decreases TGF-beta activity: Role of extracellular proteases plasmin and elastase. J. Cell. Biochem. 2006, 97, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Corchero, J.; Martín-Partido, G.; Dallas, S.L.; Fernández-Salguero, P.M. Liver portal fibrosis in dioxin receptor-null mice that overexpress the latent transforming growth factor-beta-binding protein-1. Int. J. Exp. Pathol. 2004, 85, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Gonzalez, J.M.; Mulero-Navarro, S.; Roman, A.C.; Sauzeau, V.; Merino, J.M.; Bustelo, X.R.; Fernandez-Salguero, P.M. The dioxin receptor regulates the constitutive expression of the vav3 proto-oncogene and modulates cell shape and adhesion. Mol. Biol. Cell 2009, 20, 1715–1727. [Google Scholar] [CrossRef] [Green Version]
- Carvajal-Gonzalez, J.M.; Roman, A.C.; Cerezo-Guisado, M.I.; Rico-Leo, E.M.; Martin-Partido, G.; Fernandez-Salguero, P.M. Loss of dioxin-receptor expression accelerates wound healing in vivo by a mechanism involving TGFbeta. J. Cell Sci. 2009, 122 Pt 11, 1823–1833. [Google Scholar] [CrossRef] [Green Version]
- Sarić, N.; Selby, M.; Ramaswamy, V.; Kool, M.; Stockinger, B.; Hogstrand, C.; Williamson, D.; Marino, S.; Taylor, M.D.; Clifford, S.C.; et al. The AHR pathway represses TGFβ-SMAD3 signalling and has a potent tumour suppressive role in SHH medulloblastoma. Sci. Rep. 2020, 10, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Lima, K.A.; Donate, P.B.; Talbot, J.; Davoli-Ferreira, M.; Peres, R.S.; Cunha, T.M.; Alves-Filho, J.C.; Cunha, F.Q. TGFβ1 signaling sustains aryl hydrocarbon receptor (AHR) expression and restrains the pathogenic potential of TH17 cells by an AHR-independent mechanism. Cell Death Dis. 2018, 9, 1130. [Google Scholar] [CrossRef]
- Nakano, N.; Sakata, N.; Katsu, Y.; Nochise, D.; Sato, E.; Takahashi, Y.; Yamaguchi, S.; Haga, Y.; Ikeno, S.; Motizuki, M.; et al. Dissociation of the AhR/ARNT complex by TGF-β/Smad signaling represses CYP1A1 gene expression and inhibits benze[a]pyrene-mediated cytotoxicity. J. Biol. Chem. 2020, 295, 9033–9051. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Ke, S.; Denison, M.S.; Rabson, A.B.; Gallo, M.A. Ah Receptor and NF-κB Interactions, a Potential Mechanism for Dioxin Toxicity. J. Biol. Chem. 1999, 274, 510–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Rabson, A.B.; Gallo, M.A. Ah receptor and NF-kappaB interactions: Mechanisms and physiological implications. Chem. Biol. Interact. 2002, 141, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, Q.; Ma, Y.; Li, L.; Yu, K.; Zhang, Z.; Chen, G.; Li, X.; Xiao, W.; Xu, P.; et al. Aryl Hydrocarbon Receptor Activation Modulates Intestinal Epithelial Barrier Function by Maintaining Tight Junction Integrity. Int. J. Biol. Sci. 2018, 14, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Gómez, G.; Karasová, M.; Tylichová, Z.; Kabátková, M.; Hampl, A.; Matthews, J.; Neča, J.; Ciganek, M.; Machala, M.; Vondráček, J. Aryl Hydrocarbon Receptor (AhR) Limits the Inflammatory Responses in Human Lung Adenocarcinoma A549 Cells via Interference with NF-κB Signaling. Cells 2022, 11, 707. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-H.; Chang, H.; Chang, J.T.; Lin, P. Aryl hydrocarbon receptor in association with RelA modulates IL-6 expression in non-smoking lung cancer. Oncogene 2012, 31, 2555–2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez-Acosta, O.; Vega, L.; Estrada-Muñiz, E.; Rodríguez, M.S.; Gonzalez, F.J.; Elizondo, G. Activation of aryl hydrocarbon receptor regulates the LPS/IFNγ-induced inflammatory response by inducing ubiquitin-proteosomal and lysosomal degradation of RelA/p65. Biochem. Pharmacol. 2018, 155, 141–149. [Google Scholar] [CrossRef]
- Curran, C.S.; Kopp, J.B. Aryl Hydrocarbon Receptor Mechanisms Affecting Chronic Kidney Disease. Front. Pharmacol. 2022, 13, 782199. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.F.A.; Sciullo, E.; Li, W.; Wong, P.; Lazennec, G.; Matsumura, F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol. Endocrinol. 2007, 21, 2941–2955. [Google Scholar] [CrossRef]
- Beischlag, T.V.; Wang, S.; Rose, D.W.; Torchia, J.; Reisz-Porszasz, S.; Muhammad, K.; Nelson, W.E.; Probst, M.R.; Rosenfeld, M.G.; Hankinson, O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 2002, 22, 4319–4333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinge, C.M.; Jernigan, S.C.; Risinger, K.E.; Lee, J.E.; Tyulmenkov, V.V.; Falkner, K.C.; Prough, R.A. Short heterodimer partner (SHP) orphan nuclear receptor inhibits the transcriptional activity of aryl hydrocarbon receptor (AHR)/AHR nuclear translocator (ARNT). Arch. Biochem. Biophys. 2001, 390, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hankinson, O. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J. Biol. Chem. 2002, 277, 11821–11827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Ge, K.; Roeder, R.G.; Hankinson, O. Role of Mediator in Transcriptional Activation by the Aryl Hydrocarbon Receptor. J. Biol. Chem. 2004, 279, 13593–13600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, J.; Gustafsson, J.-Å. Estrogen receptor and aryl hydrocarbon receptor signaling pathways. Nucl. Recept. Signal. 2006, 4, e016. [Google Scholar] [CrossRef] [PubMed]
- Marlowe, J.L.; Knudsen, E.S.; Schwemberger, S.; Puga, A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem. 2004, 279, 29013–29022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, L.C.; Okino, S.T.; Gonda, T.J.; Whitlock, J.P.J. Myb-binding protein 1a augments AhR-dependent gene expression. J. Biol. Chem. 2002, 277, 22515–22519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antenos, M.; Casper, R.F.; Brown, T.J. Interaction with Nedd8, a ubiquitin-like protein, enhances the transcriptional activity of the aryl hydrocarbon receptor. J. Biol. Chem. 2002, 277, 44028–44034. [Google Scholar] [CrossRef] [Green Version]
- Nacarino-Palma, A.; Rico-Leo, E.M.; Campisi, J.; Ramanathan, A.; González-Rico, F.J.; Rejano-Gordillo, C.M.; Ordiales-Talavero, A.; Merino, J.M.; Fernández-Salguero, P.M. Aryl hydrocarbon receptor blocks aging-induced senescence in the liver and fibroblast cells. Aging 2022, 14, 4281–4304. [Google Scholar] [CrossRef] [PubMed]
- Roman, A.C.; Carvajal-Gonzalez, J.M.; Rico-Leo, E.M.; Fernandez-Salguero, P.M. Dioxin receptor deficiency impairs angiogenesis by a mechanism involving VEGF-A depletion in the endothelium and transforming growth factor-beta overexpression in the stroma. J. Biol. Chem. 2009, 284, 25135–25148. [Google Scholar] [CrossRef] [PubMed]
- Tischkau, S.A.; Jaeger, C.D.; Krager, S.L. Circadian clock disruption in the mouse ovary in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Lett. 2011, 201, 116–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey-Barroso, J.; Alvarez-Barrientos, A.; Rico-Leo, E.; Contador-Troca, M.; Carvajal-Gonzalez, J.M.; Echarri, A.; Del Pozo, M.A.; Fernandez-Salguero, P.M. The Dioxin receptor modulates Caveolin-1 mobilization during directional migration: Role of cholesterol. Cell Commun. Signal. 2014, 12, 57. [Google Scholar] [CrossRef]
- Fernandez-Salguero, P.M.; Ward, J.M.; Sundberg, J.P.; Gonzalez, F.J. Lesions of Aryl-hydrocarbon Receptor–deficient Mice. Vet. Pathol. 1997, 34, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.E.; Karchner, S.I.; Shapiro, M.A.; Perera, S.A. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc. Natl. Acad. Sci. USA 1997, 94, 13743–13748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.Y.; Puga, A. Constitutive activation of the aromatic hydrocarbon receptor. Mol. Cell. Biol. 1998, 18, 525–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harper, P.A.; Riddick, D.S.; Okey, A.B. Regulating the regulator: Factors that control levels and activity of the aryl hydrocarbon receptor. Biochem. Pharmacol. 2006, 72, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Zablon, H.A.; Ko, C.-I.; Puga, A. Converging Roles of the Aryl Hydrocarbon Receptor in Early Embryonic Development, Maintenance of Stemness, and Tissue Repair. Toxicol. Sci. 2021, 182, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nacarino-Palma, A.; González-Rico, F.J.; Rejano-Gordillo, C.M.; Ordiales-Talavero, A.; Merino, J.M.; Fernández-Salguero, P.M. The aryl hydrocarbon receptor promotes differentiation during mouse preimplantational embryo development. Stem Cell Rep. 2021, 16, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Thackaberry, E.A.; Bedrick, E.J.; Goens, M.B.; Danielson, L.; Lund, A.K.; Gabaldon, D.; Smith, S.M.; Walker, M.K. Insulin regulation in AhR-null mice: Embryonic cardiac enlargement, neonatal macrosomia, and altered insulin regulation and response in pregnant and aging AhR-null females. Toxicol. Sci. 2003, 76, 407–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodale, B.C.; La Du, J.K.; Bisson, W.H.; Janszen, D.B.; Waters, K.M.; Tanguay, R.L. AHR2 mutant reveals functional diversity of aryl hydrocarbon receptors in zebrafish. PLoS ONE 2012, 7, e29346. [Google Scholar] [CrossRef] [PubMed]
- Kransler, K.M.; McGarrigle, B.P.; Olson, J.R. Comparative developmental toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in the hamster, rat and guinea pig. Toxicology 2007, 229, 214–225. [Google Scholar] [CrossRef]
- Takagi, T.N.; Matsui, K.A.; Yamashita, K.; Ohmori, H.; Yasuda, M. Pathogenesis of cleft palate in mouse embryos exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Teratog. Carcinog. Mutagen. 2000, 20, 73–86. [Google Scholar] [CrossRef]
- Nishimura, N.; Matsumura, F.; Vogel, C.F.A.; Nishimura, H.; Yonemoto, J.; Yoshioka, W.; Tohyama, C. Critical role of cyclooxygenase-2 activation in pathogenesis of hydronephrosis caused by lactational exposure of mice to dioxin. Toxicol. Appl. Pharmacol. 2008, 231, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.; Theobald, H.M.; Peterson, R.E. In utero and lactational exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin in the C57BL/6J mouse prostate: Lobe-specific effects on branching morphogenesis. Toxicol. Sci. 2002, 70, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Ricke, W.A.; Lee, C.W.; Clapper, T.R.; Schneider, A.J.; Moore, R.W.; Keil, K.P.; Abler, L.L.; Wynder, J.L.; López Alvarado, A.; Beaubrun, I.; et al. In Utero and Lactational TCDD Exposure Increases Susceptibility to Lower Urinary Tract Dysfunction in Adulthood. Toxicol. Sci. 2016, 150, 429–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vezina, C.M.; Lin, T.-M.; Peterson, R.E. AHR signaling in prostate growth, morphogenesis, and disease. Biochem. Pharmacol. 2009, 77, 566–576. [Google Scholar] [CrossRef] [Green Version]
- Bugiak, B.J.; Weber, L.P. Phenotypic anchoring of gene expression after developmental exposure to aryl hydrocarbon receptor ligands in zebrafish. Aquat. Toxicol. 2010, 99, 423–437. [Google Scholar] [CrossRef]
- Burns, F.R.; Peterson, R.E.; Heideman, W. Dioxin disrupts cranial cartilage and dermal bone development in zebrafish larvae. Aquat. Toxicol. 2015, 164, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Harstad, E.B.; Guite, C.A.; Thomae, T.L.; Bradfield, C.A. Liver deformation in Ahr-null mice: Evidence for aberrant hepatic perfusion in early development. Mol. Pharmacol. 2006, 69, 1534–1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Salguero, P.; Pineau, T.; Hilbert, D.M.; McPhail, T.; Lee, S.S.; Kimura, S.; Nebert, D.W.; Rudikoff, S.; Ward, J.M.; Gonzalez, F.J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 1995, 268, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Lahvis, G.P.; Lindell, S.L.; Thomas, R.S.; McCuskey, R.S.; Murphy, C.; Glover, E.; Bentz, M.; Southard, J.; Bradfield, C.A. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc. Natl. Acad. Sci. USA 2000, 97, 10442–10447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eti, N.A.; Flor, S.; Iqbal, K.; Scott, R.L.; Klenov, V.E.; Gibson-Corley, K.N.; Soares, M.J.; Ludewig, G.; Robertson, L.W. PCB126 induced toxic actions on liver energy metabolism is mediated by AhR in rats. Toxicology 2022, 466, 153054. [Google Scholar] [CrossRef] [PubMed]
- Jourova, L.; Anzenbacherova, E.; Dostal, Z.; Anzenbacher, P.; Briolotti, P.; Rigal, E.; Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Butyrate, a typical product of gut microbiome, affects function of the AhR gene, being a possible agent of crosstalk between gut microbiome, and hepatic drug metabolism. J. Nutr. Biochem. 2022, 107, 109042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Guo, M.; Feng, J.; Gu, Z.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Myristica fragrans Extract Regulates Gut Microbes and Metabolites to Attenuate Hepatic Inflammation and Lipid Metabolism Disorders via the AhR–FAS and NF-κB Signaling Pathways in Mice with Non-Alcoholic Fatty Liver Disease. Nutrients 2022, 14, 1699. [Google Scholar] [CrossRef] [PubMed]
- Sanmarco, L.M.; Chao, C.-C.; Wang, Y.-C.; Kenison, J.E.; Li, Z.; Rone, J.M.; Rejano-Gordillo, C.M.; Polonio, C.M.; Gutierrez-Vazquez, C.; Piester, G.; et al. Identification of environmental factors that promote intestinal inflammation. Nature 2022, 611, 801–809. [Google Scholar] [CrossRef]
- Contador-Troca, M.; Alvarez-Barrientos, A.; Merino, J.M.; Morales-Hernández, A.; Rodríguez, M.I.; Rey-Barroso, J.; Barrasa, E.; Cerezo-Guisado, M.I.; Catalina-Fernández, I.; Sáenz-Santamaría, J.; et al. Dioxin receptor regulates aldehyde dehydrogenase to block melanoma tumorigenesis and metastasis. Mol. Cancer 2015, 14, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esser, C.; Rannug, A. The Aryl Hydrocarbon Receptor in Barrier Organ Physiology, Immunology, and Toxicology. Pharmacol. Rev. 2015, 67, 259–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, C.-I.; Fan, Y.; de Gannes, M.; Wang, Q.; Xia, Y.; Puga, A. Repression of the Aryl Hydrocarbon Receptor Is Required to Maintain Mitotic Progression and Prevent Loss of Pluripotency of Embryonic Stem Cells. Stem Cells 2016, 34, 2825–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartor, M.A.; Schnekenburger, M.; Marlowe, J.L.; Reichard, J.F.; Wang, Y.; Fan, Y.; Ma, C.; Karyala, S.; Halbleib, D.; Liu, X.; et al. Genomewide Analysis of Aryl Hydrocarbon Receptor Binding Targets Reveals an Extensive Array of Gene Clusters that Control Morphogenetic and Developmental Programs. Environ. Health Perspect. 2009, 117, 1139–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rejano-Gordillo, C.M.; González-Rico, F.J.; Marín-Díaz, B.; Ordiales-Talavero, A.; Nacarino-Palma, A.; Román, Á.C.; Merino, J.M.; Fernández-Salguero, P.M. Liver regeneration after partial hepatectomy is improved in the absence of aryl hydrocarbon receptor. Sci. Rep. 2022, 12, 15446. [Google Scholar] [CrossRef] [PubMed]
- Vondráček, J.; Machala, M. Environmental Ligands of the Aryl Hydrocarbon Receptor and Their Effects in Models of Adult Liver Progenitor Cells. Stem Cells Int. 2016, 2016, 4326194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Boivin, G.P.; Knudsen, E.S.; Nebert, D.W.; Xia, Y.; Puga, A. The Aryl Hydrocarbon Receptor Functions as a Tumor Suppressor of Liver Carcinogenesis. Cancer Res. 2010, 70, 212–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Marín, N.; Barrasa, E.; Morales-Hernández, A.; Paniagua, B.; Blanco-Fernández, G.; Merino, J.M.; Fernández-Salguero, P.M. Dioxin Receptor Adjusts Liver Regeneration after Acute Toxic Injury and Protects Against Liver Carcinogenesis. Sci. Rep. 2017, 7, 10420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, D.P.; Li, H.; Mitchell, K.A.; Joshi, A.D.; Elferink, C.J. Ah Receptor–Mediated Suppression of Liver Regeneration through NC-XRE–Driven p21 Cip1 Expression. Mol. Pharmacol. 2014, 85, 533–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, K.A.; Lockhart, C.A.; Huang, G.; Elferink, C.J. Sustained Aryl Hydrocarbon Receptor Activity Attenuates Liver Regeneration. Mol. Pharmacol. 2006, 70, 163–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Tomaso Portaz, A.C.; Caimi, G.R.; Sánchez, M.; Chiappini, F.; Randi, A.S.; Kleiman de Pisarev, D.L.; Alvarez, L. Hexachlorobenzene induces cell proliferation, and aryl hydrocarbon receptor expression (AhR) in rat liver preneoplastic foci, and in the human hepatoma cell line HepG2. AhR is a mediator of ERK1/2 signaling, and cell cycle regulation in HCB-treated HepG2 cells. Toxicology 2015, 336, 36–47. [Google Scholar]
- Korecka, A.; Dona, A.; Lahiri, S.; Tett, A.J.; Al-Asmakh, M.; Braniste, V.; D’Arienzo, R.; Abbaspour, A.; Reichardt, N.; Fujii-Kuriyama, Y.; et al. Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism. Npj Biofilms Microbiomes 2016, 2, 16014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamas, B.; Natividad, J.M.; Sokol, H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 2018, 11, 1024–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goettel, J.A.; Gandhi, R.; Kenison, J.E.; Yeste, A.; Murugaiyan, G.; Sambanthamoorthy, S.; Griffith, A.E.; Patel, B.; Shouval, D.S.; Weiner, H.L.; et al. AHR Activation Is Protective against Colitis Driven by T Cells in Humanized Mice. Cell Rep. 2016, 17, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- Pernomian, L.; Duarte-Silva, M.; de Barros Cardoso, C.R. The Aryl Hydrocarbon Receptor (AHR) as a Potential Target for the Control of Intestinal Inflammation: Insights from an Immune and Bacteria Sensor Receptor. Clin. Rev. Allergy Immunol. 2020, 59, 382–390. [Google Scholar] [CrossRef]
- Schiering, C.; Wincent, E.; Metidji, A.; Iseppon, A.; Li, Y.; Potocnik, A.J.; Omenetti, S.; Henderson, C.J.; Wolf, C.R.; Nebert, D.W.; et al. Feedback control of AHR signalling regulates intestinal immunity. Nature 2017, 542, 242–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rannug, A. How the AHR Became Important in Intestinal Homeostasis-A Diurnal FICZ/AHR/CYP1A1 Feedback Controls Both Immunity and Immunopathology. Int. J. Mol. Sci. 2020, 21, 5681. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, P.J.; Nagarkatti, M.; Nagarkatti, P.S. Regulation of Intestinal Stem Cell Stemness by the Aryl Hydrocarbon Receptor and Its Ligands. Front. Immunol. 2021, 12, 638725. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Davidson, L.A.; Hensel, M.; Yoon, G.; Landrock, K.; Allred, C.; Jayaraman, A.; Ivanov, I.; Safe, S.H.; Chapkin, R.S. Loss of aryl hydrocarbon receptor promotes colon tumorigenesis in ApcS580/+; KrasG12D/+ mice. Mol. Cancer Res. 2021, 19, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011, 141, 237–248.e1. [Google Scholar] [CrossRef] [PubMed]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abron, J.D.; Singh, N.P.; Mishra, M.K.; Price, R.L.; Nagarkatti, M.; Nagarkatti, P.S.; Singh, U.P. An endogenous aryl hydrocarbon receptor ligand, ITE, induces regulatory T cells and ameliorates experimental colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G220–G230. [Google Scholar] [CrossRef]
- Gutiérrez-Vázquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauben, E.; Gregori, S.; Draghici, E.; Migliavacca, B.; Olivieri, S.; Woisetschläger, M.; Roncarolo, M.G. Activation of the aryl hydrocarbon receptor promotes allograft-specific tolerance through direct and dendritic cell–mediated effects on regulatory T cells. Blood 2008, 112, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- King, N.J.C.; Thomas, S.R. Molecules in focus: Indoleamine 2,3-dioxygenase. Int. J. Biochem. Cell Biol. 2007, 39, 2167–2172. [Google Scholar] [CrossRef] [PubMed]
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Johnson, B.P.; Burlingham, W.J.; Bradfield, C.A. An Interaction between Kynurenine and the Aryl Hydrocarbon Receptor Can Generate Regulatory T Cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.T.; Kimura, A.; Nakahama, T.; Chinen, I.; Masuda, K.; Nohara, K.; Fujii-Kuriyama, Y.; Kishimoto, T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc. Natl. Acad. Sci. USA 2010, 107, 19961–19966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haase, S.; Haghikia, A.; Wilck, N.; Müller, D.N.; Linker, R.A. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology 2018, 154, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintana, F.J.; Basso, A.S.; Iglesias, A.H.; Korn, T.; Farez, M.F.; Bettelli, E.; Caccamo, M.; Oukka, M.; Weiner, H.L. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008, 453, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 2006, 6, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Kerkvliet, N.I.; Steppan, L.B.; Vorachek, W.; Oda, S.; Farrer, D.; Wong, C.P.; Pham, D.; Mourich, D.V. Activation of aryl hydrocarbon receptor by TCDD prevents diabetes in NOD mice and increases Foxp3+ T cells in pancreatic lymph nodes. Immunotherapy 2009, 1, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.P.; Singh, U.P.; Singh, B.; Price, R.L.; Nagarkatti, M.; Nagarkatti, P.S. Activation of Aryl Hydrocarbon Receptor (AhR) Leads to Reciprocal Epigenetic Regulation of FoxP3 and IL-17 Expression and Amelioration of Experimental Colitis. PLoS ONE 2011, 6, e23522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, P.; Hu, G.; Kang, H.; Yao, H.; Kou, W.; Liu, H.; Zhang, C.; Hong, S. An aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress the Th17 response in allergic rhinitis patients. Lab. Investig. 2014, 94, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, R.; Kumar, D.; Burns, E.J.; Nadeau, M.; Dake, B.; Laroni, A.; Kozoriz, D.; Weiner, H.L.; Quintana, F.J. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell–like and Foxp3+ regulatory T cells. Nat. Immunol. 2010, 11, 846–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascanfroni, I.D.; Takenaka, M.C.; Yeste, A.; Patel, B.; Wu, Y.; Kenison, J.E.; Siddiqui, S.; Basso, A.S.; Otterbein, L.E.; Pardoll, D.M.; et al. Metabolic control of type 1 regulatory (Tr1) cell differentiation by AHR and HIF1-α. Nat. Med. 2015, 21, 638–646. [Google Scholar] [CrossRef] [PubMed]
- DiNatale, B.C.; Schroeder, J.C.; Perdew, G.H. Ah receptor antagonism inhibits constitutive and cytokine inducible IL6 production in head and neck tumor cell lines. Mol. Carcinog. 2011, 50, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollingshead, B.D.; Beischlag, T.V.; Dinatale, B.C.; Ramadoss, P.; Perdew, G.H. Inflammatory signaling and aryl hydrocarbon receptor mediate synergistic induction of interleukin 6 in MCF-7 cells. Cancer Res. 2008, 68, 3609–3617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podechard, N.; Lecureur, V.; Le Ferrec, E.; Guenon, I.; Sparfel, L.; Gilot, D.; Gordon, J.R.; Lagente, V.; Fardel, O. Interleukin-8 induction by the environmental contaminant benzo(a)pyrene is aryl hydrocarbon receptor-dependent and leads to lung inflammation. Toxicol. Lett. 2008, 177, 130–137. [Google Scholar] [CrossRef]
- Sciullo, E.M.; Dong, B.; Vogel, C.F.A.; Matsumura, F. Characterization of the pattern of the nongenomic signaling pathway through which TCDD-induces early inflammatory responses in U937 human macrophages. Chemosphere 2009, 74, 1531–1537. [Google Scholar] [CrossRef] [Green Version]
- Keshavarzi, M.; Moradbeygi, F.; Mobini, K.; Ghaffarian Bahraman, A.; Mohammadi, P.; Ghaedi, A.; Mohammadi-Bardbori, A. The interplay of aryl hydrocarbon receptor/WNT/CTNNB1/Notch signaling pathways regulate amyloid beta precursor mRNA/protein expression and effected the learning and memory of mice. Toxicol. Res. 2022, 11, 147–161. [Google Scholar] [CrossRef]
- Kimura, E.; Tohyama, C. Embryonic and Postnatal Expression of Aryl Hydrocarbon Receptor mRNA in Mouse Brain. Front. Neuroanat. 2017, 11, 4. [Google Scholar] [CrossRef] [Green Version]
- Filbrandt, C.R.; Wu, Z.; Zlokovic, B.; Opanashuk, L.; Gasiewicz, T.A. Presence and Functional Activity of the Aryl Hydrocarbon Receptor in Isolated Murine Cerebral Vascular Endothelial Cells and Astrocytes. NeuroToxicology 2004, 25, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Kubota, A.; Stegeman, J.J.; Woodin, B.R.; Iwanaga, T.; Harano, R.; Peterson, R.E.; Hiraga, T.; Teraoka, H. Role of zebrafish cytochrome P450 CYP1C genes in the reduced mesencephalic vein blood flow caused by activation of AHR2. Toxicol. Appl. Pharmacol. 2011, 253, 244–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Chen, G.; Zhao, J.; Nie, X.; Wan, C.; Liu, J.; Duan, Z.; Xu, G. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces microglial nitric oxide production and subsequent rat primary cortical neuron apoptosis through p38/JNK MAPK pathway. Toxicology 2013, 312, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Rzemieniec, J.; Litwa, E.; Wnuk, A.; Lason, W.; Krzeptowski, W.; Kajta, M. Selective Aryl Hydrocarbon Receptor Modulator 3,3′-Diindolylmethane Impairs AhR and ARNT Signaling and Protects Mouse Neuronal Cells Against Hypoxia. Mol. Neurobiol. 2016, 53, 5591–5606. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, F.J.; Fernández-Salguero, P.M.; Merino, J.M. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces apoptosis in neural growth factor (NGF)-differentiated pheochromocytoma PC12 cells. NeuroToxicology 2010, 31, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, F.J.; Fernández-Salguero, P.M.; Merino, J.M. Aryl hydrocarbon receptor-dependent induction of apoptosis by 2,3,7,8-tetrachlorodibenzo-p-dioxin in cerebellar granule cells from mouse. J. Neurochem. 2011, 118, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Juricek, L.; Coumoul, X. The Aryl Hydrocarbon Receptor and the Nervous System. Int. J. Mol. Sci. 2018, 19, 2504. [Google Scholar] [CrossRef] [Green Version]
- Rothhammer, V.; Borucki, D.M.; Tjon, E.C.; Takenaka, M.C.; Chao, C.-C.; Ardura-Fabregat, A.; de Lima, K.A.; Gutiérrez-Vázquez, C.; Hewson, P.; Staszewski, O.; et al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018, 557, 724–728. [Google Scholar] [CrossRef]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.-C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef]
- Yeste, A.; Nadeau, M.; Burns, E.J.; Weiner, H.L.; Quintana, F.J. Nanoparticle-mediated codelivery of myelin antigen and a tolerogenic small molecule suppresses experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2012, 109, 11270–11275. [Google Scholar] [CrossRef] [Green Version]
- Zamali, I.; Rekik, R.; Belhadj Hmida, N.; Ben Hmid, A.; Kammoun, O.; Barbouche, M.-R.; Ben Ahmed, M. An endogenous aryl hydrocarbon receptor ligand enhances de novo generation of regulatory T cells in humans. J. Leukoc. Biol. 2019, 105, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Dopkins, N.; Becker, W.; Miranda, K.; Walla, M.; Nagarkatti, P.; Nagarkatti, M. Tryptamine Attenuates Experimental Multiple Sclerosis Through Activation of Aryl Hydrocarbon Receptor. Front. Pharmacol. 2021, 11, 619265. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Borucki, D.M.; Kenison, J.E.; Hewson, P.; Wang, Z.; Bakshi, R.; Sherr, D.H.; Quintana, F.J. Detection of aryl hydrocarbon receptor agonists in human samples. Sci. Rep. 2018, 8, 4970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothhammer, V.; Borucki, D.M.; Garcia Sanchez, M.I.; Mazzola, M.A.; Hemond, C.C.; Regev, K.; Paul, A.; Kivisäkk, P.; Bakshi, R.; Izquierdo, G.; et al. Dynamic regulation of serum aryl hydrocarbon receptor agonists in MS. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e359. [Google Scholar] [CrossRef] [PubMed]
- Tsaktanis, T.; Beyer, T.; Nirschl, L.; Linnerbauer, M.; Grummel, V.; Bussas, M.; Tjon, E.; Mühlau, M.; Korn, T.; Hemmer, B.; et al. Aryl Hydrocarbon Receptor Plasma Agonist Activity Correlates with Disease Activity in Progressive MS. Neurol. Neuroimmunol. Neuroinflamm. 2020, 8, e933. [Google Scholar] [CrossRef]
- Kaye, J.; Piryatinsky, V.; Birnberg, T.; Hingaly, T.; Raymond, E.; Kashi, R.; Amit-Romach, E.; Caballero, I.S.; Towfic, F.; Ator, M.A.; et al. Laquinimod arrests experimental autoimmune encephalomyelitis by activating the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2016, 113, E6145–E6152. [Google Scholar] [CrossRef] [Green Version]
- Rothhammer, V.; Kenison, J.E.; Li, Z.; Tjon, E.; Takenaka, M.C.; Chao, C.-C.; de Lima, K.A.; Borucki, D.M.; Kaye, J.; Quintana, F.J. Aryl Hydrocarbon Receptor Activation in Astrocytes by Laquinimod Ameliorates Autoimmune Inflammation in the CNS. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e946. Available online: https://nn.neurology.org/content/8/2/e946 (accessed on 11 February 2022). [CrossRef]
- Rannug, A.; Rannug, U.; Rosenkranz, H.S.; Winqvist, L.; Westerholm, R.; Agurell, E.; Grafström, A.K. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 1987, 262, 15422–15427. [Google Scholar] [CrossRef]
- Tsuji, G.; Hashimoto-Hachiya, A.; Kiyomatsu-Oda, M.; Takemura, M.; Ohno, F.; Ito, T.; Morino-Koga, S.; Mitoma, C.; Nakahara, T.; Uchi, H.; et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017, 8, e2931. [Google Scholar] [CrossRef] [Green Version]
- Furue, M.; Tsuji, G.; Mitoma, C.; Nakahara, T.; Chiba, T.; Morino-Koga, S.; Uchi, H. Gene regulation of filaggrin and other skin barrier proteins via aryl hydrocarbon receptor. J. Dermatol. Sci. 2015, 80, 83–88. [Google Scholar] [CrossRef]
- Sutter, C.H.; Bodreddigari, S.; Campion, C.; Wible, R.S.; Sutter, T.R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin Increases the Expression of Genes in the Human Epidermal Differentiation Complex and Accelerates Epidermal Barrier Formation. Toxicol. Sci. 2011, 124, 128–137. [Google Scholar] [CrossRef]
- Rudyak, S.G.; Usakin, L.A.; Tverye, E.A.; Orekhov, A.S.; Belushkina, N.N.; Paus, R.; Paltsev, M.A.; Panteleyev, A.A. Retinoic acid co-treatment aggravates severity of dioxin-induced skin lesions in hairless mice via induction of inflammatory response. Biochem. Biophys. Res. Commun. 2018, 506, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.H.; Jayawickreme, C.; Rickard, D.J.; Nicodeme, E.; Bui, T.; Simmons, C.; Coquery, C.M.; Neil, J.; Pryor, W.M.; Mayhew, D.; et al. Tapinarof Is a Natural AhR Agonist that Resolves Skin Inflammation in Mice and Humans. J. Investig. Dermatol. 2017, 137, 2110–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Bogaard, E.H.; Podolsky, M.A.; Smits, J.P.; Cui, X.; John, C.; Gowda, K.; Desai, D.; Amin, S.G.; Schalkwijk, J.; Perdew, G.H.; et al. Genetic and Pharmacological Analysis Identifies a Physiological Role for the AHR in Epidermal Differentiation. J. Investig. Dermatol. 2015, 135, 1320–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rico-Leo, E.M.; Lorenzo-Martín, L.F.; Román, Á.C.; Bustelo, X.R.; Merino, J.M.; Fernández-Salguero, P.M. Aryl hydrocarbon receptor controls skin homeostasis, regeneration, and hair follicle cycling by adjusting epidermal stem cell function. Stem Cells 2021, 39, 1733–1750. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.; Weighardt, H.; Deenen, R.; Köhrer, K.; Clausen, B.; Zahner, S.; Boukamp, P.; Bloch, W.; Krutmann, J.; Esser, C. Aryl Hydrocarbon Receptor in Keratinocytes Is Essential for Murine Skin Barrier Integrity. J. Investig. Dermatol. 2016, 136, 2260–2269. [Google Scholar] [CrossRef] [Green Version]
- Uberoi, A.; Bartow-McKenney, C.; Zheng, Q.; Flowers, L.; Campbell, A.; Knight, S.A.B.; Chan, N.; Wei, M.; Lovins, V.; Bugayev, J.; et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 2021, 29, 1235–1248.e8. [Google Scholar] [CrossRef] [PubMed]
- Pollet, M.; Shaik, S.; Mescher, M.; Frauenstein, K.; Tigges, J.; Braun, S.A.; Sondenheimer, K.; Kaveh, M.; Bruhs, A.; Meller, S.; et al. The AHR represses nucleotide excision repair and apoptosis and contributes to UV-induced skin carcinogenesis. Cell Death Differ. 2018, 25, 1823–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Georges, F.; Abbas, I.; Billet, S.; Verdin, A.; Gosset, P.; Mulliez, P.; Shirali, P.; Garçon, G. Gene expression induction of volatile organic compound and/or polycyclic aromatic hydrocarbon-metabolizing enzymes in isolated human alveolar macrophages in response to airborne particulate matter (PM2.5). Toxicology 2008, 244, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Shivanna, B.; Maity, S.; Zhang, S.; Patel, A.; Jiang, W.; Wang, L.; Welty, S.E.; Belmont, J.; Coarfa, C.; Moorthy, B. Gene Expression Profiling Identifies Cell Proliferation and Inflammation as the Predominant Pathways Regulated by Aryl Hydrocarbon Receptor in Primary Human Fetal Lung Cells Exposed to Hyperoxia. Toxicol. Sci. 2016, 152, 155–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Patel, A.; Chu, C.; Jiang, W.; Wang, L.; Welty, S.E.; Moorthy, B.; Shivanna, B. Aryl hydrocarbon receptor is necessary to protect fetal human pulmonary microvascular endothelial cells against hyperoxic injury: Mechanistic roles of antioxidant enzymes and RelB. Toxicol. Appl. Pharmacol. 2015, 286, 92–101. [Google Scholar] [CrossRef]
- Luebke, R.W.; Copeland, C.B.; Daniels, M.; Lambert, A.L.; Gilmour, M.I. Suppression of Allergic Immune Responses to House Dust Mite (HDM) in Rats Exposed to 2,3,7,8-TCDD. Toxicol. Sci. 2001, 62, 71–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, A.K.; Peterson, S.L.; Timmins, G.S.; Walker, M.K. Endothelin-1–Mediated Increase in Reactive Oxygen Species and NADPH Oxidase Activity in Hearts of Aryl Hydrocarbon Receptor (AhR) Null Mice. Toxicol. Sci. 2005, 88, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, T.H.; Maggirwar, S.B.; Baglole, C.J.; Lakatos, H.F.; Gasiewicz, T.A.; Phipps, R.P.; Sime, P.J. Aryl Hydrocarbon Receptor-Deficient Mice Develop Heightened Inflammatory Responses to Cigarette Smoke and Endotoxin Associated with Rapid Loss of the Nuclear Factor-κB Component RelB. Am. J. Pathol. 2007, 170, 855–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, P.S.; Vogel, C.F.; Kokosinski, K.; Matsumura, F. Arylhydrocarbon Receptor Activation in NCI-H441 Cells and C57BL/6 Mice. Am. J. Respir. Cell Mol. Biol. 2010, 42, 210–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivanna, B.; Jiang, W.; Wang, L.; Couroucli, X.I.; Moorthy, B. Omeprazole Attenuates Hyperoxic Lung Injury in Mice via Aryl Hydrocarbon Receptor Activation and Is Associated with Increased Expression of Cytochrome P4501A Enzymes. J. Pharmacol. Exp. Ther. 2011, 339, 106–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheridan, J.A.; Zago, M.; Nair, P.; Li, P.Z.; Bourbeau, J.; Tan, W.C.; Hamid, Q.; Eidelman, D.H.; Benedetti, A.L.; Baglole, C.J. Decreased expression of the NF-κB family member RelB in lung fibroblasts from Smokers with and without COPD potentiates cigarette smoke-induced COX-2 expression. Respir. Res. 2015, 16, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorley, A.J.; Tetley, T.D. Pulmonary epithelium, cigarette smoke, and chronic obstructive pulmonary disease. Int. J. Chronic Obstr. Pulm. Dis. 2007, 2, 409–428. [Google Scholar]
- Tsai, M.-J.; Hsu, Y.-L.; Wang, T.-N.; Wu, L.-Y.; Lien, C.-T.; Hung, C.-H.; Kuo, P.-L.; Huang, M.-S. Aryl hydrocarbon receptor (AhR) agonists increase airway epithelial matrix metalloproteinase activity. J. Mol. Med. 2014, 92, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Morales-Hernández, A.; Nacarino-Palma, A.; Moreno-Marín, N.; Barrasa, E.; Paniagua-Quiñones, B.; Catalina-Fernández, I.; Alvarez-Barrientos, A.; Bustelo, X.R.; Merino, J.M.; Fernández-Salguero, P.M. Lung regeneration after toxic injury is improved in absence of dioxin receptor. Stem Cell Res. 2017, 25, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Zhang, X.; Zhang, Y.; Wu, B.; Fang, L.; Wang, N.; Yi, H.; Chang, N.; Chen, L.; Zhang, J. Aryl hydrocarbon receptor mediates Jak2/STAT3 signaling for non-small cell lung cancer stem cell maintenance. Exp. Cell Res. 2020, 396, 112288. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Wang, M.; Qian, B.; Ouyang, L.; Shi, Y.; Liu, N.; Chen, L.; Xiao, D.; Wang, X.; Cao, Y.; et al. Aryl hydrocarbon receptor activated by benzo (a) pyrene promotes SMARCA6 expression in NSCLC. Am. J. Cancer Res. 2018, 8, 1214–1227. [Google Scholar] [PubMed]
- Lin, P.; Chang, H.; Ho, W.L.; Wu, M.-H.; Su, J.-M. Association of aryl hydrocarbon receptor and cytochrome P4501B1 expressions in human non-small cell lung cancers. Lung Cancer 2003, 42, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Chang, H.; Tsai, W.-T.; Wu, M.-H.; Liao, Y.-S.; Chen, J.-T.; Su, J.-M. Overexpression of Aryl Hydrocarbon Receptor in Human Lung Carcinomas. Toxicol. Pathol. 2003, 31, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Nothdurft, S.; Thumser-Henner, C.; Breitenbücher, F.; Okimoto, R.A.; Dorsch, M.; Opitz, C.A.; Sadik, A.; Esser, C.; Hölzel, M.; Asthana, S.; et al. Functional screening identifies aryl hydrocarbon receptor as suppressor of lung cancer metastasis. Oncogenesis 2020, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-C.; Yang, W.-H.; Li, C.-H.; Cheng, Y.-W.; Tsai, C.-H.; Kang, J.-J. Ligand independent aryl hydrocarbon receptor inhibits lung cancer cell invasion by degradation of Smad4. Cancer Lett. 2016, 376, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, F.; Li, Z.; Remes-Lenicov, F.; Dávola, M.E.; Elizalde, M.; Paletta, A.; Ashkar, A.A.; Mossman, K.L.; Dugour, A.V.; Figueroa, J.M.; et al. AHR signaling is induced by infection with coronaviruses. Nat. Commun. 2021, 12, 5148. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, F.; Quintana, F.J. SARS-CoV-2-induced lung pathology: AHR as a candidate therapeutic target. Cell Res. 2021, 31, 1–2. [Google Scholar] [CrossRef] [PubMed]
| Factor | Functional Consequence | Reference |
|---|---|---|
| SRC-1, NCoA2, p300/CBP, p/CIP | Coactivators with HAT activity that interact with AHR and/or ARNT to facilitate transcriptional activation | [91] |
| SHP | Inhibits transcriptional activity of the AHR/ARNT complex | [92] |
| Brg-1 | Histone-modifying factor dependent on ATPase activity and activator of transcription mediated by AHR/ARNT | [93] |
| Med220, CDK8 | Subunits of the mediator complex involved in AHR/ARNT transcriptional activation | [94] |
| ERα | Functional interactor with AHR in gene regulation | [95] |
| RB | Direct interaction between Rb and AHR is required for maximal induction of Cyp1a1, suggesting a role of coactivator for RB | [96] |
| Mybbp1a | Associates with AHR and favors transactivation | [97] |
| Nedd8 | Interacts with AHR increasing its nuclear accumulation and transcriptional activity | [98] |
| p16, p21 | AHR transcriptionally regulates the expression of senescence-related genes | [99] |
| VEGF, HGF | AHR modulation of angiogenesis through a mechanism that requires VEGF activation in the endothelium | [100] |
| FGF | Increased fibroblast growth factor (FGF) levels with AHR overexpression. | [99] |
| Per2 y Bmal1 | Circadian rhythms-mediated interaction with AHR. Exposure to TCDD alters their expression pattern. | [101] |
| Cav-1 | AHR modulation of Caveolin-1 in cell migration | [102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rejano-Gordillo, C.M.; Marín-Díaz, B.; Ordiales-Talavero, A.; Merino, J.M.; González-Rico, F.J.; Fernández-Salguero, P.M. From Nucleus to Organs: Insights of Aryl Hydrocarbon Receptor Molecular Mechanisms. Int. J. Mol. Sci. 2022, 23, 14919. https://doi.org/10.3390/ijms232314919
Rejano-Gordillo CM, Marín-Díaz B, Ordiales-Talavero A, Merino JM, González-Rico FJ, Fernández-Salguero PM. From Nucleus to Organs: Insights of Aryl Hydrocarbon Receptor Molecular Mechanisms. International Journal of Molecular Sciences. 2022; 23(23):14919. https://doi.org/10.3390/ijms232314919
Chicago/Turabian StyleRejano-Gordillo, Claudia M., Beatriz Marín-Díaz, Ana Ordiales-Talavero, Jaime M. Merino, Francisco J. González-Rico, and Pedro M. Fernández-Salguero. 2022. "From Nucleus to Organs: Insights of Aryl Hydrocarbon Receptor Molecular Mechanisms" International Journal of Molecular Sciences 23, no. 23: 14919. https://doi.org/10.3390/ijms232314919
APA StyleRejano-Gordillo, C. M., Marín-Díaz, B., Ordiales-Talavero, A., Merino, J. M., González-Rico, F. J., & Fernández-Salguero, P. M. (2022). From Nucleus to Organs: Insights of Aryl Hydrocarbon Receptor Molecular Mechanisms. International Journal of Molecular Sciences, 23(23), 14919. https://doi.org/10.3390/ijms232314919








