Treatment with EV-miRNAs Alleviates Obesity-Associated Metabolic Dysfunction in Mice
Abstract
1. Introduction
2. Results
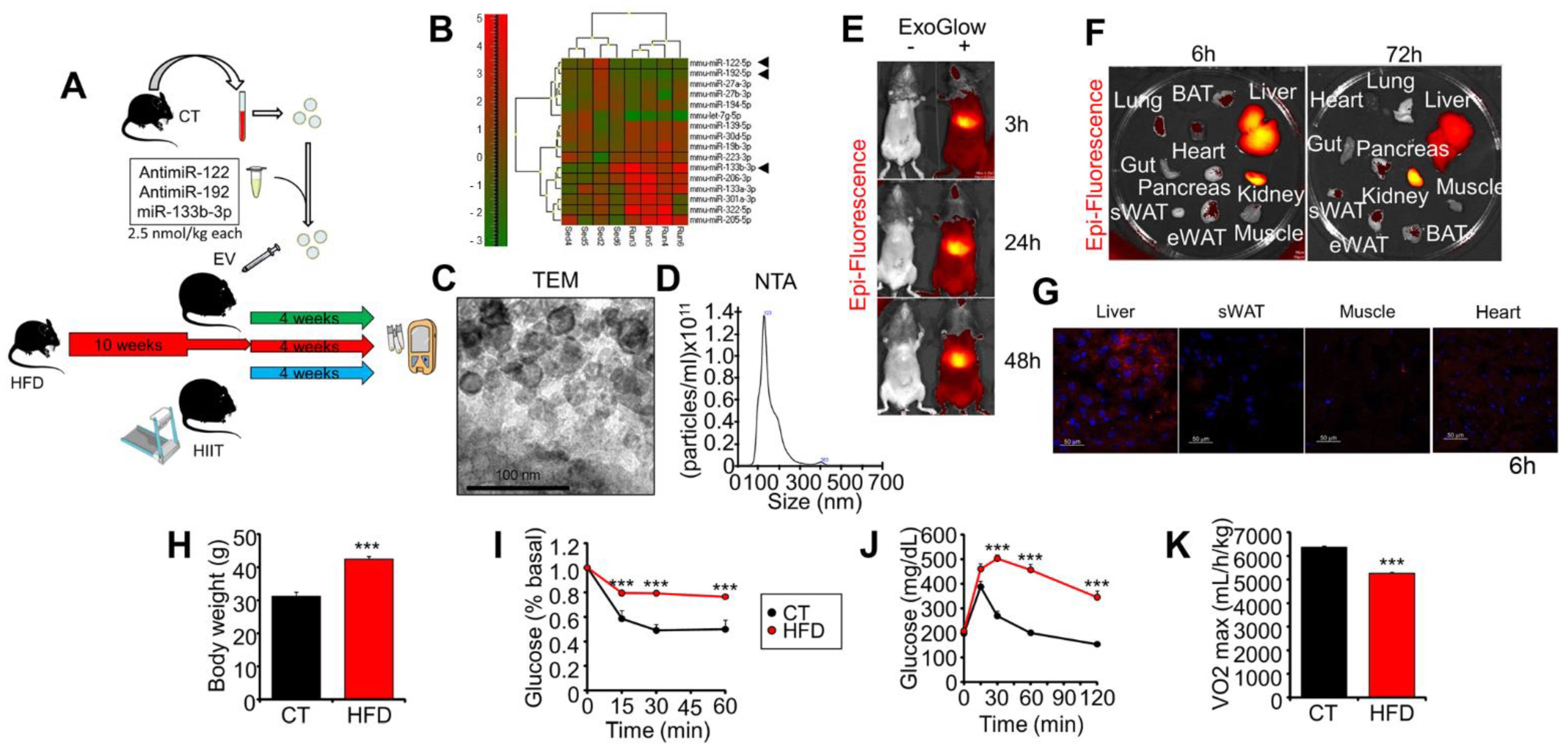
2.1. Experimental Design, Exogenous EVs’ Biodistribution, and Characterization of Obese Mice
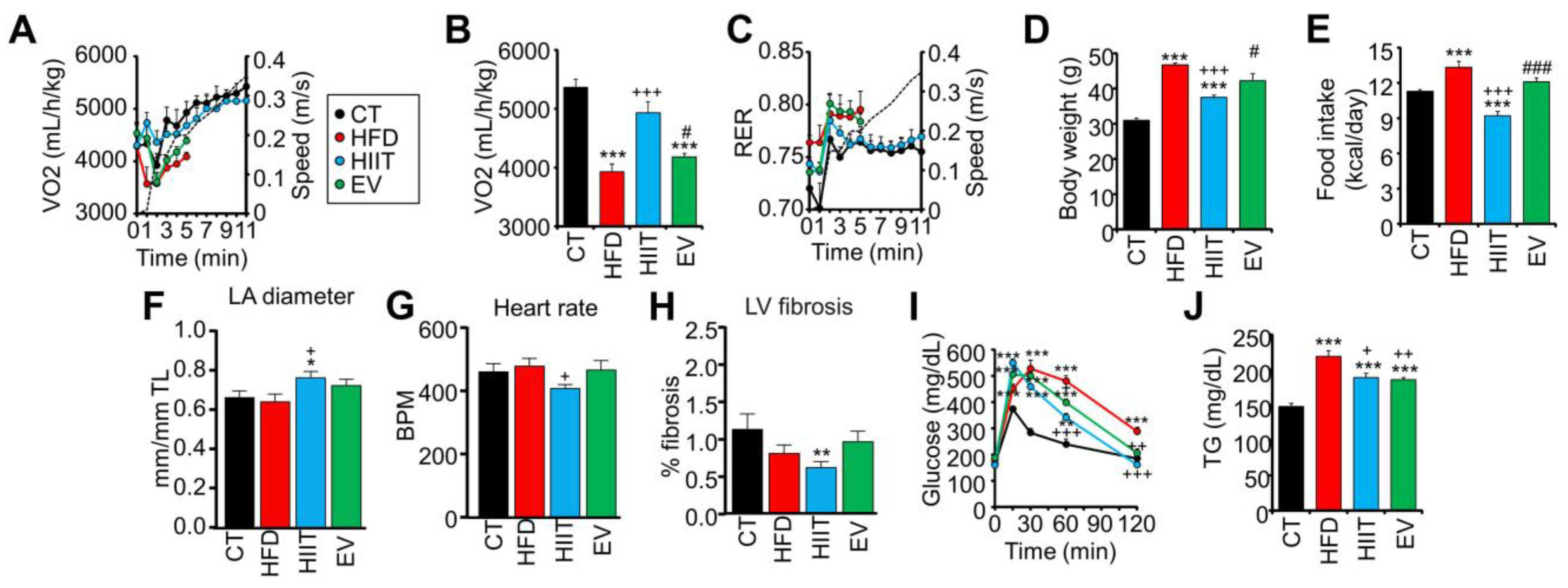
2.2. HIIT and EV-miRNAs Improve the Metabolic Profile of HFD Mice, but Only HIIT Enhances CRF and Promotes Cardiac Remodeling
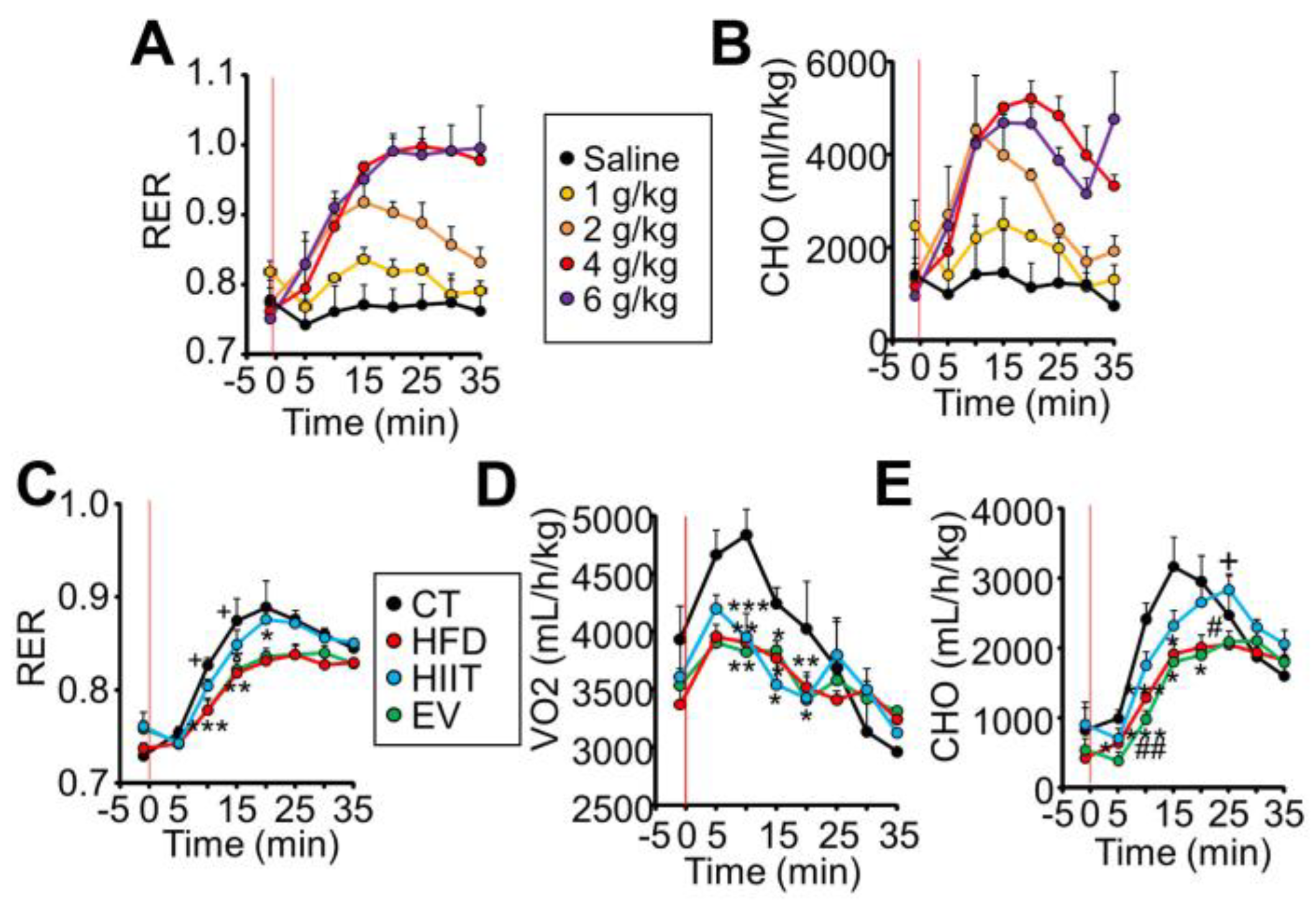
2.3. HIIT and EV-miRNAs Improve Glucose Tolerance through Different Mechanisms
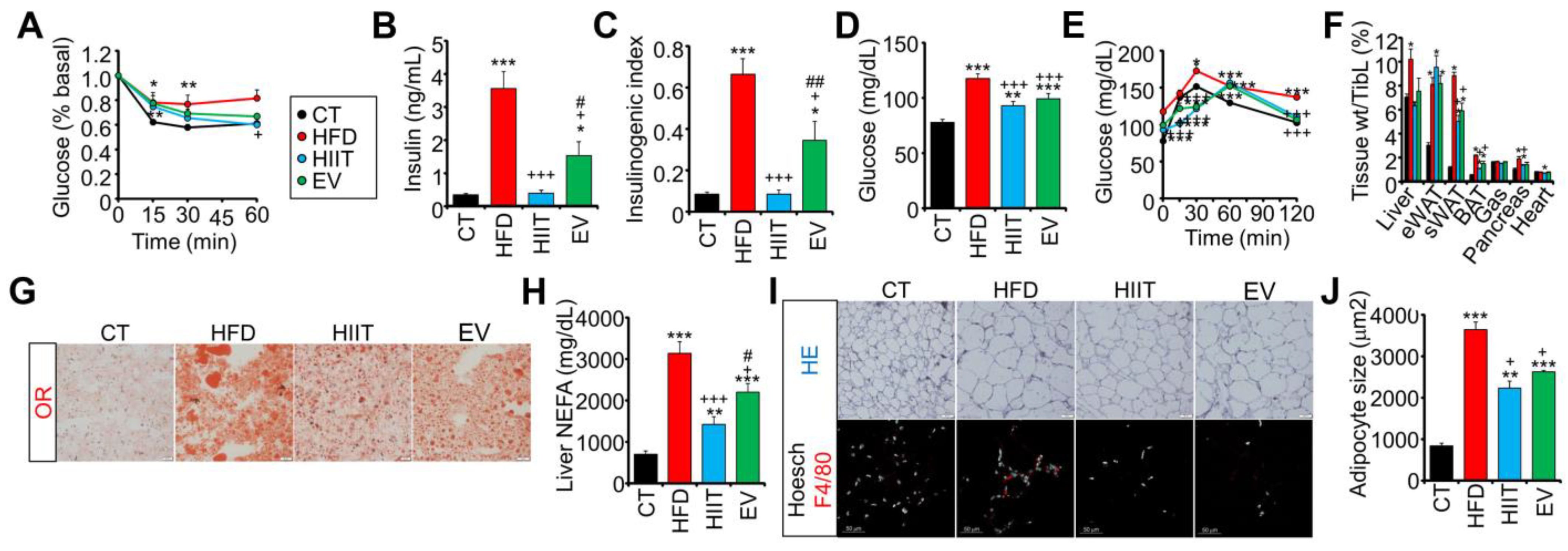
2.4. Treatment with EV-miRNAs Improves Hepatic Insulin Sensitivity and Steatosis
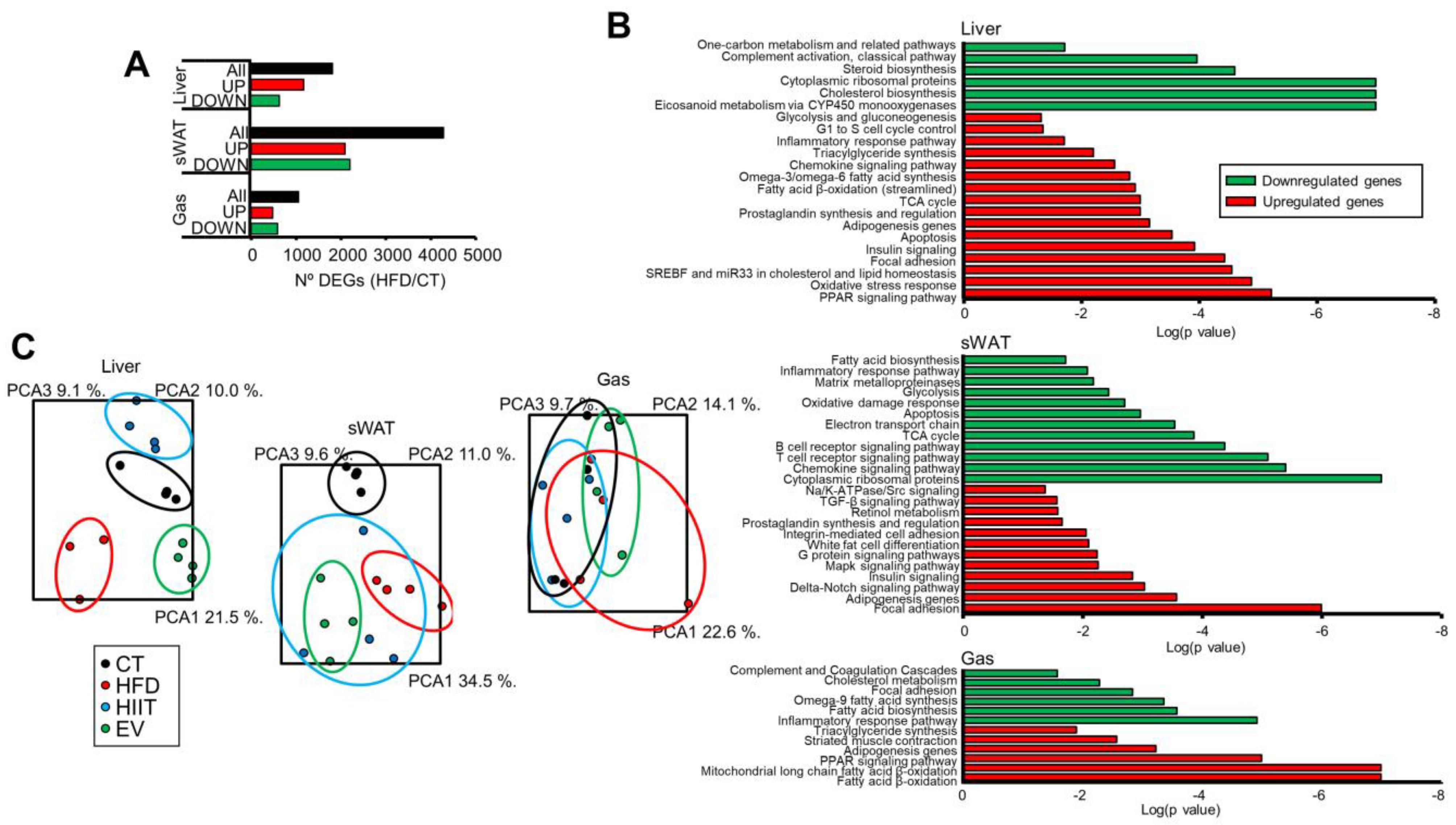
2.5. HIIT and EV-miRNAs Affect the Hepatic, sWAT, and Muscle Expression Profiles of Obese Mice
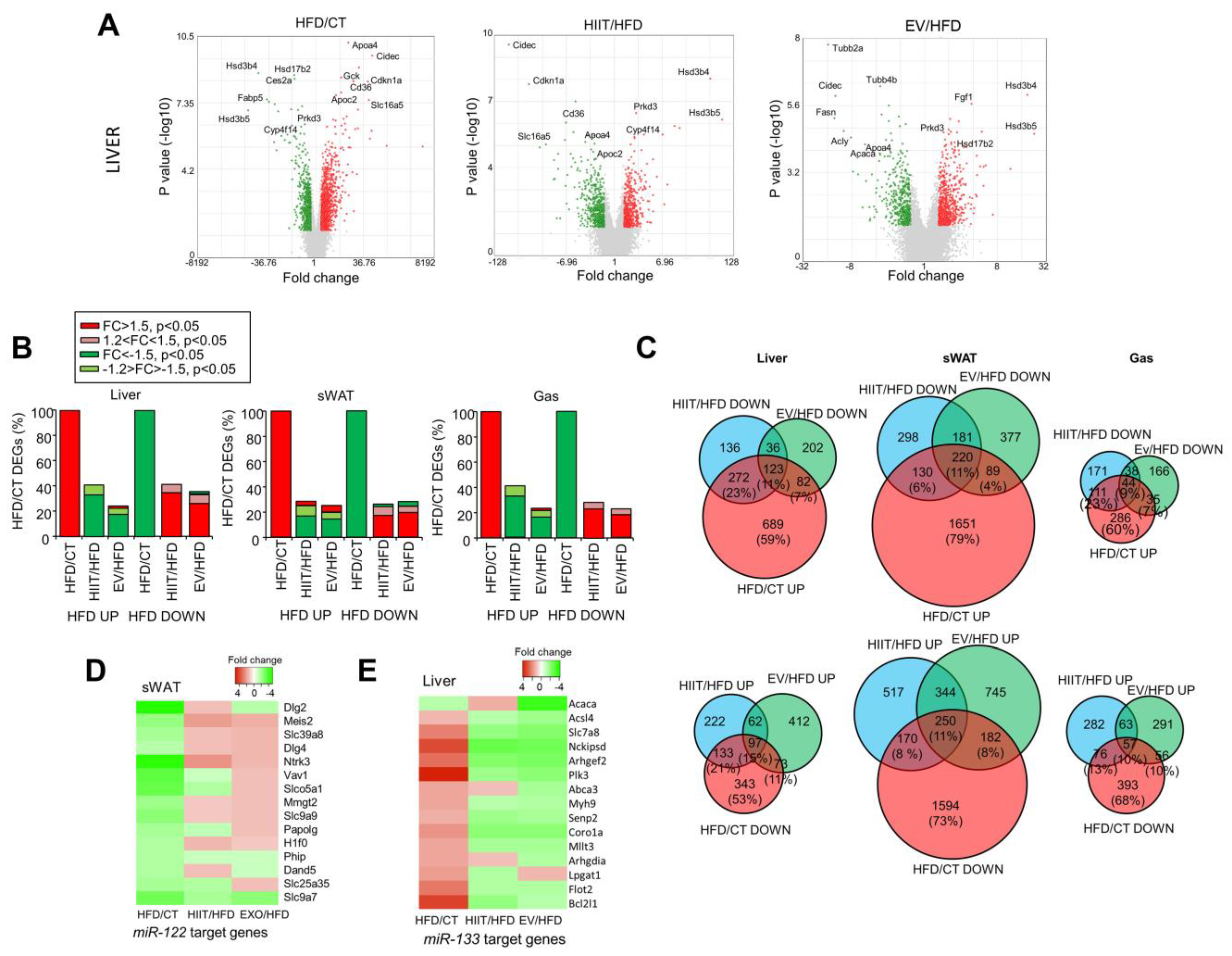
2.6. HIIT and EV-miRNAs Partially Reverse the Alterations Induced by Diet
2.7. EV-miRNAs Regulate Hepatic Lipid Metabolism to Decrease Steatosis
3. Discussion
4. Materials and Methods
4.1. Experimental Models
4.2. Indirect Calorimetry
4.3. Echocardiography and Electrocardiogram
4.4. EV Isolation, Characterization, Labeling, and Transfection
4.5. RNA Isolation and Gene Expression Analysis
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González-Muniesa, P.; Mártinez-González, M.A.; Hu, F.B.; Després, J.P.; Matsuzawa, Y.; Loos, R.J.F.; Moreno, L.A.; Bray, G.A.; Martinez, J.A. Obesity. Nat. Rev. Dis. Prim. 2017, 3, 17034. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Wang, Y.; Hou, H.; Feng, H.; Yang, H. An updated meta-analysis on the relationship between obesity and COVID-19 mortality. Metab. Clin. Exp. 2021, 122, 154820. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Lyon, C.J.; Bergin, S.; Caligiuri, M.A.; Hsueh, W.A. Obesity, Inflammation, and Cancer. Annu. Rev. Pathol. 2016, 11, 421–449. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017, 127, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Goodpaster, B.H.; Sparks, L.M. Metabolic Flexibility in Health and Disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Hivert, M.-F.; Christophi, C.A.; Franks, P.W.; Jablonski, K.A.; Ehrmann, D.A.; Kahn, S.E.; Horton, E.S.; Pollin, T.I.; Mather, K.J.; Perreault, L.; et al. Lifestyle and Metformin Ameliorate Insulin Sensitivity Independently of the Genetic Burden of Established Insulin Resistance Variants in Diabetes Prevention Program Participants. Diabetes 2016, 65, 520–526. [Google Scholar] [CrossRef]
- Zaccardi, F.; O’Donovan, G.; Webb, D.R.; Yates, T.; Kurl, S.; Khunti, K.; Davies, M.J.; Laukkanen, J.A. Cardiorespiratory fitness and risk of type 2 diabetes: A 23-year cohort study and a meta-analysis of prospective studies. Atherosclerosis 2015, 243, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Brugnara, L.; Murillo, S.; Novials, A.; Rojo-Martínez, G.; Soriguer, F.; Goday, A.; Calle-Pascual, A.; Castaño, L.; Gaztambide, S.; Valdés, S.; et al. Low physical activity and its association with diabetes and other cardiovascular risk factors. PLoS ONE 2016, 11, e0160959. [Google Scholar] [CrossRef] [PubMed]
- Joyner, M.J.; Green, D.J. Exercise protects the cardiovascular system: Effects beyond traditional risk factors. J. Physiol. 2009, 587, 5551–5558. [Google Scholar] [CrossRef] [PubMed]
- Astorino, T.A.; Schubert, M.M. Changes in fat oxidation in response to various regimes of high intensity interval training (HIIT). Eur. J. Appl. Physiol. 2018, 118, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Karstoft, K.; Pedersen, B.K. Skeletal muscle as a gene regulatory endocrine organ. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Trovato, E.; Di Felice, V.; Barone, R. Extracellular Vesicles: Delivery Vehicles of Myokines. Front. Physiol. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Whitham, M.; Parker, B.L.; Friedrichsen, M.; Hingst, J.R.; Hjorth, M.; Hughes, W.E.; Egan, C.L.; Cron, L.; Watt, K.I.; Kuchel, R.P.; et al. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 2018, 27, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Garai, K.; Adam, Z.; Herczeg, R.; Banfai, K.; Gyebrovszki, A.; Gyenesei, A.; Pongracz, J.E.; Wilhelm, M.; Kvell, K. Physical Activity as a Preventive Lifestyle Intervention Acts Through Specific Exosomal miRNA Species-Evidence From Human Short- and Long-Term Pilot Studies. Front. Physiol. 2021, 12, 658218. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Roles for microRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Castaño, C.; Novials, A.; Párrizas, M. Exosomes and diabetes. Diabetes/Metab. Res. Rev. 2019, 35, e3107. [Google Scholar] [CrossRef] [PubMed]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Mirasierra, M.; Vallejo, M.; Novials, A.; Párrizas, M. Delivery of muscle-derived exosomal miRNAs induced by HIIT improves insulin sensitivity through down-regulation of hepatic FoxO1 in mice. Proc. Natl. Acad. Sci. USA 2020, 117, 30335–30343. [Google Scholar] [CrossRef]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e5. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef]
- Gallo-Villegas, J.; Aristizabal, J.C.; Estrada, M.; Valbuena, L.H.; Narvaez-Sanchez, R.; Osorio, J.; Aguirre-Acevedo, D.C.; Calderón, J.C. Efficacy of high-intensity, low-volume interval training compared to continuous aerobic training on insulin resistance, skeletal muscle structure and function in adults with metabolic syndrome: Study protocol for a randomized controlled clinical trial. Trials 2018, 19, 144. [Google Scholar] [CrossRef]
- Wahl, P.; Bloch, W.; Proschinger, S. The Molecular Signature of High-intensity Training in the Human Body. Int. J. Sport. Med. 2021, 43, 195–205. [Google Scholar] [CrossRef]
- Gerosa-Neto, J.; Panissa, V.L.G.; Monteiro, P.A.; Inoue, D.S.; Ribeiro, J.P.J.; Figueiredo, C.; Zagatto, A.M.; Little, J.P.; Lira, F.S. High- or moderate-intensity training promotes change in cardiorespiratory fitness, but not visceral fat, in obese men: A randomised trial of equal energy expenditure exercise. Respir. Physiol. Neurobiol. 2019, 266, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, R.S.; Fitzpatrick, B.; Fitzpatrick, S.; McDermott, G.; Brick, N.; McClean, C.; Davison, G.W. Extremely short duration interval exercise improves 24-h glycaemia in men with type 2 diabetes. Eur. J. Appl. Physiol. 2018, 118, 2551–2562. [Google Scholar] [CrossRef]
- Dela, F.; Ingersen, A.; Andersen, N.B.; Nielsen, M.B.; Petersen, H.H.H.; Hansen, C.N.; Larsen, S.; Wojtaszewski, J.; Helge, J.W. Effects of one-legged high-intensity interval training on insulin-mediated skeletal muscle glucose homeostasis in patients with type 2 diabetes. Acta Physiol. 2019, 226, e13245. [Google Scholar] [CrossRef] [PubMed]
- Jelleyman, C.; Yates, T.; O’Donovan, G.; Gray, L.J.; King, J.A.; Khunti, K.; Davies, M.J. The effects of high-intensity interval training on glucose regulation and insulin resistance: A meta-analysis. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2015, 16, 942–961. [Google Scholar] [CrossRef] [PubMed]
- Paramanantham, A.; Asfiya, R.; Das, S.; McCully, G.; Srivastava, S. Extracellular Vesicle (EVs) Associated Non-Coding RNAs in Lung Cancer and Therapeutics. Int. J. Mol. Sci. 2022, 23, 13637. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Varshney, A.; Bajaj, R.; Pokharkar, V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules 2022, 27, 7289. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Yu, P.; Li, F.; Jiang, X.; Jiao, X.; Shen, Y.; Lai, X. Human Umbilical Cord-Derived Mesenchymal Stem Cell-Exosomal MiR-627-5p Ameliorates Non-Alcoholic Fatty Liver Disease by Repressing FTO Expression. Hum. Cell 2021, 34, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Wang, T.; Wang, Z.; Wang, F.; Huang, D.; Sun, H.; Liu, H. Adipose-Derived Mesenchymal Stem Cell-Secreted Extracellular Vesicles Alleviate Non-Alcoholic Fatty Liver Disease via Delivering MiR-223-3p. Adipocyte 2022, 11, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Luo, Q.; Wang, Y.; Ma, Y.; Chen, F.; Zhu, X.; Shi, J. Adipose Mesenchymal Stem Cell-Derived Extracellular Vesicles Containing MicroRNA-26a-5p Target TLR4 and Protect against Diabetic Nephropathy. J. Biol. Chem. 2020, 295, 12868–12884. [Google Scholar] [CrossRef]
- Wang, J.; Wu, H.; Peng, Y.; Zhao, Y.; Qin, Y.; Zhang, Y.; Xiao, Z. Hypoxia Adipose Stem Cell-Derived Exosomes Promote High-Quality Healing of Diabetic Wound Involves Activation of PI3K/Akt Pathways. J. Nanobiotechnol. 2021, 19, 202. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, D.; Liu, Z.; Zhou, Y.; Chu, D.; Li, X.; Jiang, X.; Hou, D.; Chen, X.; Chen, Y.; et al. Targeted Exosome-Mediated Delivery of Opioid Receptor Mu SiRNA for the Treatment of Morphine Relapse. Sci. Rep. 2015, 5, 17543. [Google Scholar] [CrossRef]
- Sanz-de la Garza, M.; Rubies, C.; Batlle, M.; Bijnens, B.H.; Mont, L.; Sitges, M.; Guasch, E. Severity of structural and functional right ventricular remodeling depends on training load in an experimental model of endurance exercise. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H459–H468. [Google Scholar] [CrossRef]
- Cavalcanti-de-Albuquerque, J.P.; Bober, J.; Zimmer, M.R.; Dietrich, M.O. Regulation of substrate utilization and adiposity by Agrp neurons. Nat. Commun. 2019, 10, 311. [Google Scholar] [CrossRef]
- Frayn, K.N. Calculation of substrate oxidation rates in vivo from gaseous exchange. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1983, 55, 628–634. [Google Scholar] [CrossRef]
- Gonzalez-Franquesa, A.; Gama-Perez, P.; Kulis, M.; Szczepanowska, K.; Dahdah, N.; Moreno-Gomez, S.; Latorre-Pellicer, A.; Fernández-Ruiz, R.; Aguilar-Mogas, A.; Hoffman, A.; et al. Remission of obesity and insulin resistance is not sufficient to restore mitochondrial homeostasis in visceral adipose tissue. Redox Biol. 2022, 54, 102353. [Google Scholar] [CrossRef]
- Samms, R.J.; Zhang, G.; He, W.; Ilkayeva, O.; Droz, B.A.; Bauer, S.M.; Stutsman, C.; Pirro, V.; Collins, K.A.; Furber, E.C.; et al. Tirzepatide induces a thermogenic-like amino acid signature in brown adipose tissue. Mol. Metab. 2022, 64, 101550. [Google Scholar] [CrossRef] [PubMed]
- Mathus-Vliegen, E.M.H. Obesity and the elderly. J. Clin. Gastroenterol. 2012, 46, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Wall, C.E.; Yu, R.T.; Atkins, A.R.; Downes, M.; Evans, R.M. Nuclear receptors and AMPK: Can exercise mimetics cure diabetes? J. Mol. Endocrinol. 2016, 57, R49–R58. [Google Scholar] [CrossRef] [PubMed]
- Prattichizzo, F.; Matacchione, G.; Giuliani, A.; Sabbatinelli, J.; Olivieri, F.; de Candia, P.; De Nigris, V.; Ceriello, A. Extracellular vesicle-shuttled miRNAs: A critical appraisal of their potential as nano-diagnostics and nano-therapeutics in type 2 diabetes mellitus and its cardiovascular complications. Theranostics 2021, 11, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- de Abreu, R.C.; Fernandes, H.; da Costa Martins, P.A.; Sahoo, S.; Emanueli, C.; Ferreira, L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat. Rev. Cardiol. 2020, 17, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Milbank, E.; Dragano, N.R.V.; González-García, I.; Garcia, M.R.; Rivas-Limeres, V.; Perdomo, L.; Hilairet, G.; Ruiz-Pino, F.; Mallegol, P.; Morgan, D.A.; et al. Small extracellular vesicle-mediated targeting of hypothalamic AMPKα1 corrects obesity through BAT activation. Nat. Metab. 2021, 3, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–435. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019, 30, 114–127. [Google Scholar]
- Wiklander, O.P.B.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with “antagomirs”. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Párrizas, M.; Brugnara, L.; Esteban, Y.; González-Franquesa, A.; Canivell, S.; Murillo, S.; Gordillo-Bastidas, E.; Cussó, R.; Cadefau, J.A.; García-Roves, P.M. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J. Clin. Endocrinol. Metab. 2015, 100, E407–E415. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Murthy, V.; Pacold, M.; Danielson, K.; Tanriverdi, K.; Larson, M.G.; Hanspers, K.; Pico, A.; Mick, E.; Reis, J.; et al. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care 2017, 40, 546–553. [Google Scholar] [CrossRef]
- Pirola, C.J.; Gianotti, T.F.; Castaño, G.O.; Mallardi, P.; Martino, J.S.; Ledesma, M.M.G.L.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef]
- Long, J.-K.; Dai, W.; Zheng, Y.-W.; Zhao, S.-P. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol. Med. 2019, 25, 26. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Chen, J.-X.; Ren, Y.; Fan, L.-C.; Xiang, W.; He, X.-J. Exosomal miR-122 promotes adipogenesis and aggravates obesity through the VDR/SREBF1 axis. Obesity 2022, 30, 666–679. [Google Scholar] [CrossRef]
- Ding, R.-B.; Bao, J.; Deng, C.-X. Emerging roles of SIRT1 in fatty liver diseases. Int. J. Biol. Sci. 2017, 13, 852–867. [Google Scholar] [CrossRef]
- Kolb, H.; Stumvoll, M.; Kramer, W.; Kempf, K.; Martin, S. Insulin translates unfavourable lifestyle into obesity. BMC Med. 2018, 16, 232. [Google Scholar] [CrossRef]
- Qu, P.; Wang, L.; Min, Y.; McKennett, L.; Keller, J.R.; Lin, P.C. Vav1 Regulates Mesenchymal Stem Cell Differentiation Decision Between Adipocyte and Chondrocyte via Sirt1. Stem Cells 2016, 34, 1934–1946. [Google Scholar] [CrossRef]
- Bové, M.; Monto, F.; Guillem-Llobat, P.; Ivorra, M.D.; Noguera, M.A.; Zambrano, A.; Sirerol-Piquer, M.S.; Requena, A.C.; García-Alonso, M.; Tejerina, T.; et al. NT3/TrkC Pathway Modulates the Expression of UCP-1 and Adipocyte Size in Human and Rodent Adipose Tissue. Front. Endocrinol. 2021, 12, 630097. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, S.; Rébillard, A.; Muti, P.; Friedenreich, C.M.; Brenner, D.R. A review of physical activity and circulating miRNA expression: Implications in cancer risk and progression. Cancer Epidemiol. Biomark. Prev. 2018, 27, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhou, H.; Tang, Q. miR-133: A Suppressor of Cardiac Remodeling? Front. Pharmacol. 2018, 9, 903. [Google Scholar] [CrossRef] [PubMed]
- Habibi, P.; Alihemmati, A.; Ahmadiasl, N.; Fateh, A.; Anvari, E. Exercise training attenuates diabetes-induced cardiac injury through increasing miR-133a and improving pro-apoptosis/anti-apoptosis balance in ovariectomized rats. Iran. J. Basic Med. Sci. 2020, 23, 79–85. [Google Scholar] [PubMed]
- Dou, X.; Zhou, W.-Y.; Ding, M.; Ma, Y.-J.; Yang, Q.-Q.; Qian, S.-W.; Tang, Y.; Tang, Q.Q.; Liu, Y. The protease SENP2 controls hepatic gluconeogenesis by regulating the SUMOylation of the fuel sensor AMPKα. J. Biol. Chem. 2022, 298, 101544. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-W.; Addy, C.; Kusunoki, J.; Anderson, N.; Deja, S.; Fu, X.; Burgess, S.C.; Li, C.; Ruddy, M.; Chakravarthy, M.; et al. Acetyl CoA Carboxylase Inhibition Reduces Hepatic Steatosis but Elevates Plasma Triglycerides in Mice and Humans: A Bedside to Bench Investigation. Cell Metab. 2017, 26, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Wang, Z.; Duan, R.; Yang, C.; Zhao, R.; Feng, Q.; Qin, Y.; Jiang, J.; Gu, S.; Lv, K.; et al. Therapeutic targeting of hepatic ACSL4 ameliorates NASH in mice. Hepatology 2022, 75, 140–153. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Y.; Ma, Y.; Wen, D. High-intensity interval versus moderate-intensity continuous training: Superior metabolic benefits in diet-induced obesity mice. Life Sci. 2017, 191, 122–131. [Google Scholar] [CrossRef]
- Marcinko, K.; Sikkema, S.R.; Samaan, M.C.; Kemp, B.E.; Fullerton, M.D.; Steinberg, G.R. High intensity interval training improves liver and adipose tissue insulin sensitivity. Mol. Metab. 2015, 4, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Huenchullan, S.F.; Ban, L.A.; Olaya-Agudo, L.F.; Maharjan, B.R.; Williams, P.F.; Tam, C.S.; Mclennan, S.V.; Twigg, S.M. Constant-Moderate and High-Intensity Interval Training Have Differential Benefits on Insulin Sensitive Tissues in High-Fat Fed Mice. Front. Physiol. 2019, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Booth, F.W.; Laye, M.J. Lack of adequate appreciation of physical exercise’s complexities can pre-empt appropriate design and interpretation in scientific discovery. J. Physiol. 2009, 587 (Pt 23), 5527–5539. [Google Scholar] [CrossRef]
- Van de Water, A.; Verheyen, J.; Xhonneux, R.; Reneman, R.S. An improved method to correct the QT interval of the electrocardiogram for changes in heart rate. J. Pharmacol. Methods 1989, 22, 207–217. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaño, C.; Meza-Ramos, A.; Batlle, M.; Guasch, E.; Novials, A.; Párrizas, M. Treatment with EV-miRNAs Alleviates Obesity-Associated Metabolic Dysfunction in Mice. Int. J. Mol. Sci. 2022, 23, 14920. https://doi.org/10.3390/ijms232314920
Castaño C, Meza-Ramos A, Batlle M, Guasch E, Novials A, Párrizas M. Treatment with EV-miRNAs Alleviates Obesity-Associated Metabolic Dysfunction in Mice. International Journal of Molecular Sciences. 2022; 23(23):14920. https://doi.org/10.3390/ijms232314920
Chicago/Turabian StyleCastaño, Carlos, Aline Meza-Ramos, Montserrat Batlle, Eduard Guasch, Anna Novials, and Marcelina Párrizas. 2022. "Treatment with EV-miRNAs Alleviates Obesity-Associated Metabolic Dysfunction in Mice" International Journal of Molecular Sciences 23, no. 23: 14920. https://doi.org/10.3390/ijms232314920
APA StyleCastaño, C., Meza-Ramos, A., Batlle, M., Guasch, E., Novials, A., & Párrizas, M. (2022). Treatment with EV-miRNAs Alleviates Obesity-Associated Metabolic Dysfunction in Mice. International Journal of Molecular Sciences, 23(23), 14920. https://doi.org/10.3390/ijms232314920






