Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress
Abstract
:1. Introduction
2. Results
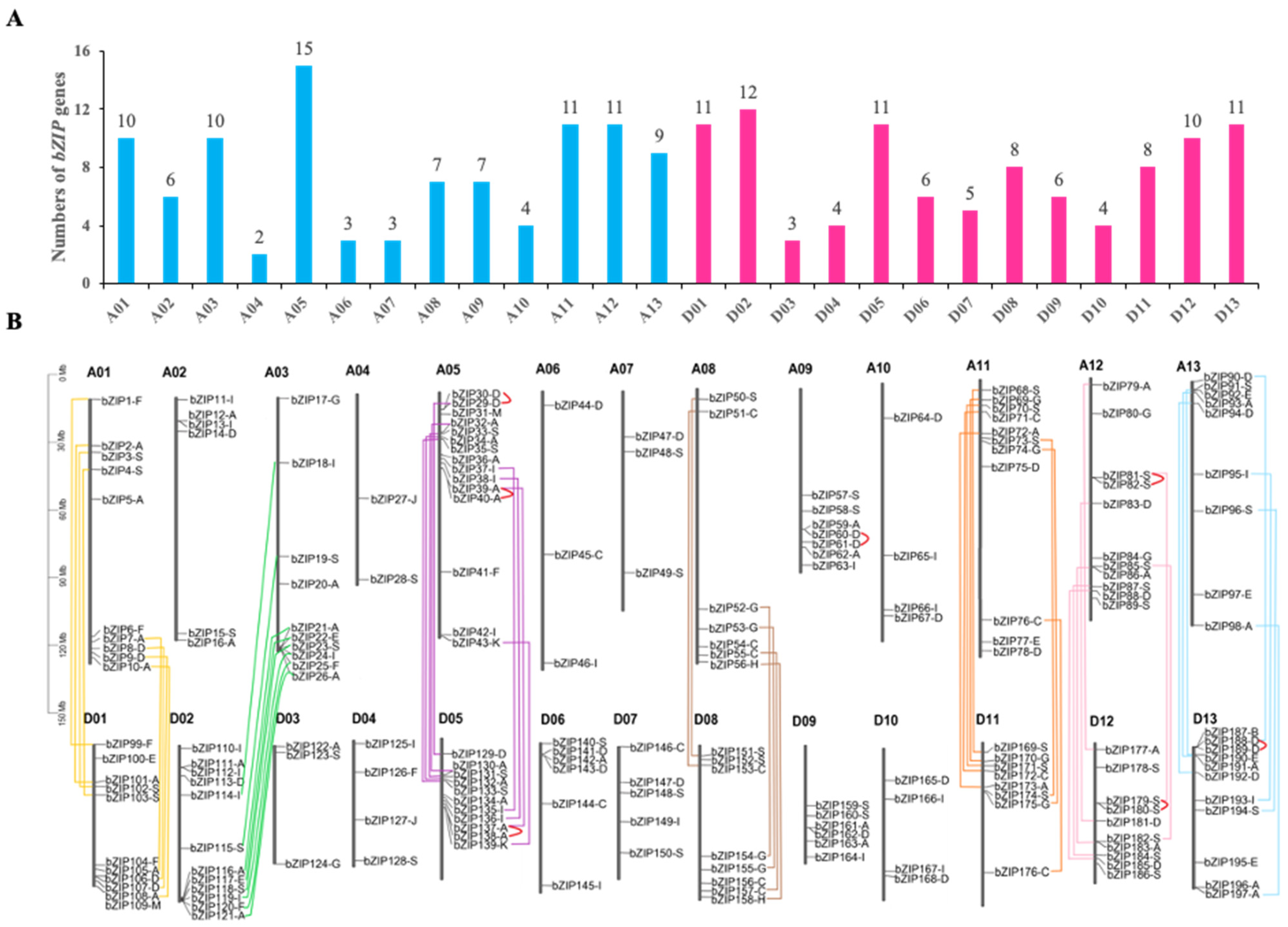
2.1. Identification of bZIP Transcription Factors in G. Hirsutum
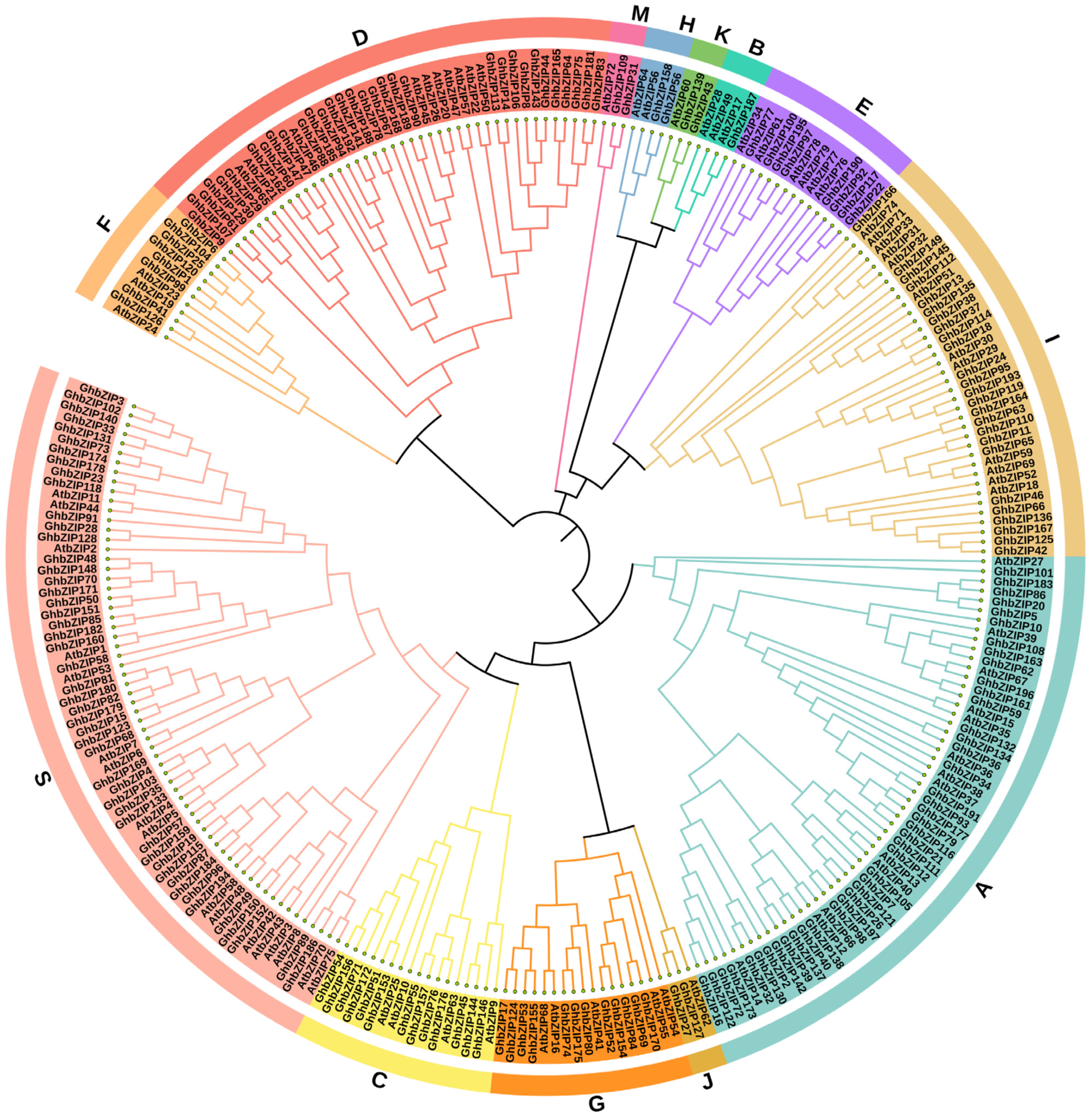
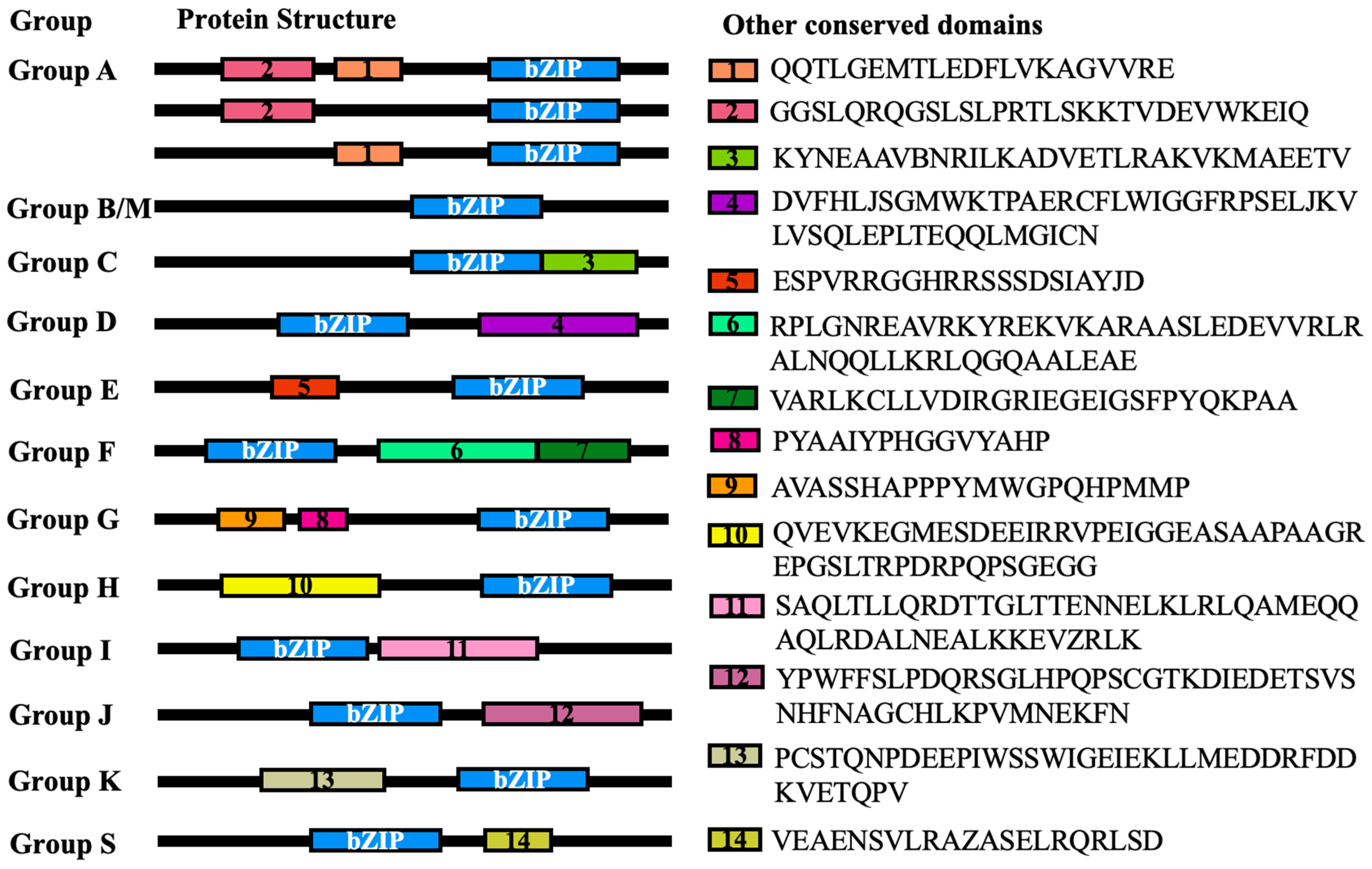
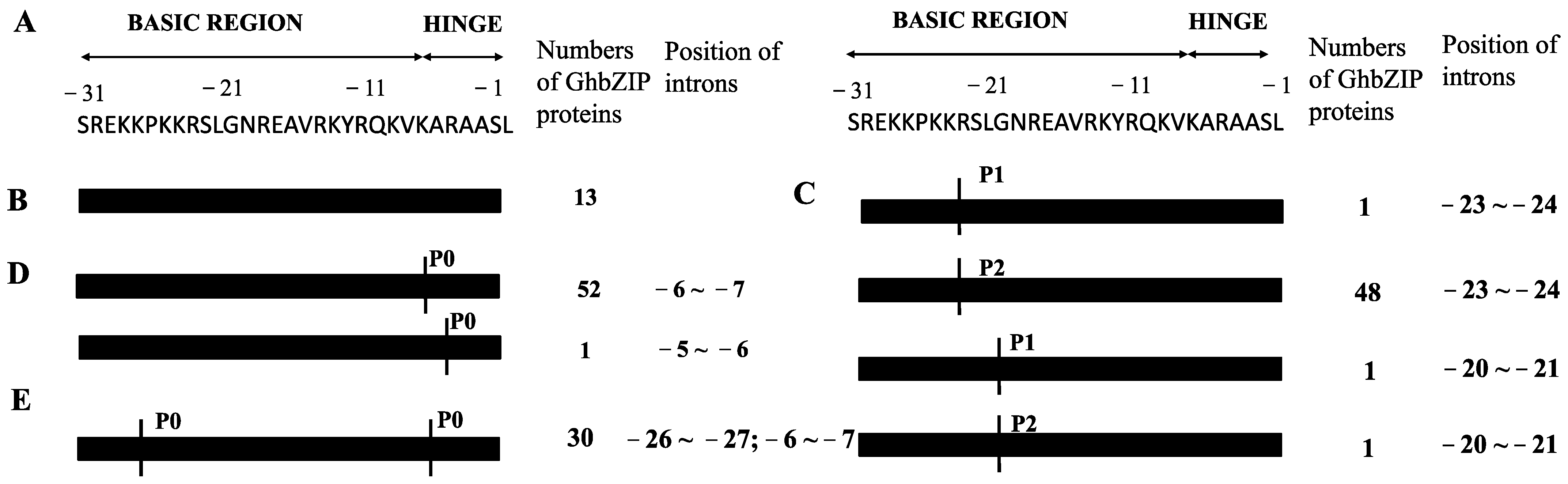
2.2. Phylogenetic Analysis, Prediction of Conserved Motifs, and Display of Gene Structure
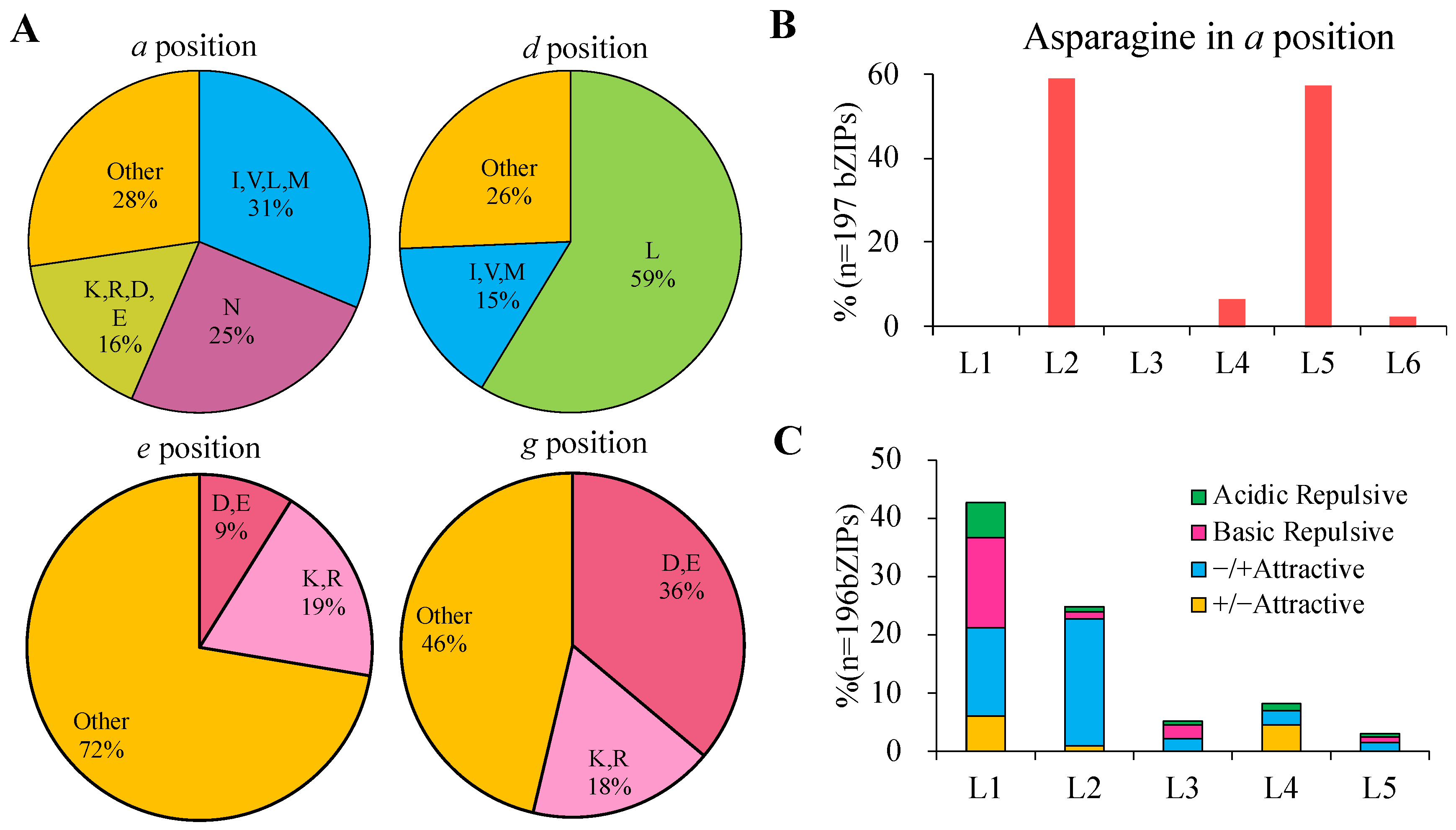
2.3. DNA-Binding Site Specificity Prediciton of GhbZIP Transcription Factors
2.4. Dimerization Properties Prediction of bZIP Transcription Factors
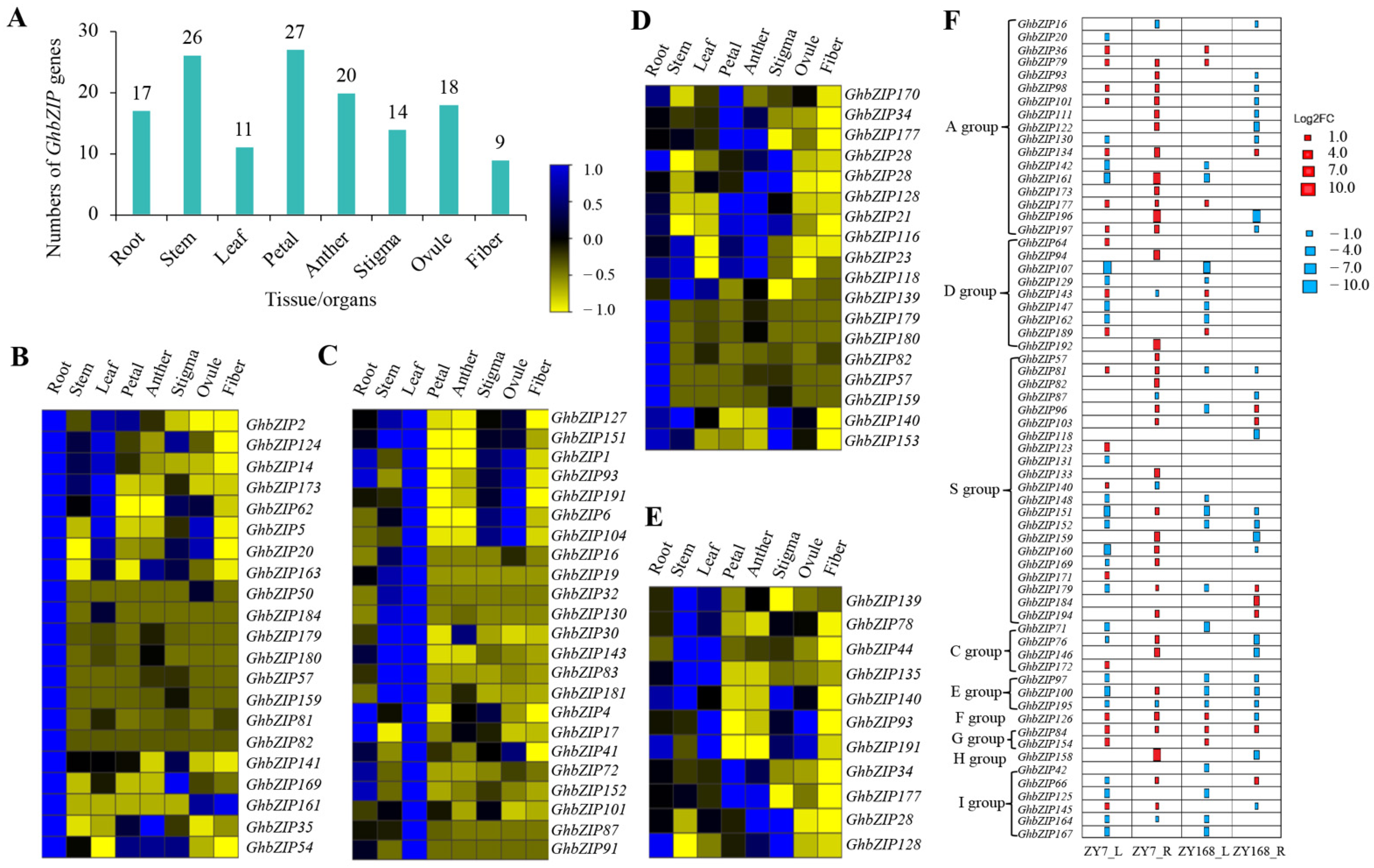
2.5. Expression Analysis of GhbZIP Family Members in Cotton under Drought Treatments
2.6. GhbZIP177 Conferred Drought Resistance in Cotton
3. Discussion
4. Materials and Method
4.1. Identification of bZIP Family Genes in Gossypium spp
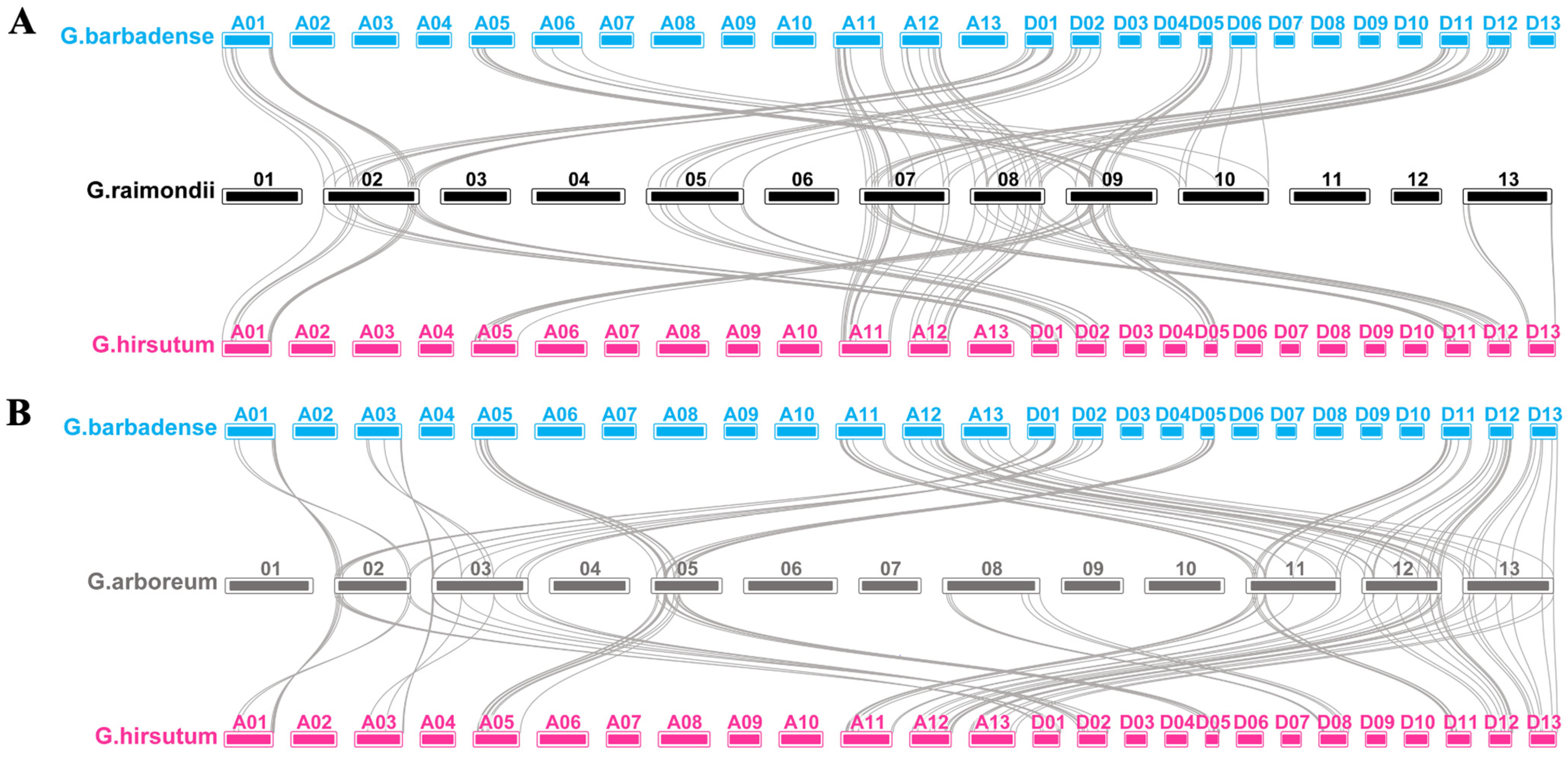
4.2. Chromosomal Locations and Gene Duplication Analysis
4.3. Phylogeny Analysis bZIP
4.4. Sequence Analysis
4.5. Expression Profiles
4.6. Functional Analysis of GhbZIP177 in Cotton Response to Drought Stress
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurst, H.C. Transcription factors. 1: bZIP proteins. Protein Profile 1994, 1, 123–168. [Google Scholar] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinson, C.; Acharya, A.; Taparowsky, E.J. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta. 2006, 1759, 4–12. [Google Scholar] [CrossRef] [Green Version]
- Deppmann, C.D.; Acharya, A.; Rishi, V.; Wobbes, B.; Smeekens, S.; Taparowsky, E.J.; Vinson, C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004, 32, 3435–3445. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.; Izawa, T.; Chua, N.H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994, 8, 192–200. [Google Scholar] [CrossRef]
- Corrêa, L.G.; Riaño-Pachón, D.M.; Schrago, C.G.; dos Santos, R.V.; Mueller-Roeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Pourabed, E.; Ghane Golmohamadi, F.; Soleymani Monfared, P.; Razavi, S.M.; Shobbar, Z.S. Basic leucine zipper family in barley: Genome-wide characterization of members and expression analysis. Mol. Biotechnol. 2015, 57, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-wide identification and structural analysis of bZIP transcription factor genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolly, N.K.; Imran, Q.M.; Shahid, M.; Imran, M.; Khan, M.; Lee, S.U.; Hussain, A.; Lee, I.J.; Yun, B.W. Drought-induced AtbZIP62 transcription factor regulates drought stress response in Arabidopsis. Plant Physiol. Biochem. 2020, 156, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Pedrotti, L.; Weiste, C.; Nägele, T.; Wolf, E.; Lorenzin, F.; Dietrich, K.; Mair, A.; Weckwerth, W.; Teige, M.; Baena-González, E.; et al. Snf1-RELATED KINASE1-controlled C/S(1)-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 2018, 30, 495–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, R.; Oñate-Sánchez, L.; Weltmeier, F.; Ehlert, A.; Diaz, I.; Dietrich, K.; Vicente-Carbajosa, J.; Dröge-Laser, W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.G.; Price, J.; Lin, P.C.; Hong, J.C.; Jang, J.C. The arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 2010, 3, 361–373. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Howell, S.H. Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 2008, 31, 1735–1743. [Google Scholar] [CrossRef]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Liu, J.; Zhao, T.; Gomez, A.; Li, C.; Yu, C.; Li, H.; Lin, J.; Yang, Y.; Liu, B.; et al. A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol. 2016, 171, 334–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñiz García, M.N.; Stritzler, M.; Capiati, D.A. Heterologous expression of Arabidopsis ABF4 gene in potato enhances tuberization through ABA-GA crosstalk regulation. Planta 2014, 239, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Meng, Z.; Meng, Z.; Malik, W.; Yan, R.; Lwin, K.M.; Lin, F.; Wang, Y.; Sun, G.; Zhou, T.; et al. GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 2016, 6, 35040. [Google Scholar] [CrossRef] [Green Version]
- Kerr, T.C.C.; Abdel-Mageed, H.; Aleman, L.; Lee, J.; Payton, P.; Cryer, D.; Allen, R.D. Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum). Plant Cell Environ. 2018, 41, 898–907. [Google Scholar] [CrossRef]
- Herath, V.; Verchot, J. Insight into the bZIP gene family in Solanum tuberosum: Genome and transcriptome analysis to understand the roles of gene diversification in spatiotemporal gene expression and function. Int. J. Mol. Sci. 2021, 22, 253. [Google Scholar] [CrossRef]
- Bi, C.; Yu, Y.; Dong, C.; Yang, Y.; Zhai, Y.; Du, F.; Xia, C.; Ni, Z.; Kong, X.; Zhang, L. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 2021, 19, 209–211. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [Green Version]
- Stolz, M.L.; McCormick, C. The bZIP Proteins of oncogenic viruses. Viruses 2020, 12, 757. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Malik, W.A.; Chen, X.; Wang, J.; Wang, D.; Wang, S.; Chen, C.; Guo, L.; Ye, W. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int. J. Biol. Macromol. 2020, 146, 569–578. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Shabalina, S.A.; Ogurtsov, A.Y.; Spiridonov, A.N.; Novichkov, P.S.; Spiridonov, N.A.; Koonin, E.V. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 2010, 27, 1745–1749. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, P.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct. Integr. Genom. 2008, 8, 69–78. [Google Scholar] [CrossRef]
- Niu, X.; Renshaw-Gegg, L.; Miller, L.; Guiltinan, M.J. Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol. Biol. 1999, 41, 1–13. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Zandkarimi, H.; Ebadi, A.; Salami, S.A.; Alizade, H.; Baisakh, N. Analyzing the Expression Profile of AREB/ABF and DREB/CBF Genes under Drought and Salinity Stresses in Grape (Vitis vinifera L.). PLoS ONE 2015, 10, e0134288. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, L.; Sun, W.; Wu, D.; Wang, M.; Yu, Y.; Chen, G.; Yang, W.; Lin, Z.; Zhang, X.; et al. Phenomics-based GWAS analysis reveals the genetic architecture for drought resistance in cotton. Plant Biotechnol. J. 2020, 18, 2533–2544. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, X.; Nie, Y.; Guo, X.; Liang, S.; Zhu, H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 2006, 50, 519–524. [Google Scholar] [CrossRef]

| Group | No. of Members | Characteristic Features of bZIP Domain | Putative Binding Site |
|---|---|---|---|
| A | 41 | *********MIK************QAY*** | ABREs with the core ACGT or others containing GCGT/AAGT |
| B | 1 | **********RNRESAQLSR********** | G- and C-boxes with equal affinity |
| C | 13 | ***********************QAHLEE* | Hybrid ACGT elements such as G/C, G/A, and C/G boxes |
| D | 32 | ********LAQN**AA*KSR***KAYVQQ* | C-box sequence |
| E | 8 | *********A**************QYIS(/A)E* | Unknown |
| F/K | 10 | **************A**************** | C-box elements |
| G/M/S | 62 | ***********N**SA**SR********** | G-box and/or G-box-like sequences |
| H | 2 | ***********NRVSAQQAR********** | G-box-like sequences |
| I | 25 | *******************K********** | Sequences other than those containing a palindromic ACGT core, such as CCA/TGG repeats |
| J | 2 | *******************I********** | Unknown |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Feng, C.; Chen, L.; Li, B.; Zhang, X.; Yang, X. Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress. Int. J. Mol. Sci. 2022, 23, 14894. https://doi.org/10.3390/ijms232314894
Zhang B, Feng C, Chen L, Li B, Zhang X, Yang X. Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress. International Journal of Molecular Sciences. 2022; 23(23):14894. https://doi.org/10.3390/ijms232314894
Chicago/Turabian StyleZhang, Boyang, Cheng Feng, Lin Chen, Baoqi Li, Xianlong Zhang, and Xiyan Yang. 2022. "Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress" International Journal of Molecular Sciences 23, no. 23: 14894. https://doi.org/10.3390/ijms232314894
APA StyleZhang, B., Feng, C., Chen, L., Li, B., Zhang, X., & Yang, X. (2022). Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress. International Journal of Molecular Sciences, 23(23), 14894. https://doi.org/10.3390/ijms232314894





