Transient Retention of Photoreceptor Outer Segments in Matrigel-Embedded Retinal Organoids
Abstract
:1. Introduction
2. Results
2.1. Loss of Photoreceptor OSs during Retinal Organoid Processing
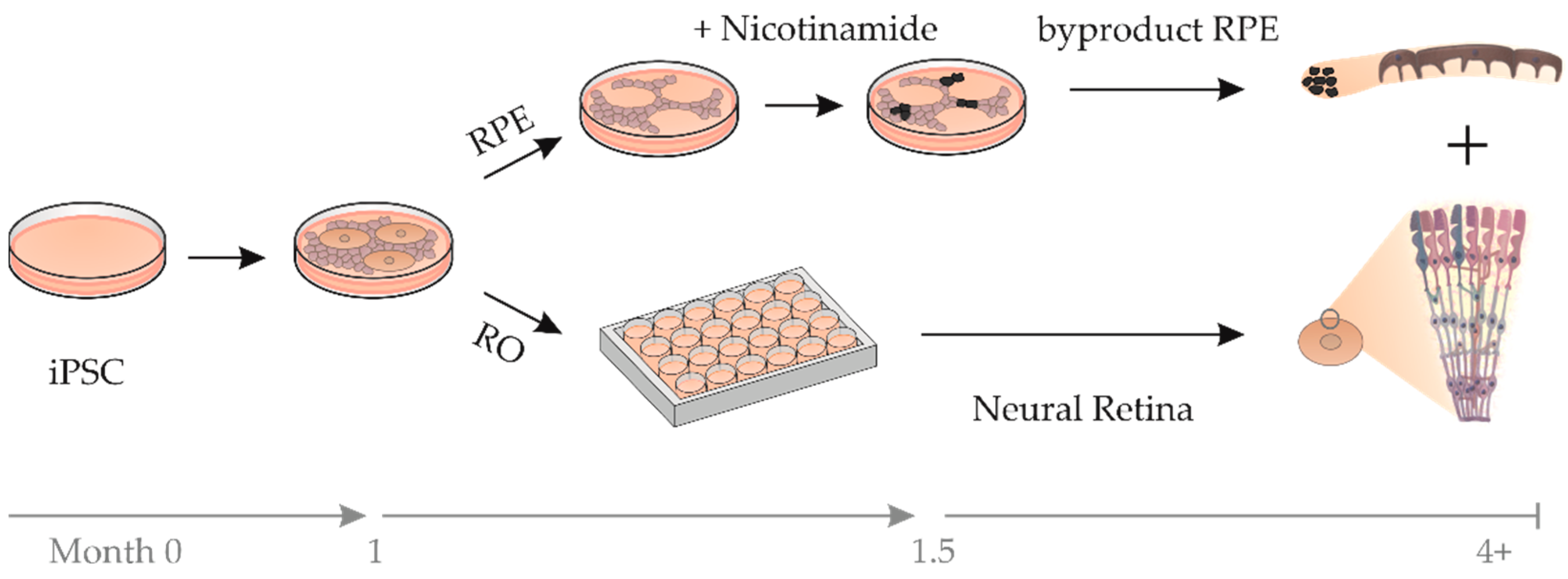
2.2. RPE Cells as a Byproduct during RO Differentiation
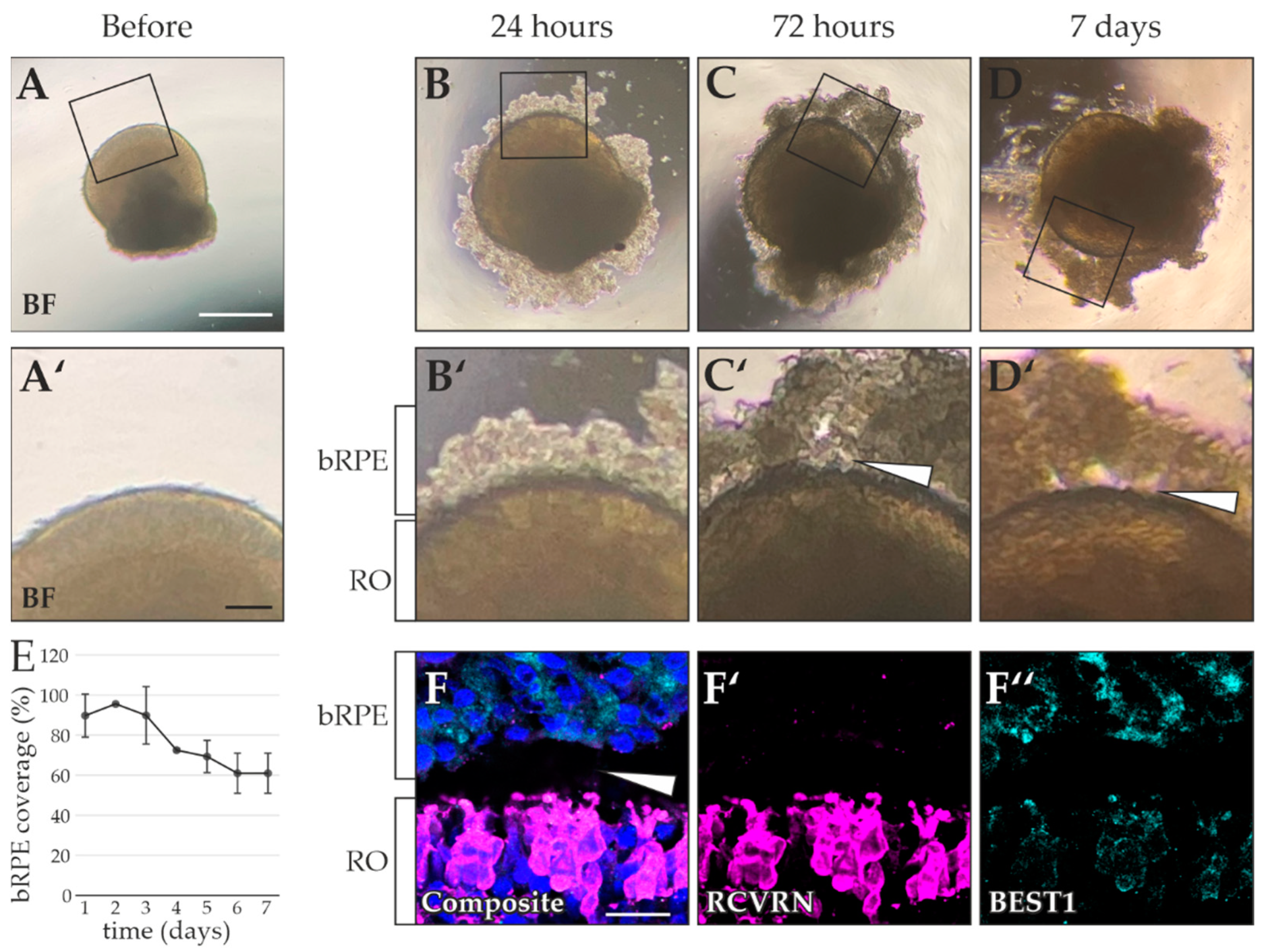
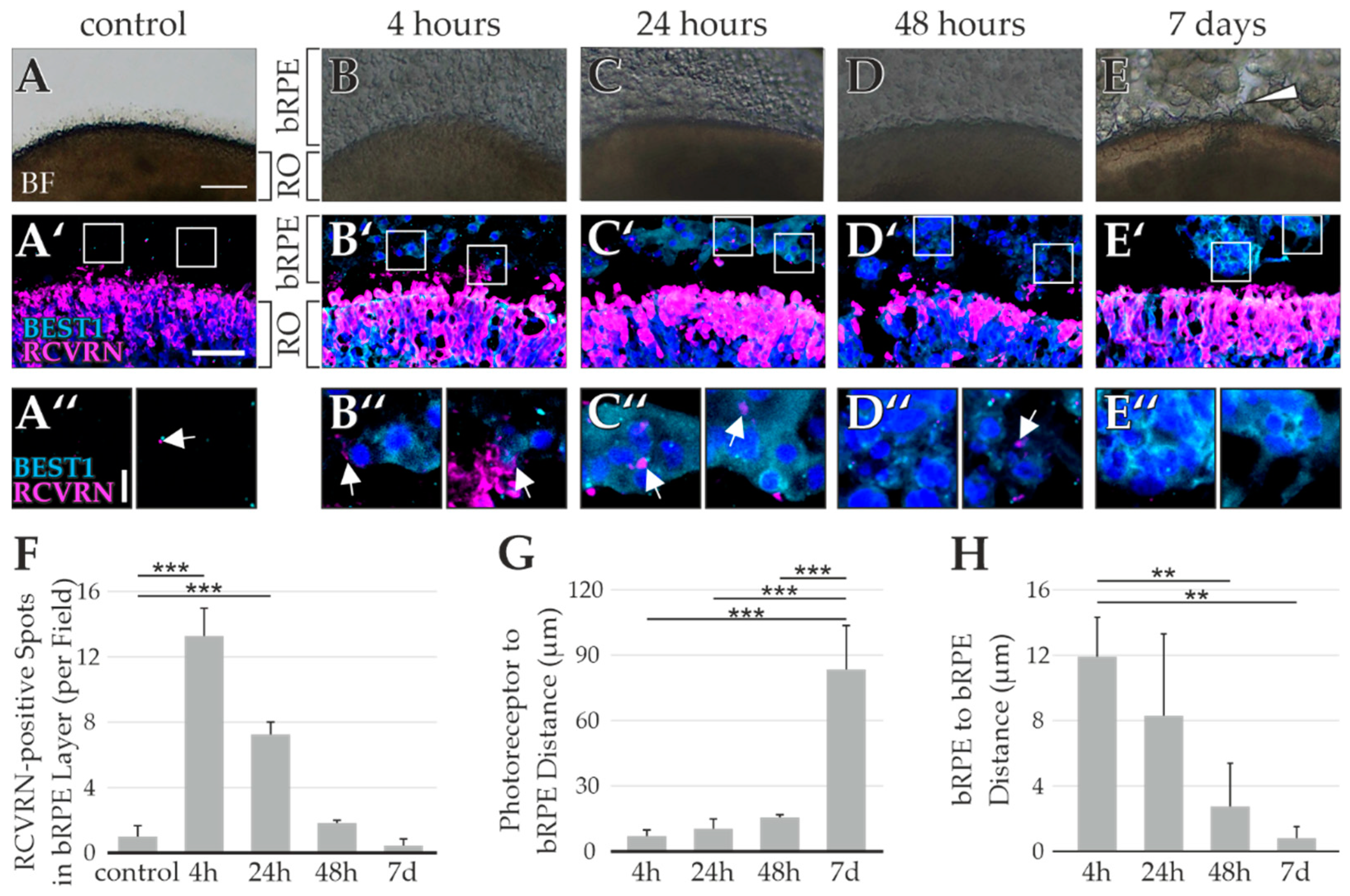
2.3. Testing of Multiple bRPE-RO Coculture Techniques
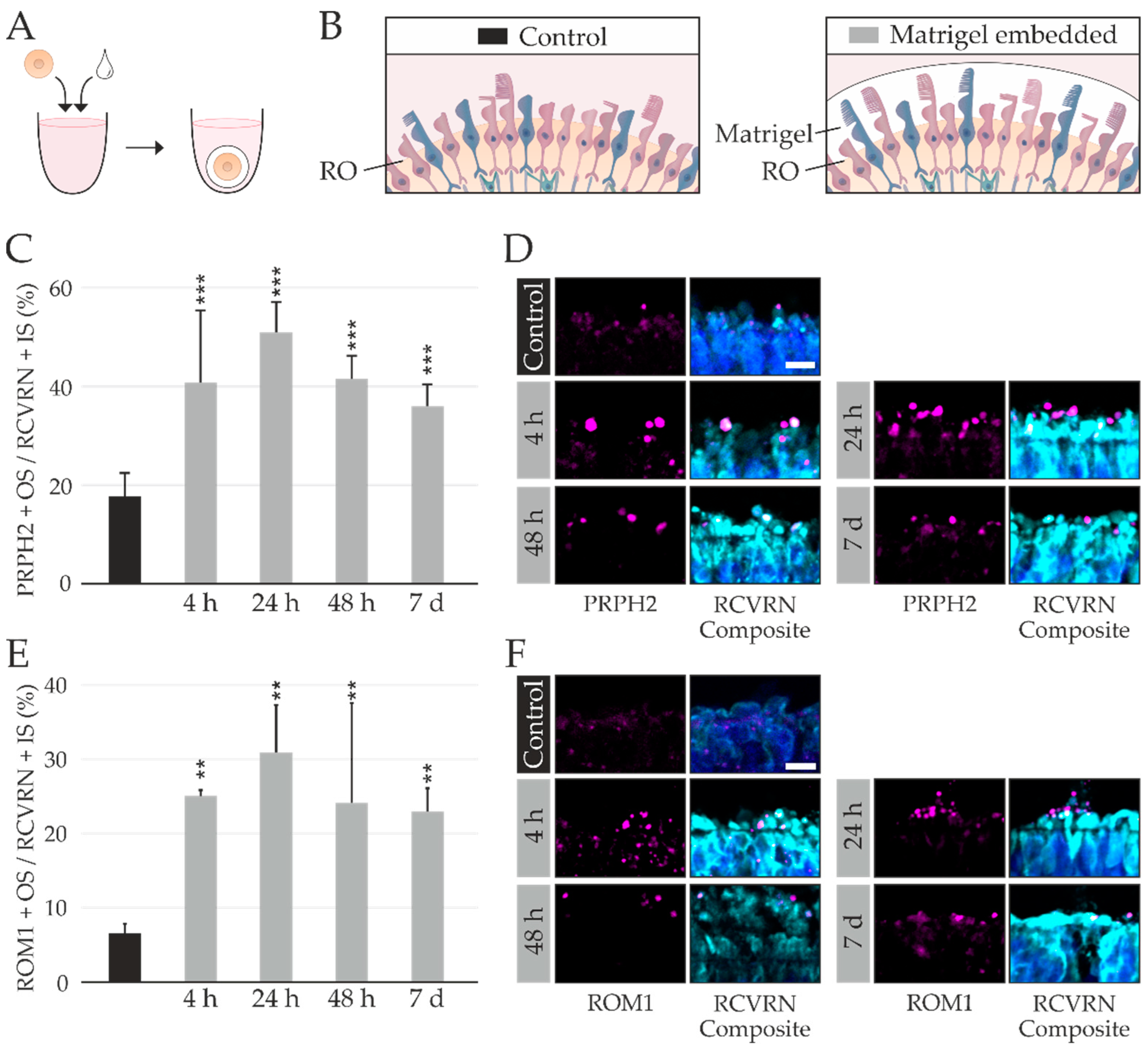
2.4. RO Embedding Improves OS Retention
3. Discussion
4. Materials and Methods
4.1. Sampling of Biomaterials, iPSC Reprogramming and Maintenance
4.2. Differentiation and Characterization of RO and bRPE
4.3. RNA Isolation and Reverse Transcription
4.4. qRT-PCR
4.5. Immunocytochemistry
4.6. Photoreceptor OS Phagocytosis Assay
4.7. bRPE and RO Coculture Techniques
- (1)
- bRPE cells were dissociated with TrypLETM Select and suspended in maintenance medium without nicotinamide [20]. The bRPE were counted using a CASY® Cell Counter and Analyzer (Innovatis Roche AG, Bielefeld, Germany). One RO and 100,000 bRPE cells were added to one well of a Costar® Ultra-Low Attachment 96-Well Plate with round bottoms (Corning). Co-cultures were imaged after 1 day, 3 days, and 7 days using a Nikon Eclipse TE2 microscope. After 14 days, the co-cultures were cryosectioned and immunostained as described previously.
- (2)
- Co-culture was performed as described in (1) and treated with the following additives, which were applied when the bRPE were introduced to the RO. Co-cultures were treated with ApInh: 5 µM Blebbistatin (Cayman Chemical, Michigan, USA) or with PrECM: 5 µL/mL Fibronectin (Life Technologies), 47.5 µg/mL Laminin (Cayman Chemical), and 16.6 µL/mL vitronectin (kindly provided by Fabiola Biasella University of Regensburg, Regensburg, Germany and purified according to [42]). The PrECM components were chosen based on Ref. [43]. The co-cultures were harvested after 48 h, cryosectioned and immunostained as described previously.
- (3)
- For the co-culture with bRPE suspended in pure Matrigel GFR, 500,000 bRPE were dissociated using TrypLETM Select and suspended in 10 µL cold Matrigel GFR (based on ref [44]). The RO was transferred to sterile Bemis™ Parafilm™ M Laboratory Wrapping Film molds (Fisher Scientific GmbH), embedded in the bRPE–Matrigel GFR mixture and incubated for 20 min at 37 °C. After 20 min 10 µL cold Matrigel GFR was added, and the cultures were incubated at 37 °C for 10 min. ROs were harvested after 4 h, 24 h, 48 h, or 1 week. Control ROs were not embedded in the bRPE–Matrigel mixture.
- (4)
- Co-culturing was conducted as described in (1) and incubated for 48 h. Brightfield images were taken using a Nikon Eclipse TE2 microscope. The co-culture was transferred to sterile Bemis™ Parafilm™ M Laboratory Wrapping Film molds, embedded in the Matrigel GFR mixture and incubated for 30 min at 37 °C. After embedding, brightfield images were taken using a Nikon Eclipse TE2 microscope.
- (5)
- For the co-cultures on Transwell filter inserts, bRPE were cultured as described previously. The ROs were gently added to the Transwell filter inserts and incubated for at least 2 days. The ROs spontaneously detached from the inserts while handling, during medium changes, and/or during fixation in 4% PFA. After detachment, the Transwell filter inserts were immunostained to analyze OS uptake by the bRPE.
- (6)
- Optionally, 200 µL Matrigel GFR (Corning, New York, USA) or 200 µL hyaluronic acid-based hydrogel HyStem®-C (Sigma-Aldrich, St. Louis, USA) was added after plating the ROs on the bRPE lawn (based on ref. [32]).
4.8. Quantification of bRPE Coverage
4.9. Interaction Analysis of bRPE-RO Cocultures
4.10. Matrigel Embedding and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donovan, S.L.; Schweers, B.; Martins, R.; Johnson, D.; Dyer, M.A. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.Y.; Kelley, R.A.; Li, T.; Swaroop, A. Primary cilia biogenesis and associated retinal ciliopathies. Semin. Cell Dev. Biol. 2021, 110, 70–88. [Google Scholar] [CrossRef] [PubMed]
- Bujakowska, K.M.; Liu, Q.; Pierce, E.A. Photoreceptor Cilia and Retinal Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028274. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.K.; Lewis, G.P.; Linberg, K.A.; Verardo, M.R. Cellular remodeling in mammalian retina: Results from studies of experimental retinal detachment. Prog. Retin. Eye Res. 2005, 24, 395–431. [Google Scholar] [CrossRef]
- Cook, B.; Lewis, G.P.; Fisher, S.K.; Adler, R. Apoptotic photoreceptor degeneration in experimental retinal detachment. Investig. Ophthalmol. Vis. Sci. 1995, 36, 990–996. [Google Scholar]
- Foltz, L.P.; Clegg, D.O. Patient-derived induced pluripotent stem cells for modelling genetic retinal dystrophies. Prog. Retin. Eye Res. 2019, 68, 54–66. [Google Scholar] [CrossRef]
- Pașca, S.P.; Arlotta, P.; Bateup, H.S.; Camp, J.G.; Cappello, S.; Gage, F.H.; Knoblich, J.A.; Kriegstein, A.R.; Lancaster, M.A.; Ming, G.-L.; et al. A nomenclature consensus for nervous system organoids and assembloids. Nature 2022, 609, 907–910. [Google Scholar] [CrossRef] [PubMed]
- Kruczek, K.; Swaroop, A. Pluripotent stem cell-derived retinal organoids for disease modeling and development of therapies. Stem Cells 2020, 38, 1206–1215. [Google Scholar] [CrossRef]
- Singh, R.; Cuzzani, O.; Binette, F.; Sternberg, H.; West, M.D.; Nasonkin, I.O. Pluripotent Stem Cells for Retinal Tissue Engineering: Current Status and Future Prospects. Stem Cell Rev. Rep. 2018, 14, 463–483. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Gutierrez, C.; Xue, T.; Hampton, C.; Vergara, M.N.; Cao, L.-H.; Peters, A.; Park, T.S.; Zambidis, E.T.; Meyer, J.S.; et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014, 5, 4047. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, R.; Wen, Y.-T.; Ding, D.-C.; Tsai, R.-K. Aberrant hiPSCs-Derived from Human Keratinocytes Differentiates into 3D Retinal Organoids that Acquire Mature Photoreceptors. Cells 2019, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.; Harris, R.; Bhansali, P.; Cvekl, A.; Liu, W. Intercellular Adhesion-Dependent Cell Survival and ROCK-Regulated Actomyosin-Driven Forces Mediate Self-Formation of a Retinal Organoid. Stem Cell Rep. 2016, 6, 743–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichman, S.; Terray, A.; Slembrouck, A.; Nanteau, C.; Orieux, G.; Habeler, W.; Nandrot, E.F.; Sahel, J.A.; Monville, C.; Goureau, O. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc. Natl. Acad. Sci. USA 2014, 111, 8518–8523. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Jin, Z.-B. Directed Induction of Retinal Organoids from Human Pluripotent Stem Cells. J. Vis. Exp. 2021, 170, e62298. [Google Scholar] [CrossRef] [PubMed]
- Cowan, C.S.; Renner, M.; De Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krol, J.; Szikra, T.; et al. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 2020, 182, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, A.; Hoshino, A.; Finkbeiner, C.R.; Chitsazan, A.; Dai, L.; Haugan, A.K.; Eschenbacher, K.M.; Jackson, D.L.; Trapnell, C.; Bermingham-McDonogh, O.; et al. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 2020, 30, 1644–1659. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, P.E.; Heredero Berzal, A.; Boon, C.J.F.; Quinn, P.M.J.; ten Asbroek, A.L.M.A.; Bergen, A.A. The Role of Small Molecules and Their Effect on the Molecular Mechanisms of Early Retinal Organoid Development. Int. J. Mol. Sci. 2021, 22, 7081. [Google Scholar] [CrossRef]
- Wagstaff, P.E.; ten Asbroek, A.L.M.A.; ten Brink, J.B.; Jansonius, N.M.; Bergen, A.A.B. An alternative approach to produce versatile retinal organoids with accelerated ganglion cell development. Sci. Rep. 2021, 11, 1101. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Berber, P.; Milenkovic, A.; Michaelis, L.; Weber, B.H.F. Retinal organoid differentiation methods determine organoid cellular composition. J. Transl. Genet. Genom. 2021, 5, 292–303. [Google Scholar] [CrossRef]
- Welby, E.; Lakowski, J.; Di Foggia, V.; Budinger, D.; Gonzalez-Cordero, A.; Lun, A.T.L.; Epstein, M.; Patel, A.; Cuevas, E.; Kruczek, K.; et al. Isolation and Comparative Transcriptome Analysis of Human Fetal and iPSC-Derived Cone Photoreceptor Cells. Stem Cell Rep. 2017, 9, 1898–1915. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.Q.; Barnstable, C.J. Pluripotent Stem Cells as Models of Retina Development. Mol. Neurobiol. 2019, 56, 6056–6070. [Google Scholar] [CrossRef] [PubMed]
- O’Hara-Wright, M.; Gonzalez-Cordero, A. Retinal organoids: A window into human retinal development. Development 2020, 147, dev189746. [Google Scholar] [CrossRef] [PubMed]
- Wahlin, K.J.; Maruotti, J.A.; Sripathi, S.R.; Ball, J.; Angueyra, J.M.; Kim, C.; Grebe, R.; Li, W.; Jones, B.W.; Zack, D.J. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci. Rep. 2017, 7, 766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capowski, E.E.; Samimi, K.; Mayerl, S.J.; Phillips, M.J.; Pinilla, I.; Howden, S.E.; Saha, J.; Jansen, A.D.; Edwards, K.L.; Jager, L.D.; et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 2019, 146, dev171686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idelson, M.; Alper, R.; Obolensky, A.; Ben-Shushan, E.; Hemo, I.; Yachimovich-Cohen, N.; Khaner, H.; Smith, Y.; Wiser, O.; Gropp, M.; et al. Directed Differentiation of Human Embryonic Stem Cells into Functional Retinal Pigment Epithelium Cells. Cell Stem Cell 2009, 5, 396–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandl, C.; Zimmermann, S.J.; Milenkovic, V.M.; Rosendahl, S.M.G.; Grassmann, F.; Milenkovic, A.; Hehr, U.; Federlin, M.; Wetzel, C.H.; Helbig, H.; et al. In-Depth Characterisation of Retinal Pigment Epithelium (RPE) Cells Derived from Human Induced Pluripotent Stem Cells (hiPSC). NeuroMolecular Med. 2014, 16, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Marmorstein, A.D.; Marmorstein, L.Y.; Rayborn, M.; Wang, X.; Hollyfield, J.G.; Petrukhin, K. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2000, 97, 12758–12763. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, B.R.; Siliciano, J.D.; Mooseker, M.S.; Goodenough, D.A. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986, 103, 755–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgiadis, A.; Tschernutter, M.; Bainbridge, J.W.B.; Balaggan, K.S.; Mowat, F.; West, E.L.; Munro, P.M.G.; Thrasher, A.J.; Matter, K.; Balda, M.S.; et al. The Tight Junction Associated Signalling Proteins ZO-1 and ZONAB Regulate Retinal Pigment Epithelium Homeostasis in Mice. PLoS ONE 2010, 5, e15730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, P.M.J.; Wijnholds, J. Retinogenesis of the Human Fetal Retina: An Apical Polarity Perspective. Genes 2019, 10, 987. [Google Scholar] [CrossRef] [PubMed]
- Achberger, K.; Probst, C.; Haderspeck, J.; Bolz, S.; Rogal, J.; Chuchuy, J.; Nikolova, M.; Cora, V.; Antkowiak, L.; Haq, W.; et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. eLife 2019, 8, 8–11. [Google Scholar] [CrossRef] [PubMed]
- West, E.L.; Majumder, P.; Naeem, A.; Fernando, M.; O’Hara-Wright, M.; Lanning, E.; Kloc, M.; Ribeiro, J.; Ovando-Roche, P.; Shum, I.O.; et al. Antioxidant and lipid supplementation improve the development of photoreceptor outer segments in pluripotent stem cell-derived retinal organoids. Stem Cell Rep. 2022, 17, 775–788. [Google Scholar] [CrossRef]
- Burnside, B.; Laties, A.M. Actin filaments in apical projections of the primate pigmented epithelial cell. Investig. Ophthalmol. 1976, 15, 570–575. [Google Scholar]
- Shrestha, R.; Wen, Y.-T.; Tsai, R.-K. Effective Differentiation and Biological Characterization of Retinal Pigment Epithelium Derived from Human Induced Pluripotent Stem Cells. Curr. Eye Res. 2020, 45, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, T.; Xie, H.; Khan, M.I.; Zhao, H.; Bao, J.; Zhang, M.; Xue, T. Accelerated photoreceptor differentiation of hiPSC-derived retinal organoids by contact co-culture with retinal pigment epithelium. Stem Cell Res. 2019, 39, 101491. [Google Scholar] [CrossRef]
- Nachtigal, A.-L.; Milenkovic, A.; Brandl, C.; Schulz, H.L.; Duerr, L.M.J.; Lang, G.E.; Reiff, C.; Herrmann, P.; Kellner, U.; Weber, B.H.F. Mutation-Dependent Pathomechanisms Determine the Phenotype in the Bestrophinopathies. Int. J. Mol. Sci. 2020, 21, 1597. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fisher, R.A. Statistical Methods for Research Workers; Kotz, S., Johnson, N.L., Eds.; Springer: New York, NY, USA, 1992; pp. 66–70. [Google Scholar]
- Bonferroni, C. Teoria statistica delle classi e calcolo delle probabilita. Pubbl. Del R Ist. Super. Di Sci. Econ. E Commericiali Di Firenze 1936, 8, 3–62. [Google Scholar]
- Plössl, K.; Webster, E.; Kiel, C.; Grassmann, F.; Brandl, C.; Weber, B.H.F. In vitro modeling of the complex retinal condition age-related macular degeneration. J. Transl. Genet. Genom. 2022, 6, 46–62. [Google Scholar] [CrossRef]
- Biasella, F.; Plössl, K.; Karl, C.; Weber, B.H.F.; Friedrich, U. Altered Protein Function Caused by AMD-associated Variant rs704 Links Vitronectin to Disease Pathology. Investig. Opthalmology Vis. Sci. 2020, 61, 2. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Sawada, Y.; Yoshitomi, T. Structure and function of the interphotoreceptor matrix surrounding retinal photoreceptor cells. Exp. Eye Res. 2015, 133, 3–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagley, J.A.; Reumann, D.; Bian, S.; Lévi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Massey, F.J. The Kolmogorov-Smirnov Test for Goodness of Fit. J. Am. Stat. Assoc. 1951, 46, 68–78. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berber, P.; Bondarenko, S.; Michaelis, L.; Weber, B.H.F. Transient Retention of Photoreceptor Outer Segments in Matrigel-Embedded Retinal Organoids. Int. J. Mol. Sci. 2022, 23, 14893. https://doi.org/10.3390/ijms232314893
Berber P, Bondarenko S, Michaelis L, Weber BHF. Transient Retention of Photoreceptor Outer Segments in Matrigel-Embedded Retinal Organoids. International Journal of Molecular Sciences. 2022; 23(23):14893. https://doi.org/10.3390/ijms232314893
Chicago/Turabian StyleBerber, Patricia, Sofiia Bondarenko, Lisa Michaelis, and Bernhard Heinrich Friedrich Weber. 2022. "Transient Retention of Photoreceptor Outer Segments in Matrigel-Embedded Retinal Organoids" International Journal of Molecular Sciences 23, no. 23: 14893. https://doi.org/10.3390/ijms232314893
APA StyleBerber, P., Bondarenko, S., Michaelis, L., & Weber, B. H. F. (2022). Transient Retention of Photoreceptor Outer Segments in Matrigel-Embedded Retinal Organoids. International Journal of Molecular Sciences, 23(23), 14893. https://doi.org/10.3390/ijms232314893






