miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach
Abstract
:1. Introduction
2. Results
2.1. Biometric and Metabolic Parameters of Pregnant and Offspring Rats
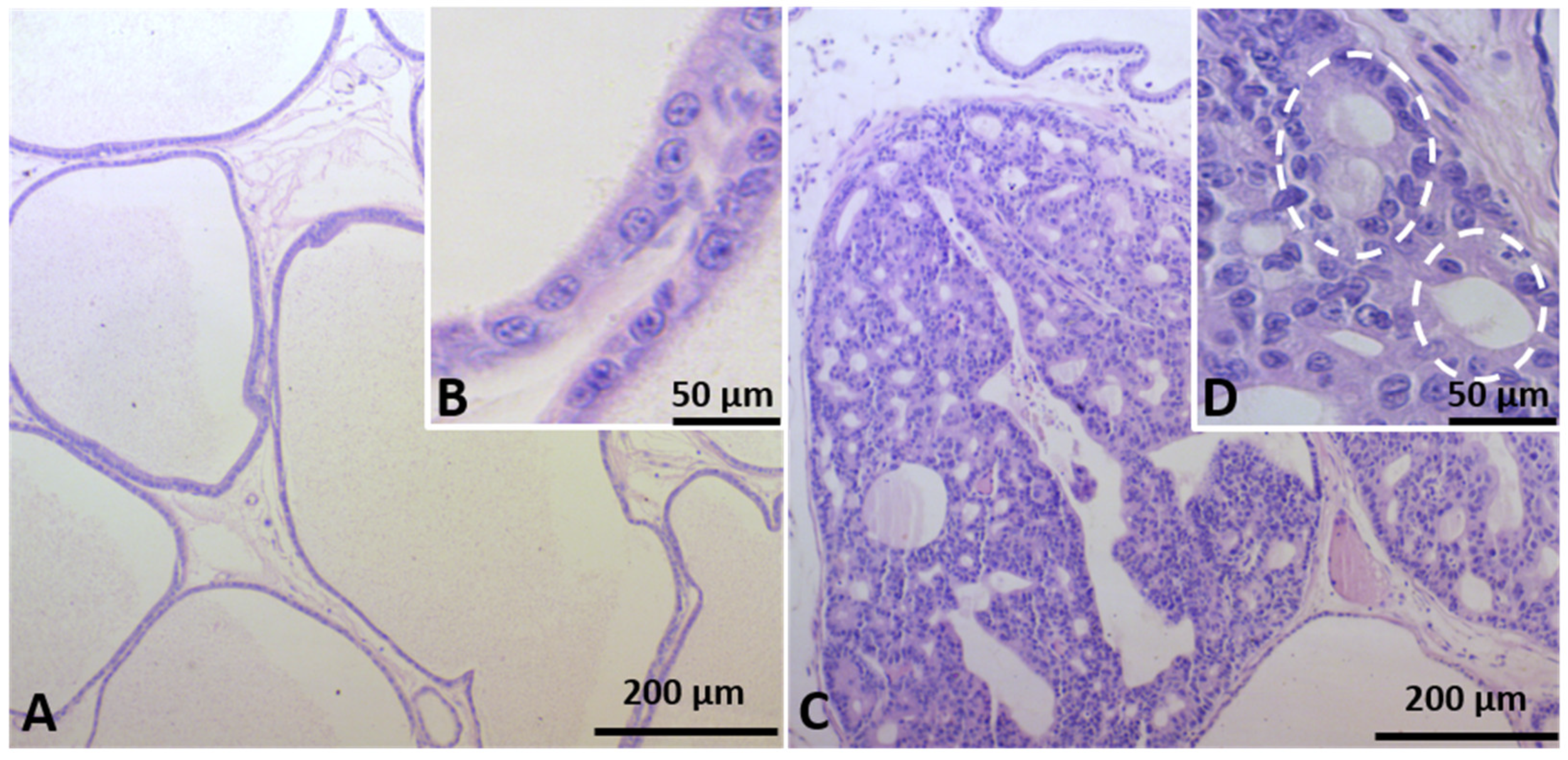
2.2. Identification of Prostate Carcinogenesis in Offspring Submitted to Maternal Malnutrition
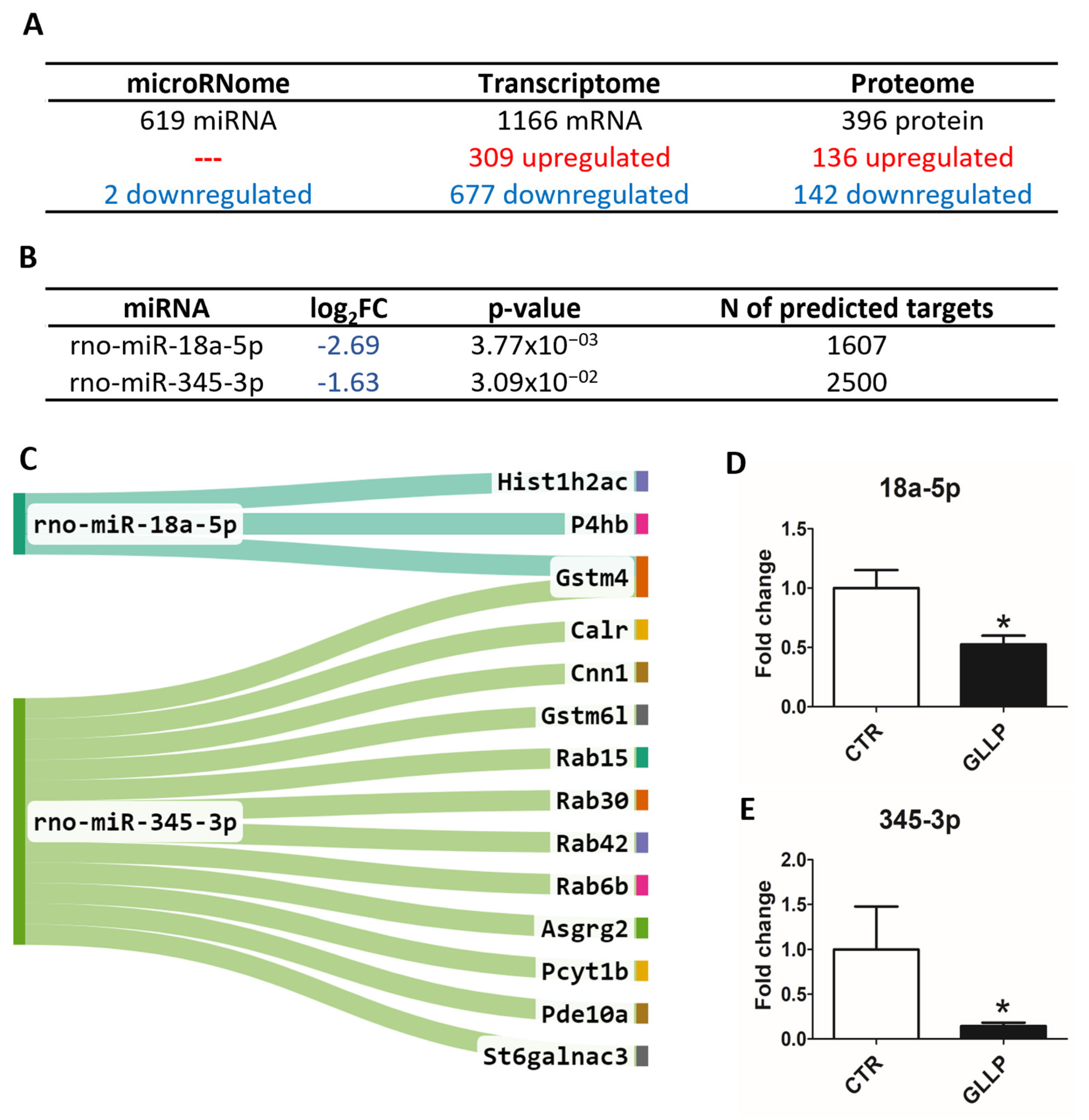
2.3. Multiomic Analysis Identified Molecular Markers Associated with Prostate Carcinogenesis in the VP of Maternally Malnourished Offspring
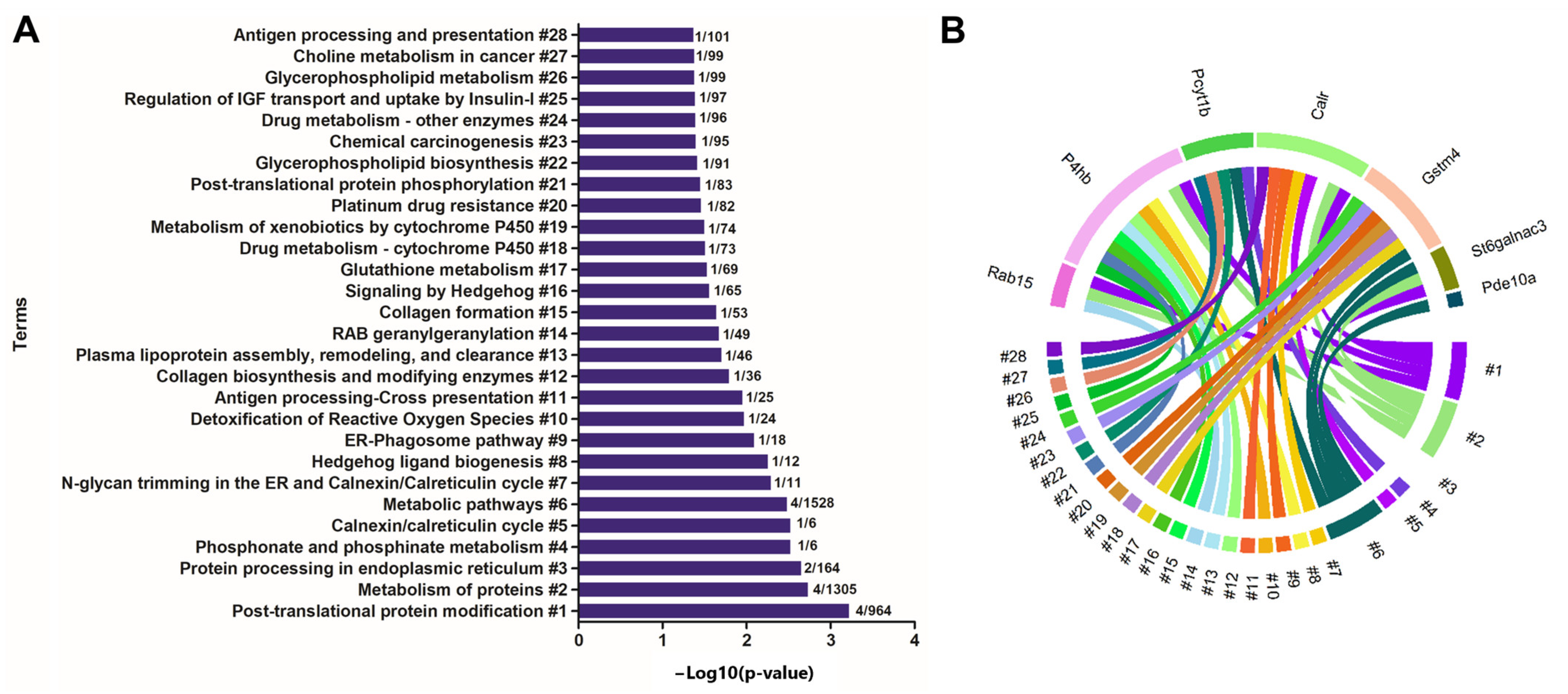
2.4. Enrichment of Ontological Terms Related to DE miRNAs-Networks
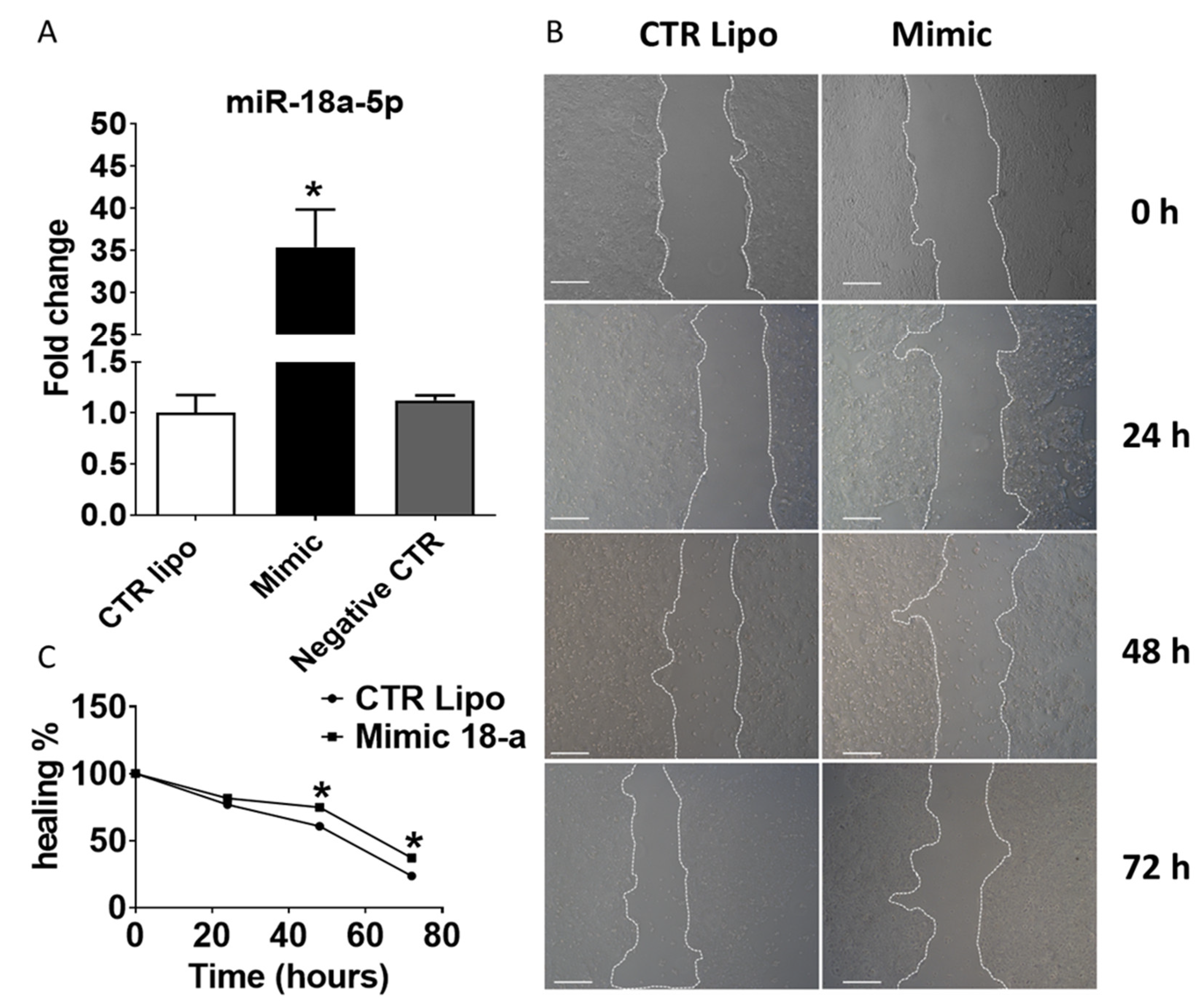
2.5. Effects of Transient Transfection of Human PNT-2 Cells on miR-18a-5p Expression and Wound Healing Capacity
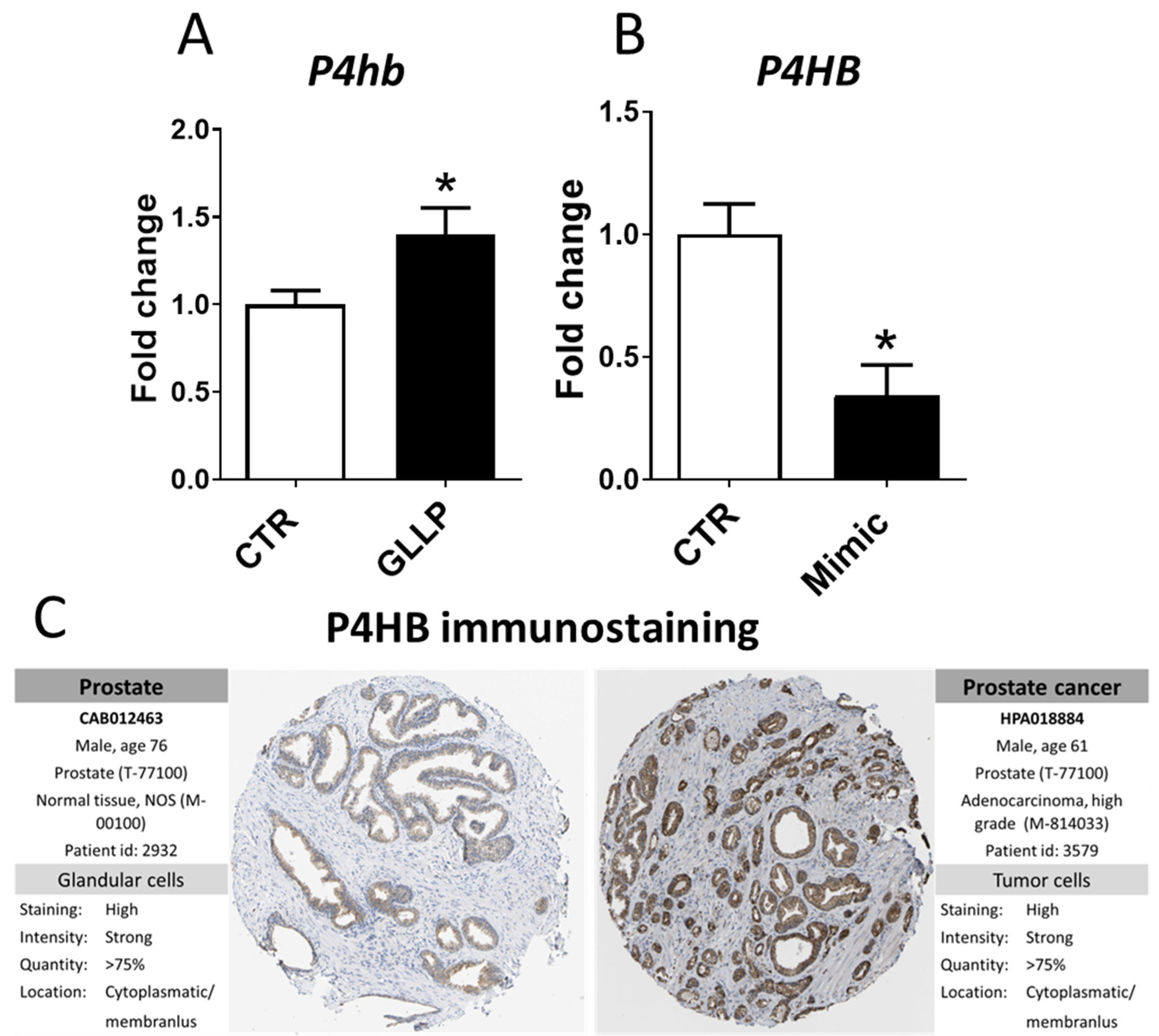
2.6. In Vitro Modulation of miR-18a-5p Confirms the Results Observed in the VP of Older Rats
2.7. Potential Modulation of the Estrogen Receptors by miR-18a-5p in the Prostate
3. Discussion
4. Material and Methods
4.1. Animals
4.2. Hormonal Analysis
4.3. Ventral Prostate Histopathology
4.4. RNA Purification, Library Construction, and Sequencing
4.5. miRNA Sequencing Analysis
4.6. mRNA Sequencing Analysis
4.7. Prediction, Filtering, and Enrichment Analyses of the DE miRNAs’ Predicted Targets
4.8. Validation of Selected miRNA-mRNA Networks in the Offspring VP by RT-qPCR
4.9. Functional Validation of Selected miRNA in a Transfected Human Prostatic Cell Line
4.10. miRNA Transfection and Cell Viability Assays
4.11. Wound Healing Assay
4.12. In Vitro and In Silico Validation of Selected miRNA-mRNA Networks
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chatterji, S.; Byles, J.; Cutler, D.; Seeman, T.; Verdes, E. Health, functioning, and disability in older adults—Present status and future implications. Lancet 2015, 385, 563–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, C.B.; Preiss, D.; Tobert, J.A.; Jacobson, T.A.; Page, I.R.L.; Goldstein, L.B.; Chin, C.; Tannock, L.R.; Miller, M.; Raghuveer, G.; et al. Statin Safety and Associated Adverse Events: A Scientific Statement From the American Heart Association. Arter. Thromb. Vasc. Biol. 2019, 39, e38–e81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, D.J.; Osmond, C.; Golding, J.; Kuh, D.; Wadsworth, M.E. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ 1989, 298, 564–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, F.M.; Pyone, T.; Broek, N.V.D. Experiences from the field: Maternal, reproductive and child health data collection in humanitarian and emergency situations. Int. Health 2015, 8, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKerracher, L.; Moffat, T.; Barker, M.; McConnell, M.; Atkinson, S.A.; Murray-Davis, B.; McDonald, S.D.; Sloboda, D.M. Knowledge about the Developmental Origins of Health and Disease is independently associated with variation in diet quality during pregnancy. Matern. Child Nutr. 2019, 16, e12891. [Google Scholar] [CrossRef] [Green Version]
- Strachan, D.P.; Hart, J.T. Fetal and placental size and risk of hypertension in adult life. BMJ 1990, 301, 552. [Google Scholar] [CrossRef] [Green Version]
- Lumey, L.H.; Van Poppel, F.W.A. The Dutch Famine of 1944-45: Mortality and Morbidity in Past and Present Generations. Soc. Hist. Med. 1994, 7, 229–246. [Google Scholar] [CrossRef]
- Kahn, H.S.; Stein, A.D.; Lumey, L.H. Prenatal environmental exposures that may influence β-cell function or insulin sensitivity in middle age. J. Dev. Orig. Health Dis. 2010, 1, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Hoek, H.W.; Susser, E.; A Buck, K.; Lumey, L.H.; Lin, S.P.; Gorman, J.M. Schizoid personality disorder after prenatal exposure to famine. Am. J. Psychiatry 1996, 153, 1637–1639. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.; Osmond, C.; Thornburg, K.L.; Kajantie, E.; Eriksson, J.G. A possible link between the pubertal growth of girls and prostate cancer in their sons. Am. J. Hum. Biol. 2012, 24, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Keinan-Boker, L.; Vin-Raviv, N.; Liphshitz, I.; Linn, S.; Barchana, M. Cancer Incidence in Israeli Jewish Survivors of World War II. JNCI J. Natl. Cancer Inst. 2009, 101, 1489–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A A Santos, S.; Camargo, A.C.; Constantino, F.B.; Colombelli, K.T.; Mani, F.; Rinaldi, J.C.; Franco, S.; Portela, L.M.F.; Duran, B.O.S.; Scarano, W.R.; et al. Maternal Low-Protein Diet Impairs Prostate Growth in Young Rat Offspring and Induces Prostate Carcinogenesis With Aging. J. Gerontol. Ser. A 2019, 74, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Scarano, W.R.; Bedrat, A.; Alonso-Costa, L.G.; Aquino, A.M.; E A Fantinatti, B.; A Justulin, L.; Barbisan, L.F.; Freire, P.P.; A Flaws, J.; Lemos, B. Exposure to an Environmentally Relevant Phthalate Mixture During Prostate Development Induces MicroRNA Upregulation and Transcriptome Modulation in Rats. Toxicol. Sci. 2019, 171, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Varuzza, M.B.; Zapaterini, J.R.; Colombelli, K.T.; Barquilha, C.N.; Justulin, L.A.; Muñoz-De-Toro, M.; Kass, L.; Barbisan, L.F. Impact of gestational low protein diet and postnatal bisphenol A exposure on chemically induced mammary carcinogenesis in female offspring rats. Environ. Toxicol. 2019, 34, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.L.; Ho, S.-M. Developmental reprogramming of cancer susceptibility. Nat. Cancer 2012, 12, 479–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, S.G.; Peeters, P.H.M.; E Grobbee, D.; Noord, P.A.H.V. Breast Cancer Risk After Caloric Restriction during the 1944–1945 Dutch Famine. JNCI J. Natl. Cancer Inst. 2004, 96, 539–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinaldi, J.C.; Justulin, L.A.; Lacorte, L.M.; Sarobo, C.; Boer, P.A.; Scarano, W.R.; Felisbino, S.L. Implications of intrauterine protein malnutrition on prostate growth, maturation and aging. Life Sci. 2013, 92, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinho, C.F.; Ribeiro, M.A.; Rinaldi, J.C.; Felisbino, S.L.; Pinheiro, P.F.; Domeniconi, R.F.; Fochi, R.A.; Boer, P.A.; Scarano, W.R. Gestational protein restriction delays prostate morphogenesis in male rats. Reprod. Fertil. Dev. 2014, 26, 967–973. [Google Scholar] [CrossRef]
- Santos, S.A.A.; Camargo, A.C.L.; Constantino, F.B.; Colombelli, K.T.; Portela, L.M.F.; Fioretto, M.N.; Vieira, J.C.S.; Padilha, P.M.; De Oliveira, M.B.; Felisbino, S.L.; et al. Identification of potential molecular pathways involved in prostate carcinogenesis in offspring exposed to maternal malnutrition. Aging 2020, 12, 19954–19978. [Google Scholar] [CrossRef]
- Portela, L.M.; Santos, S.A.; Constantino, F.B.; Camargo, A.C.; Colombelli, K.T.; Fioretto, M.N.; Barquilha, C.N.; Périco, L.L.; Hiruma-Lima, C.A.; Scarano, W.R.; et al. Increased oxidative stress and cancer biomarkers in the ventral prostate of older rats submitted to maternal malnutrition. Mol. Cell. Endocrinol. 2021, 523, 111148. [Google Scholar] [CrossRef]
- Ganu, R.; A Harris, R.; Collins, K.K.; Aagaard, K. Maternal diet: A modulator for epigenomic regulation during development in nonhuman primates and humans. Int. J. Obes. Suppl. 2012, 2, S14–S18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao-Lei, L.; Veru, F.; Elgbeili, G.; Szyf, M.; Laplante, D.P.; King, S. DNA methylation mediates the effect of exposure to prenatal maternal stress on cytokine production in children at age 13½ years: Project Ice Storm. Clin. Epigenetics 2016, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, M.; Saito, K.; Jia, H.; Kato, H. Maternal Protein Restriction and Post-Weaning High-Fat Feeding Alter Plasma Amino Acid Profiles and Hepatic Gene Expression in Mice Offspring. Foods 2022, 11, 753. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Simino, L.A.P.; Panzarin, C.; Fontana, M.F.; de Fante, T.; Geraldo, M.V.; Ignácio-Souza, L.M.; Milanski, M.; Torsoni, M.A.; Ross, M.G.; Desai, M.; et al. MicroRNA Let-7 targets AMPK and impairs hepatic lipid metabolism in offspring of maternal obese pregnancies. Sci. Rep. 2021, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Almeida-Faria, J.; Duque-Guimarães, D.E.; Ong, T.P.; Pantaleão, L.C.; Carpenter, A.A.; Loche, E.; Kusinski, L.C.; Ashmore, T.J.; Antrobus, R.; Bushell, M.; et al. Maternal obesity during pregnancy leads to adipose tissue ER stress in mice via miR-126-mediated reduction in Lunapark. Diabetologia 2021, 64, 890–902. [Google Scholar] [CrossRef]
- Ramanathan, K.; Padmanabhan, G. MiRNAs as potential biomarker of kidney diseases: A review. Cell Biochem. Funct. 2020, 38, 990–1005. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Goffin, V.; Bernichtein, S.; Touraine, P.; Kelly, P.A. Development and Potential Clinical Uses of Human Prolactin Receptor Antagonists. Endocr. Rev. 2005, 26, 400–422. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Suarez, J.A.; Brandhorst, S.; Balasubramanian, P.; Cheng, C.-W.; Madia, F.; Fontana, L.; Mirisola, M.G.; Guevara-Aguirre, J.; Wan, J.; et al. Low Protein Intake Is Associated with a Major Reduction in IGF-1, Cancer, and Overall Mortality in the 65 and Younger but Not Older Population. Cell Metab. 2014, 19, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.N.; Wlodek, M.E.; Moritz, K.M.; Cuffe, J. Programming of maternal and offspring disease: Impact of growth restriction, fetal sex and transmission across generations. J. Physiol. 2016, 594, 4727–4740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prins, G.S. Endocrine disruptors and prostate cancer risk. Endocr. -Relat. Cancer 2008, 15, 649–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, I.J.; Meyskens, F.L. African American men and hereditary/familial prostate cancer: Intermediate-risk populations for chemoprevention trials. Urology 2001, 57, 178–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Guo, C.; Zheng, W.; Wang, Q.; Zhou, L. Exosome-Mediated Transfer of miR-1323 from Cancer-Associated Fibroblasts Confers Radioresistance of C33A Cells by Targeting PABPN1 and Activating Wnt/β-Catenin Signaling Pathway in Cervical Cancer. Reprod. Sci. 2022, 29, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhu, C.; Ruan, Y.; Fan, L.; Ruan, Z.; Chen, Q.; Yuan, J.; Xu, Y.; Wang, H.; Wei, Q. hsa-miR-206b Involves in the Development of Papillary Thyroid Carcinoma via Targeting LMX1B. BioMed Res. Int. 2022, 2022, 7488708 . [Google Scholar] [CrossRef]
- Moro, M.; Fortunato, O.; Bertolini, G.; Mensah, M.; Borzi, C.; Centonze, G.; Andriani, F.; Di Paolo, D.; Perri, P.; Ponzoni, M.; et al. MiR-486-5p Targets CD133+ Lung Cancer Stem Cells through the p85/AKT Pathway. Pharmaceuticals 2022, 15, 297. [Google Scholar] [CrossRef] [PubMed]
- Dinami, R.; Petti, E.; Porru, M.; Rizzo, A.; Ganci, F.; Sacconi, A.; Ostano, P.; Chiorino, G.; Trusolino, L.; Blandino, G.; et al. TRF2 cooperates with CTCF for controlling the oncomiR-193b-3p in colorectal cancer. Cancer Lett. 2022, 533, 215607. [Google Scholar] [CrossRef]
- Klicka, K.; Grzywa, T.; Klinke, A.; Mielniczuk, A.; Włodarski, P. The Role of miRNAs in the Regulation of Endometrial Cancer Invasiveness and Metastasis—A Systematic Review. Cancers 2021, 13, 3393. [Google Scholar] [CrossRef] [PubMed]
- Namløs, H.M.; Meza-Zepeda, L.A.; Barøy, T.; Østensen, I.H.G.; Kresse, S.H.; Kuijjer, M.; Serra, M.; Bürger, H.; Cleton-Jansen, A.-M.; Myklebost, O. Modulation of the Osteosarcoma Expression Phenotype by MicroRNAs. PLoS ONE 2012, 7, e48086. [Google Scholar] [CrossRef]
- Hsu, T.-I.; Hsu, C.-H.; Lee, K.-H.; Lin, J.-T.; Chen, C.-S.; Chang, K.-C.; Su, C.-Y.; Hsiao, M.; Lu, P.-J. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 2014, 3, e99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Cai, X.; Liu, E.; Tian, X.; Tian, C. MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget 2017, 8, 68263–68269. [Google Scholar] [CrossRef] [Green Version]
- Yau, T.; Wu, C.W.; Dong, Y.; Tang, C.-M.; Ng, S.S.M.; Chan, F.K.; Sung, J.J.Y.; Yu, J. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br. J. Cancer 2014, 111, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Zhang, Y.; Tu, W.; Guo, Y. Integrated miRNA profiling and bioinformatics analyses reveal upregulated miRNAs in gastric cancer. Oncol. Lett. 2019, 18, 1979–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grassi, A.; Perilli, L.; Albertoni, L.; Tessarollo, S.; Mescoli, C.; Urso, E.D.L.; Fassan, M.; Rugge, M.; Zanovello, P. A coordinate deregulation of microRNAs expressed in mucosa adjacent to tumor predicts relapse after resection in localized colon cancer. Mol. Cancer 2018, 17, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, X.-X.; Yi, H.; Qu, J.-Q.; He, Q.-Y.; Xiao, Z.-Q. Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues. Oncol. Rep. 2015, 34, 2585–2601. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Shen, H.; Liu, L.; Xu, J.; Xu, J.; Shu, Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J. Cancer Res. Clin. Oncol. 2011, 137, 557–566. [Google Scholar] [CrossRef]
- Song, Y.; Wang, P.; Zhao, W.; Yao, Y.; Liu, X.; Ma, J.; Xue, Y.; Liu, Y. MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp. Cell Res. 2014, 324, 54–64. [Google Scholar] [CrossRef]
- Castellano, L.; Giamas, G.; Jacob, J.; Coombes, R.C.; Lucchesi, W.; Thiruchelvam, P.; Barton, G.; Jiao, L.R.; Wait, R.; Waxman, J.; et al. The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. USA 2009, 106, 15732–15737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morimura, R.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Nagata, H.; Konishi, H.; Shiozaki, A.; Ikoma, H.; Okamoto, K.; et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br. J. Cancer 2011, 105, 1733–1740. [Google Scholar] [CrossRef]
- Xi, T.; Zhang, G. Epigenetic regulation on the gene expression signature in esophagus adenocarcinoma. Pathol. Res. Pract. 2017, 213, 83–88. [Google Scholar] [CrossRef]
- Zhou, L.; Li, Z.; Pan, X.; Lai, Y.; Quan, J.; Zhao, L.; Xu, J.; Xu, W.; Guan, X.; Li, H.; et al. Identification of miR-18a-5p as an oncogene and prognostic biomarker in RCC. Am. J. Transl. Res. 2018, 10, 1874–1886. [Google Scholar] [PubMed]
- Wang, X.; Jiang, L.; Liu, Q. miR-18a-5p derived from mesenchymal stem cells-extracellular vesicles inhibits ovarian cancer cell proliferation, migration, invasion, and chemotherapy resistance. J. Transl. Med. 2022, 20, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Quiñones-Díaz, B.I.; Reyes-González, J.M.; Sánchez-Guzmán, V.; Moral, I.C.-D.; Valiyeva, F.; Santiago-Sánchez, G.S.; Vivas-Mejía, P.E. MicroRNA-18a-5p Suppresses Tumor Growth via Targeting Matrix Metalloproteinase-3 in Cisplatin-Resistant Ovarian Cancer. Front. Oncol. 2020, 10, 602670. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, L.; Li, D.; Xu, Y.; Zhang, L.; Niu, K.; Kong, R.; Gu, J.; Xu, Z.; Chen, Z.; et al. Radiosensitizing effects of miR-18a-5p on lung cancer stem-like cells via downregulating both ATM and HIF-1α. Cancer Med. 2018, 7, 3834–3847. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Qi, X.; Bian, C.; Yang, F.; Lin, X.; Zhou, S.; Xie, C.; Zhao, X.; Yi, T. MicroRNA-18a inhibits ovarian cancer growth via directly targeting TRIAP1 and IPMK. Oncol. Lett. 2017, 13, 4039–4046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolenda, T.; Guglas, K.; Kopczyńska, M.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Good or not good: Role of miR-18a in cancer biology. Rep. Pract. Oncol. Radiother. 2020, 25, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Cao, Z.; Zhu, R.; You, L.; Zhang, T. The dual functional role of MicroRNA-18a (miR-18a) in cancer development. Clin. Transl. Med. 2019, 8, 32. [Google Scholar] [CrossRef] [Green Version]
- Parakh, S.; Atkin, J.D. Novel roles for protein disulphide isomerase in disease states: A double edged sword? Front. Cell Dev. Biol. 2015, 3, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Guo, S.; Wu, Y.; Zheng, Z.-C.; Wang, Y.; Zhao, Y. P4HB, a Novel Hypoxia Target Gene Related to Gastric Cancer Invasion and Metastasis. BioMed Res. Int. 2019, 2019, 9749751 . [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, J.; Zhang, Q.; Xu, Q.; Lu, L.; Wang, J.; Xia, W. P4HB knockdown induces human HT29 colon cancer cell apoptosis through the generation of reactive oxygen species and inactivation of STAT3 signaling. Mol. Med. Rep. 2019, 19, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; He, A.; Lv, T.; Xu, C.; Lin, L.; Lin, J. Overexpression of P4HB is correlated with poor prognosis in human clear cell renal cell carcinoma. Cancer Biomark. 2019, 26, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Sun, S.; Zhang, X.-Q.; De Zhang, P.; Ho, A.S.; Kiang, K.M.; Fung, C.F.; Lui, W.M.; Leung, G.K. MicroRNA-210 and Endoplasmic Reticulum Chaperones in the Regulation of Chemoresistance in Glioblastoma. J. Cancer 2015, 6, 227–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Lee, D.; Ho, A.S.; Pu, J.K.; Zhang, X.; Lee, N.P.; Day, P.J.; Lui, W.; Fung, C.; Leung, G.K. Inhibition of prolyl 4-hydroxylase, beta polypeptide (P4HB) attenuates temozolomide resistance in malignant glioma via the endoplasmic reticulum stress response (ERSR) pathways. Neuro-Oncol. 2013, 15, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Cheong, A.; Zhang, X.; Cheung, Y.-Y.; Tang, W.-Y.; Chen, J.; Ye, S.-H.; Medvedovic, M.; Leung, Y.-K.; Prins, G.S.; Ho, S.-M. DNA methylome changes by estradiol benzoate and bisphenol A links early-life environmental exposures to prostate cancer risk. Epigenetics 2016, 11, 674–689. [Google Scholar] [CrossRef]
- Prins, G.S.; Korach, K.S. The role of estrogens and estrogen receptors in normal prostate growth and disease. Steroids 2008, 73, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Barker, D.J.P.; Osmond, C.; Thornburg, K.; Kajantie, E.; Eriksson, J.G. A possible link between the pubertal growth of girls and ovarian cancer in their daughters. Am. J. Hum. Biol. 2008, 20, 659–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zambrano, E.; Rodríguez-González, G.L.; Guzmán, C.; García-Becerra, R.; Boeck, L.; Díaz, L.; Menjivar, M.; Larrea, F.; Nathanielsz, P.W. A maternal low protein diet during pregnancy and lactation in the rat impairs male reproductive development. J. Physiol. 2005, 563, 275–284. [Google Scholar] [CrossRef]
- Prins, G.S.; Birch, L.; Tang, W.-Y.; Ho, S.-M. Developmental estrogen exposures predispose to prostate carcinogenesis with aging. Reprod. Toxicol. 2007, 23, 374–382. [Google Scholar] [CrossRef] [Green Version]
- Guzman, C.; Cabrera, R.; Cardenas, M.; Larrea, F.; Nathanielsz, P.W.; Zambrano, E. Protein restriction during fetal and neonatal development in the rat alters reproductive function and accelerates reproductive ageing in female progeny. J. Physiol. 2006, 572, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Bonkhoff, H. Estrogen receptor signaling in prostate cancer: Implications for carcinogenesis and tumor progression. Prostate 2018, 78, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Grindstad, T.; Skjefstad, K.; Andersen, S.; Ness, N.; Nordby, Y.; Al-Saad, S.; Fismen, S.; Donnem, T.; Khanehkenari, M.R.; Busund, L.-T.; et al. Estrogen receptors α and β and aromatase as independent predictors for prostate cancer outcome. Sci. Rep. 2016, 6, 33114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, M.; Prabhu, J.S.; Korlimarla, A.; Rajarajan, S.; Hari, P.S.; Kaul, R.; Alexander, A.; Raghavan, R.; Srintah, B.S.; Srintah, T.S. miR-18a activates Wnt pathway in ER-positive breast cancer and is associated with poor prognosis. Cancer Med. 2020, 9, 5587–5597. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-H.; Yeh, S.-H.; Lu, C.-C.; Yu, S.-L.; Chen, H.-Y.; Lin, C.-Y.; Chen, D.-S.; Chen, P.-J. MicroRNA-18a Prevents Estrogen Receptor-α Expression, Promoting Proliferation of Hepatocellular Carcinoma Cells. Gastroenterology 2009, 136, 683–693. [Google Scholar] [CrossRef] [PubMed]
- McPherson, S.J.; Ellem, S.J.; Risbridger, G.P. Estrogen-regulated development and differentiation of the prostate. Differentiation 2008, 76, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Reeves, F.H.; Nielsen, G.; Fahey, C. AIN-93 Purified Diets for Laboratory Rodents: Final Report of the American Institute of Nutrition Ad Hoc Writing Committee on the Reformulation of the AIN-76A Rodent Diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef] [Green Version]
- Sadowska, J.; Rygielska, M. The effect of high fructose corn syrup on the plasma insulin and leptin concentration, body weight gain and fat accumulation in rat. Adv. Clin. Exp. Med. 2019, 28, 879–884. [Google Scholar] [CrossRef]
- Puchtler, H.; Waldrop, F.S.; Meloan, S.N.; Terry, M.S.; Conner, H.M. Methacarn (methanol-Carnoy) fixation. Histochem. Cell Biol. 1970, 21, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Edgar, R. [19] Gene Expression Omnibus: Microarray Data Storage, Submission, Retrieval, and Analysis. Methods Enzymol. 2006, 411, 352–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Pena, J.G.; Treviño, V. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2015, 33, e179. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.; Tan, A. Characterization of the Cellular Reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular Localization, Substrate Dependence, and Involvement of Mitochondrial Electron Transport in MTT Reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef] [PubMed]

| Parameters | CTR | GLLP | p-Value |
|---|---|---|---|
| Pregnant body weight at gestational day 21 (g) | 368.00 ± 16.49 | 343.80 ± 14.49 * | 0.004 |
| Pregnant energy intake (Kj/day) | 310.30 ± 22.01 | 326.20 ± 61.38 | 0.3651 |
| Female offspring BW at PND 1 (g) | 7.2 ± 0.52 | 5.9 ±0.35 * | <0.0001 |
| Male offspring BW at PND 1 (g) | 7.367± 0.52 | 6.307 ± 0.49 * | <0.0001 |
| Number of pups per litter | 10.50 ± 1.87 | 10.29 ± 1.38 | 0.9264 |
| Litter male/female ratio | 0.97 ± 0.21 | 1.08 ± 0.26 | 0.1648 |
| Female offspring BW at PND 21 (g) | 47.4 ± 4.30 | 22.3 ± 4.5 | <0.0001 |
| Male offspring BW at PND 21 (g) | 37.23 ± 6.57 | 20.03 ± 3.88 * | <0.0001 |
| Absolute VP weight at PND 21 (g) | 0.32 ± 0.08 | 0.14 ± 0.03 * | <0.0001 |
| Relative VP weight at PND 21 | 0.92 ± 0.19 | 0.7893 ± 0.16 * | 0.0398 |
| Testosterone at PND 21 (ng/mL) | 0.72 ± 0.32 | 1.84 ± 0.77 * | 0.0024 |
| Estrogen at PND 21(pg/mL) | 18.28 ± 1.15 | 20.1 ± 3.35 * | 0.0177 |
| Male offspring BW at PND 540 (g) | 428.30 ± 9.73 | 376.80 ± 6.34 * | <0.0001 |
| Absolute VP weight at PND 540 (g) | 0.62 ± 0.03 | 0.65 ± 0.04 | 0.6686 |
| Relative VP weight at PND 540 | 1.44 ± 0.40 | 1.73 ± 0.60 | 0.0512 |
| Testosterone at PND 540 (ng/mL) | 2.73 ± 0.38 | 1.68 ± 0.18 * | <0.0001 |
| Estrogen at PND 540 (pg/mL) | 14.64 ± 4.76 | 35.88 ± 4.42 * | 0.0023 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, S.A.A.; Portela, L.M.F.; Camargo, A.C.L.; Constantino, F.B.; Colombelli, K.T.; Fioretto, M.N.; Mattos, R.; de Almeida Fantinatti, B.E.; Denti, M.A.; Piazza, S.; et al. miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach. Int. J. Mol. Sci. 2022, 23, 14855. https://doi.org/10.3390/ijms232314855
Santos SAA, Portela LMF, Camargo ACL, Constantino FB, Colombelli KT, Fioretto MN, Mattos R, de Almeida Fantinatti BE, Denti MA, Piazza S, et al. miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach. International Journal of Molecular Sciences. 2022; 23(23):14855. https://doi.org/10.3390/ijms232314855
Chicago/Turabian StyleSantos, Sergio Alexandre Alcantara, Luiz Marcos Frediani Portela, Ana Carolina Lima Camargo, Flavia Bessi Constantino, Ketlin Thassiani Colombelli, Matheus Naia Fioretto, Renato Mattos, Bruno Evaristo de Almeida Fantinatti, Michela Alessandra Denti, Silvano Piazza, and et al. 2022. "miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach" International Journal of Molecular Sciences 23, no. 23: 14855. https://doi.org/10.3390/ijms232314855







