Chemistry towards Biology—Instruct: Snapshot †
Abstract
:1. Introduction
2. Similarity-Mediated Property Profiling in Drug Design
3. Potential of Langmuir Balance and Isotherms in Research and Development of New Pharmaceuticals
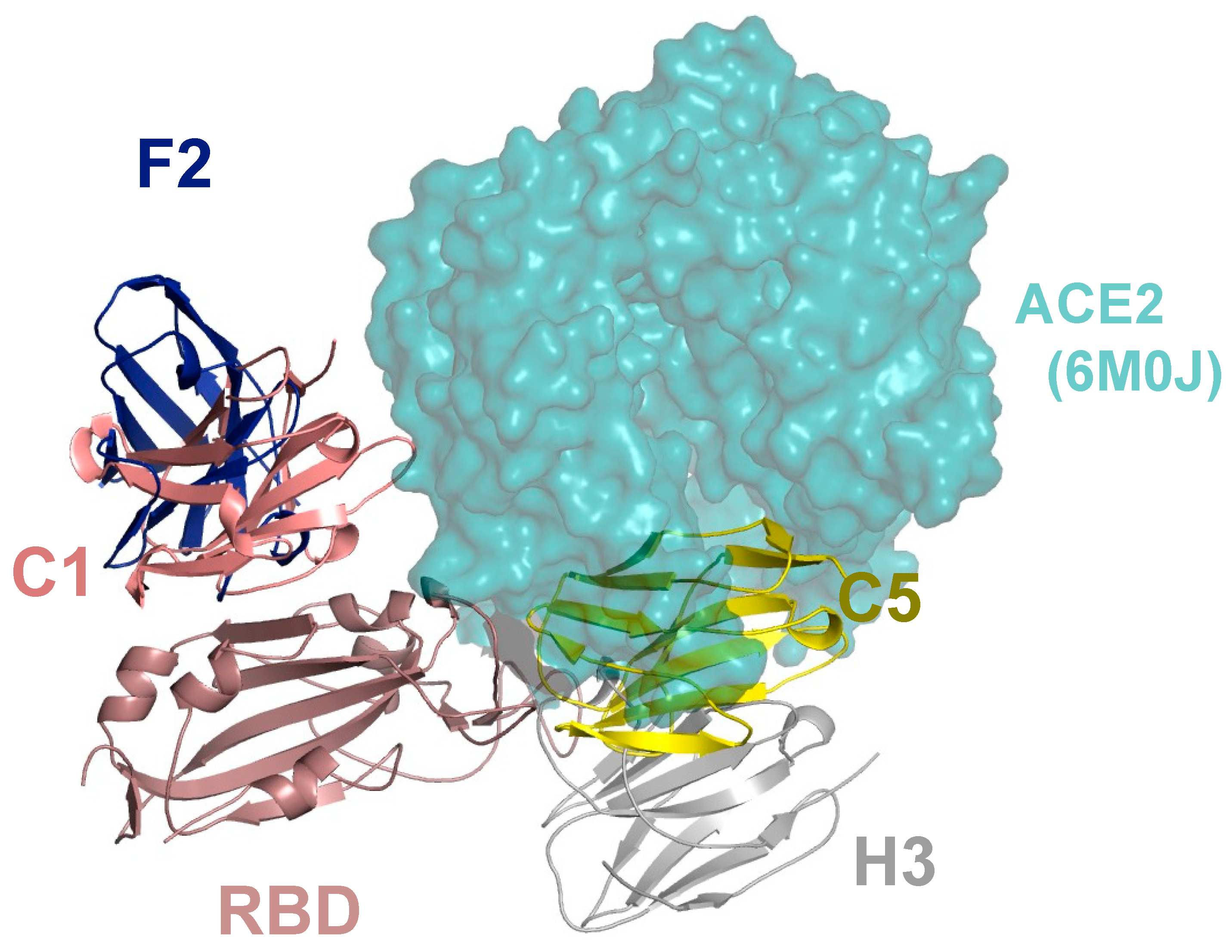
4. Structural and Functional Analysis of Nanobodies to the Spike Protein of SARS-CoV-2
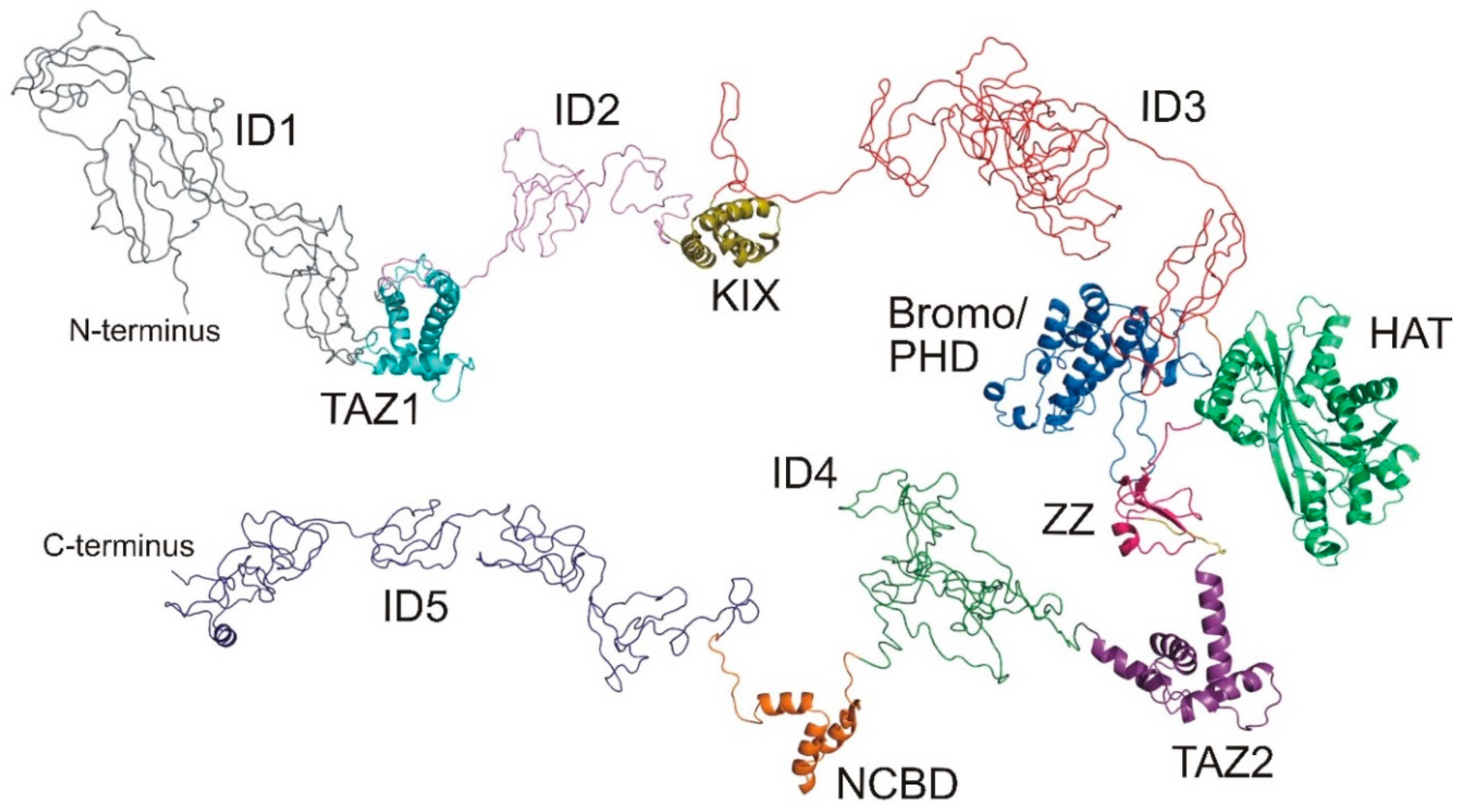
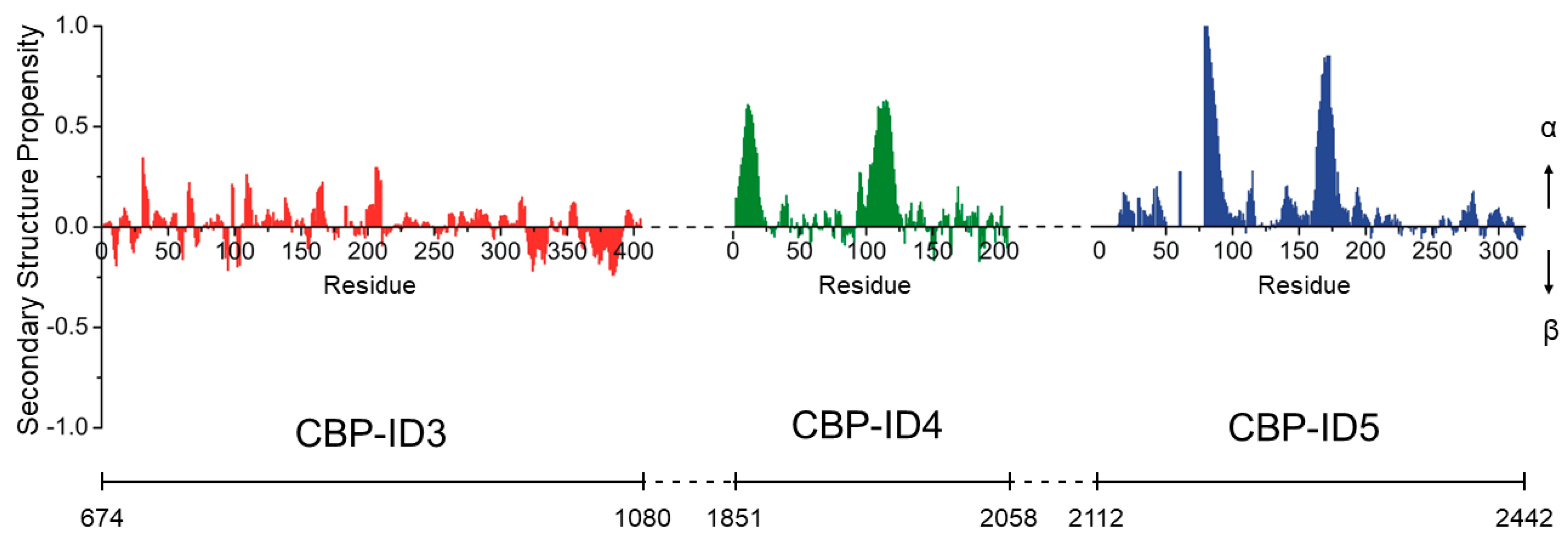
5. Just Flexible Linkers?
6. Changes of SERCA Protein after Ligand Binding
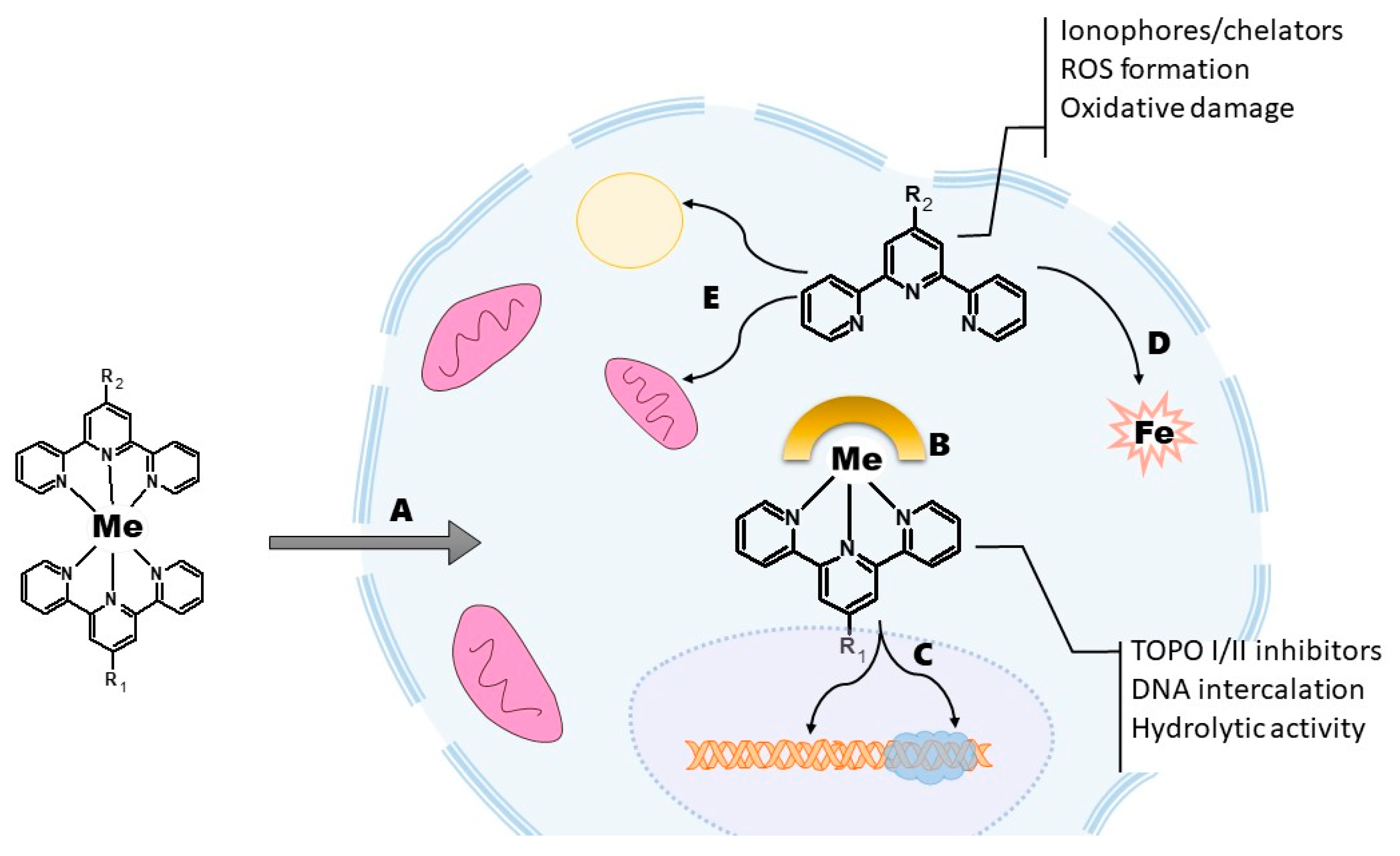
7. Should We Have Complexes with Terpyridines?
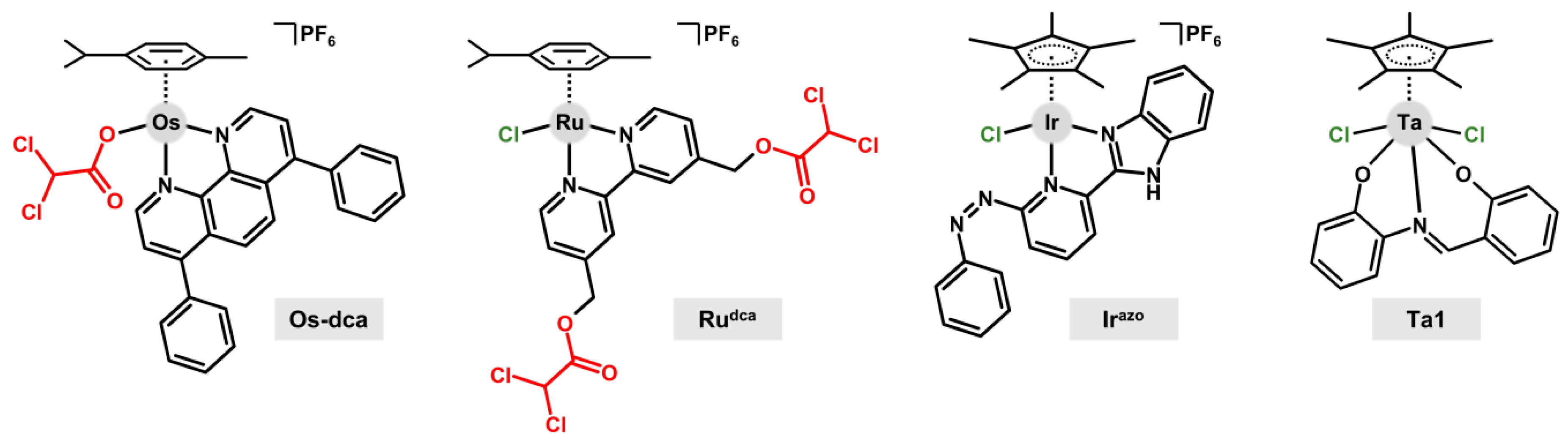
8. Transition Metal Complexes for Cancer Therapy
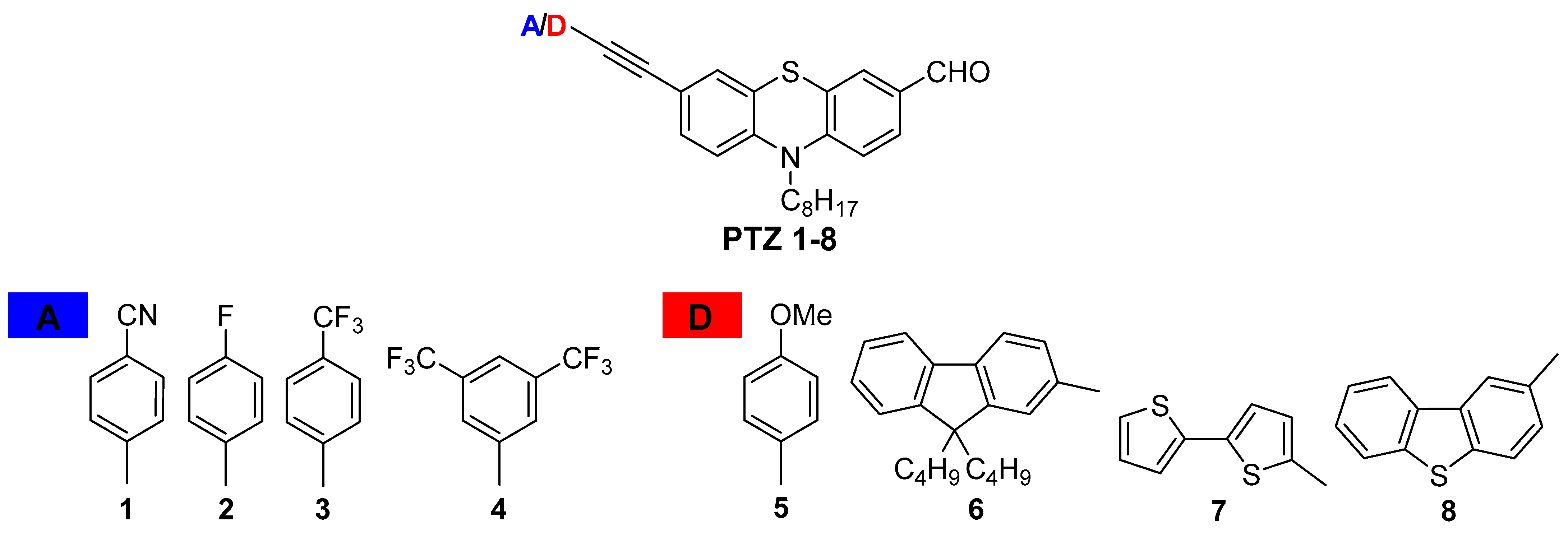
9. Effect of Donor/Acceptor (D/A) Terminal Substituents on Photophysical and Biological Properties of Phenothiazine Derivatives
10. Preparation and Characterization of Pullulan-Enriched Polymer-Ceramic Composites
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buehler, L.K. An Introduction to Molecular Interaction in Biological Systems. Available online: http://www.whatislife.com/reader/interaction-reader.html (accessed on 16 September 2022).
- Williams, L.D. Molecular Interactions and the Behaviors of Biological Macromolecules. Available online: https://williams.chemistry.gatech.edu/structure/molecular_interactions/mol_int.html (accessed on 16 September 2022).
- Chemistry towards Biology Conference Series. Available online: http://www-phch.chem.elte.hu (accessed on 16 September 2022).
- The European Research Infrastructure Consortium for Structural Biology Research. Available online: www.instruct-eric.org (accessed on 11 September 2022).
- Chemistry towards Biology 10—Instruct. Available online: https://www.instruct.sav.sk/index.html (accessed on 16 September 2022).
- Bak, A.; Kozik, V.; Walczak, M.; Fraczyk, J.; Kaminski, Z.; Kolesinska, B.; Smolinski, A.; Jampilek, J. Towards intelligent drug design system: Application of artificial dipeptide receptor library in QSAR-oriented studies. Molecules 2018, 23, 1964. [Google Scholar] [CrossRef] [Green Version]
- Van de Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef]
- Rykowski, S.; Gurda-Woźna, D.; Orlicka-Płocka, M.; Fedoruk-Wyszomirska, A.; Giel-Pietraszuk, M.; Wyszko, E.; Kowalczyk, A.; Stączek, P.; Bak, A.; Kiliszek, A.; et al. Design, synthesis, and evaluation of novel 3-carboranyl-1,8-naphthalimide derivatives as potential anticancer agents. Int. J. Mol. Sci. 2021, 22, 2772. [Google Scholar] [CrossRef]
- Bak, A.; Kozik, V.; Smolinski, A.; Jampilek, J. Multidimensional (3D/4D-QSAR) probability-guided pharmacophore mapping: Investigation of activity profile for a series of drug absorption promoters. RSC Adv. 2016, 6, 76183–76205. [Google Scholar] [CrossRef]
- Empel, A.; Bak, A.; Kozik, V.; Latocha, M.; Cizek, A.; Jampilek, J.; Suwinska, K.; Sochanik, A.; Zieba, A. Towards property profiling: Synthesis and SAR probing of new tetracyclic diazaphenothiazine analogues. Int. J. Mol. Sci. 2021, 22, 12826. [Google Scholar] [CrossRef]
- Kos, J.; Bak, A.; Kozik, V.; Jankech, T.; Strharsky, T.; Swietlicka, A.; Michnova, H.; Hosek, J.; Smolinski, A.; Oravec, M.; et al. Biological activities and ADMET-related properties of novel set of cinnamanilides. Molecules 2020, 25, 4121. [Google Scholar] [CrossRef] [PubMed]
- Chrobak, E.; Marciniec, K.; Dąbrowska, A.; Pęcak, P.; Bębenek, E.; Kadela-Tomanek, M.; Bak, A.; Jastrzębska, M.; Boryczka, S. New phosphorus analogs of bevirimat: Synthesis, evaluation of anti-HIV-1 activity and molecular docking study. Int. J. Mol. Sci. 2019, 20, 5209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggiora, G.M.; Shanmugasundaram, V. Molecular similarity measures. Methods Mol. Biol. 2011, 672, 39–100. [Google Scholar]
- Bak, A.; Kos, J.; Michnova, H.; Gonec, T.; Pospisilova, S.; Kozik, V.; Cizek, A.; Smolinski, A.; Jampilek, J. Consensus-based pharmacophore mapping for new set of N-(disubstituted-phenyl)-3-hydroxyl-naphthalene-2-carboxamides. Int. J. Mol. Sci. 2020, 21, 6583. [Google Scholar] [CrossRef]
- Michnová, H.; Pospíšilová, Š.; Goněc, T.; Kapustíková, I.; Kollár, P.; Kozik, V.; Musioł, R.; Jendrzejewska, I.; Vančo, J.; Trávníček, Z.; et al. Bioactivity of methoxylated and methylated 1-hydroxynaphthalene-2-carboxanilides: Comparative molecular surface analysis. Molecules 2019, 24, 2991. [Google Scholar] [CrossRef] [Green Version]
- Polanski, J.; Bak, A.; Gieleciak, R.; Magdziarz, T. Self-organizing neural networks for modeling robust 3D and 4D QSAR: Application to dihydrofolate reductase inhibitors. Molecules 2004, 9, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Polanski, J.; Bak, A.; Gieleciak, R.; Magdziarz, T. Modeling robust QSAR. J. Chem. Inf. Model. 2003, 46, 2310–2318. [Google Scholar] [CrossRef]
- Potemkin, V.; Grishina, M. Principles for 3D/4D QSAR classification of drugs. Drug Discov. Today 2008, 13, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Polanski, J.; Bak, A. Modeling steric and electronic effects in 3D- and 4D-QSAR schemes: Predicting benzoic pKa values and steroid CBG binding affinities. J. Chem. Inf. Comput. Sci. 2003, 43, 2081–2092. [Google Scholar] [CrossRef] [PubMed]
- Bak, A.; Polanski, J. A 4D-QSAR study on anti-HIV HEPT analogues. Bioorg. Med. Chem. 2006, 14, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Bak, A.; Polanski, J. Modeling Robust QSAR 3: SOM-4D-QSAR with iterative variable elimination IVE-PLS: Application to steroid, azo dye, and benzoic acid series. J. Chem. Inf. Model. 2007, 47, 1469–1480. [Google Scholar] [CrossRef]
- Bak, A. Two Decades of 4D-QSAR: A dying art or staging a comeback? Int. J. Mol. Sci. 2021, 22, 5212. [Google Scholar] [CrossRef]
- Bak, A.; Kozik, V.; Kozakiewicz, D.; Gajcy, K.; Strub, D.J.; Swietlicka, A.; Stepankova, S.; Imramovsky, A.; Polanski, J.; Smolinski, A.; et al. Novel Benzene-Based Carbamates for AChE/BChE Inhibition: Synthesis and Ligand/Structure-Oriented SAR Study. Int. J. Mol. Sci. 2019, 20, 1524. [Google Scholar] [CrossRef] [Green Version]
- Bak, A.; Pizova, H.; Kozik, V.; Vorcakova, K.; Kos, J.; Treml, J.; Odehnalova, K.; Oravec, M.; Imramovsky, A.; Bobal, P.; et al. SAR-mediated similarity assessment of the property profile for new, silicon-based AChE/BChE inhibitors. Int. J. Mol. Sci. 2019, 20, 5385. [Google Scholar] [CrossRef] [Green Version]
- Kos, J.; Kozik, V.; Pindjakova, D.; Jankech, T.; Smolinski, A.; Stepankova, S.; Hosek, J.; Oravec, M.; Jampilek, J.; Bak, A. Synthesis and hybrid SAR property modeling of novel cholinesterase inhibitors. Int. J. Mol. Sci. 2021, 22, 3444. [Google Scholar] [CrossRef]
- Pedrosa, M.; Maldonado-Valderrama, J.; Gálvez-Ruiz, M.J. Interactions between curcumin and cell membrane models by Langmuir monolayers. Colloids Surf. B Biointerfaces 2022, 217, 112636. [Google Scholar] [CrossRef] [PubMed]
- Scholl, F.A.; Siqueira, J.R.; Caseli, L. Graphene oxide modulating the bioelectronic properties of penicillinase immobilized in lipid Langmuir–Blodgett films. Langmuir 2022, 38, 2372–2378. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, L.; Hirvonen, J. Physicochemical characterization of nano-and microparticles. Curr. Nanosci. 2008, 4, 101–107. [Google Scholar] [CrossRef]
- Havre, T.; Ese, M.H.; Sjöblom, J.; Blokhus, A. Langmuir films of naphthenic acids at different pH and electrolyte concentrations. Colloid Polym. Sci. 2002, 280, 647–652. [Google Scholar] [CrossRef]
- de Souza, K.D.; Perez, K.R.; Duran, N.; Justo, G.Z.; Caseli, L. Interaction of violacein in models for cellular membranes: Regulation of the interaction by the lipid composition at the air-water interface. Colloids Surf. B Biointerfaces 2017, 160, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Olżyńska, A.; Wizert, A.; Štefl, M.; Iskander, D.R.; Cwiklik, L. Mixed polar-nonpolar lipid films as minimalistic models of tear film lipid layer: A Langmuir trough and fluorescence microscopy study. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183300. [Google Scholar] [CrossRef] [PubMed]
- Cañadas, O.; García-García, A.; Prieto, M.A.; Pérez-Gil, J. Polyhydroxyalkanoate nanoparticles for pulmonary drug delivery: Interaction with lung surfactant. Nanomaterials 2021, 11, 1482. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Le Bas, A.; Ruza, R.R.; Duyvesteyn, H.M.; Mikolajek, H.; Malinauskas, T.; Tan, T.K.; Rijal, P.; Dumoux, M.; Ward, P.N.; et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct Mol Biol 2020, 27, 846–854. [Google Scholar] [CrossRef]
- Mikolajek, H.; Weckner, M.; Brotzakis, Z.; Huo, J.; Dalietou, E.; Le Bas, A.; Sormanni, P.; Harrison, P.J.; Ward, P.N.; Truong, S.; et al. Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes. Proc. Natl. Acad. Sci. USA 2022, 119, e2205412119. [Google Scholar] [CrossRef]
- Bonomi, M.; Pellarin, R.; Vendruscolo, M. Simultaneous determination of protein structure and dynamics using cryo-electron microscopy. Biophys. J. 2018, 114, 1604–1613. [Google Scholar] [CrossRef]
- Bonomi, M.; Vendruscolo, M. Determination of protein structural ensembles usingcryo-electron microscopy. Curr.Opin. Struct. Biol. 2019, 56, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Girt, G.C.; Lakshminarayan, A.; Huo, J.; Dormon, J.; Norman, C.; Afrough, B.; Harding, A.; James, W.; Owens, R.J.; Naismith, J.H. The use of nanobodies in a sensitive ELISA test for SARS-CoV-2 Spike 1 protein R. Soc. Open Sci. 2021, 8, 211016. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Mikolajek, H.; Le Bas, A.; Clark, J.L.; Sharma, P.; Kipar, A.; Dormon, J.; Norman, C.; Weckener, M.; Clare, D.K.; et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021, 12, 5469. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Role of intrinsic protein disorder in the function and interactions of the transcriptional coactivators CREB-binding Protein (CBP) and p300. J. Biol. Chem. 2016, 291, 6714–6722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The IntFOLD Integrated Protein Structure and Function Prediction Server. Available online: https://www.reading.ac.uk/bioinf/IntFOLD/ (accessed on 16 September 2022).
- Gunasekaran, K.; Tsai, C.J.; Kumar, S.; Zanuy, D.; Nussinov, R. Extended disordered proteins: Targeting function with less scaffold. Trends Biochem. Sci. 2005, 28, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Piai, A.; Calçada, E.O.; Tarenzi, T.; del Grande, A.; Varadi, M.; Tompa, P.; Felli, I.C.; Pierattelli, R. Just a Flexible Linker? The structural and dynamic properties of CBP-ID4 revealed by NMR spectroscopy. Biophys. J. 2016, 110, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Martos, S.; Piai, A.; Kosol, S.; Varadi, M.; Bekesi, A.; Lebrun, P.; Volkov, A.N.; Gevaert, K.; Pierattelli, R.; Felli, I.C.; et al. Linking functions: An additional role for an intrinsically disordered linker domain in the transcriptional coactivator CBP. Sci. Rep. 2017, 7, 4676. [Google Scholar] [CrossRef] [Green Version]
- Kosol, S.; Contreras-Martos, S.; Piai, A.; Varadi, M.; Lazar, T.; Bekesi, A.; Lebrun, P.; Felli, I.C.; Pierattelli, R.; Tompa, P. Interaction between the scaffold proteins CBP by IQGAP1 provides an interface between gene expression and cytoskeletal activity. Sci. Rep. 2020, 10, 5753. [Google Scholar] [CrossRef] [Green Version]
- Murrali, M.G.; Felli, I.C.; Pierattelli, R. Adenoviral E1A exploits flexibility and disorder to target cellular proteins. Biomolecules 2020, 10, 1541. [Google Scholar] [CrossRef]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [Green Version]
- Marsh, J.A.; Singh, V.K.; Jia, Z.; Forman-Kay, J.D. Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 2006, 15, 2795–2804. [Google Scholar] [CrossRef] [PubMed]
- Neighbor Corrected Structural Propensity Calculator. Available online: https://st-protein02.chem.au.dk/ncSPC/ (accessed on 16 September 2022).
- Periasamy, M.; Kalyanasundaram, A. SERCA pump isoforms: Their role in calcium transport and disease. Muscle Nerve 2007, 35, 430–442. [Google Scholar] [CrossRef]
- Ikeda, Y. Modification of sarco-endoplasmic reticulum Ca(2+)-ATPase in the failing cardiomyocyte. Clin. Calcium 2013, 23, 535–542. [Google Scholar]
- Brini, M.; Calì, T.; Ottolini, D.; Carafoli, E. The plasma membrane calcium pump in health and disease. FEBS J. 2013, 280, 5385–5397. [Google Scholar] [CrossRef]
- Marambaud, P.; Dreses-Werringloer, U.; Vingtdeux, V. Calcium signaling in neurodegeneration. Mol. Neurodegener. 2009, 4, 20–28. [Google Scholar] [CrossRef]
- Viskupicova, J.; Majekova, M.; Horakova, L. Inhibition of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA1) by rutin derivatives. J. Muscle Res. Cell Motil. 2015, 36, 183–194. [Google Scholar] [CrossRef]
- Kang, S.; Dahl, R.; Hsieh, W.; Shin, A.; Zsebo, K.M.; Buettner, C.; Hajjar, R.J.; Lebeche, D. Small molecular allosteric activator of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) attenuates diabetes and metabolic disorders. J. Biol. Chem. 2016, 291, 5185–5198. [Google Scholar] [CrossRef] [Green Version]
- YASARA Biosciences GmbH, Vienna, Austria. Available online: www.yasara.org (accessed on 16 September 2022).
- Rodríguez, Y.; Májeková, M. Structural changes of sarco/endoplasmic reticulum Ca2+-ATPase induced by rutin arachidonate: A molecular dynamics study. Biomolecules 2020, 10, 214. [Google Scholar] [CrossRef] [Green Version]
- Clausen, J.D.; McIntosh, D.B.; Woolley, D.G.; Andersen, J.P. Modulatory ATP binding affinity in intermediate 978 states of E2P dephosphorylation of sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 2011, 286, 11792–11802. [Google Scholar] [CrossRef] [Green Version]
- Clausen, J.D.; Andersen, J.P. Glutamate 90 at the luminal ion gate of sarcoplasmic reticulum Ca2+-ATPase is 981 critical for Ca2+ binding on both sides of the membrane. J. Biol. Chem. 2010, 285, 20780–20792. [Google Scholar] [CrossRef] [Green Version]
- Mrozek-Wilczkiewicz, A.; Spaczynska, E.; Malarz, K.; Cieslik, W.; Rams-Baron, M.; Krystof, V.; Musiol, R. Design, Synthesis and in vitro activity of anticancer styrylquinolines. The p53 independent mechanism of action. PLoS ONE 2015, 10, e0142678. [Google Scholar] [CrossRef] [Green Version]
- Krawczyk, M.; Pastuch-Gawolek, G.; Mrozek-Wilczkiewicz, A.; Kuczak, M.; Skonieczna, M.; Musiol, R. Synthesis of 8-hydroxyquinoline glycoconjugates and preliminary assay of their beta1,4-GalT inhibitory and anti-cancer properties. Bioorg. Chem. 2019, 84, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Mrozek-Wilczkiewicz, A.; Kalinowski, D.S.; Musiol, R.; Finster, J.; Szurko, A.; Serafin, K.; Knas, M.; Kamalapuram, S.K.; Kovacevic, Z.; Jampilek, J.; et al. Investigating the anti-proliferative activity of styrylazanaphthalenes and azanaphthalenediones. Bioorg. Med. Chem. 2010, 18, 2664–2671. [Google Scholar] [CrossRef]
- Mularski, J.; Malarz, K.; Pacholczyk, M.; Musiol, R. The p53 stabilizing agent CP-31398 and multi-kinase inhibitors. Designing, synthesizing and screening of styrylquinazoline series. Eur. J. Med. Chem. 2019, 163, 610–625. [Google Scholar] [CrossRef]
- Malarz, K.; Mularski, J.; Pacholczyk, M.; Musiol, R. The landscape of the anti-kinase activity of the IDH1 inhibitors. Cancers 2020, 12, 536. [Google Scholar] [CrossRef] [Green Version]
- Malarz, K.; Mularski, J.; Kuczak, M.; Mrozek-Wilczkiewicz, A.; Musiol, R. Novel benzenesulfonate scaffolds with a high anticancer activity and G2/M cell cycle arrest. Cancers 2021, 13, 1790. [Google Scholar] [CrossRef] [PubMed]
- Serda, M.; Kalinowski, D.S.; Rasko, N.; Potuckova, E.; Mrozek-Wilczkiewicz, A.; Musiol, R.; Malecki, J.G.; Sajewicz, M.; Ratuszna, A.; Muchowicz, A.; et al. Exploring the anti-cancer activity of novel thiosemicarbazones generated through the combination of retro-fragments: Dissection of critical structure-activity relationships. PLoS ONE 2014, 9, e110291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malarz, K.; Mrozek-Wilczkiewicz, A.; Serda, M.; Rejmund, M.; Polanski, J.; Musiol, R. The role of oxidative stress in activity of anticancer thiosemicarbazones. Oncotarget 2018, 9, 17689–17710. [Google Scholar] [CrossRef] [Green Version]
- Rejmund, M.; Mrozek-Wilczkiewicz, A.; Malarz, K.; Pyrkosz-Bulska, M.; Gajcy, K.; Sajewicz, M.; Musiol, R.; Polanski, J. Piperazinyl fragment improves anticancer activity of triapine. PLoS ONE 2018, 13, e0188767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mrozek-Wilczkiewicz, A.; Malarz, K.; Rejmund, M.; Polanski, J.; Musiol, R. Anticancer activity of the thiosemicarbazones that are based on di-2-pyridine ketone and quinoline moiety. Eur. J. Med. Chem. 2019, 171, 180–194. [Google Scholar] [CrossRef]
- Musiol, R.; Malecki, P.; Pacholczyk, M.; Mularski, J. Terpyridines as promising antitumor agents: An overview of their discovery and development. Expert Opin. Drug Discov. 2022, 17, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; He, Y.; Shi, X.; Song, Z. Terpyridine-metal complexes: Applications in catalysis and supramolecular chemistry. Coord. Chem. Rev. 2019, 385, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Hasslauer, I.; Kurth, D.G. From terpyridine-based assemblies to metallo-supramolecular polyelectrolytes (MEPEs). Adv. Colloid. Interface Sci. 2014, 207, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Saccone, D.; Magistris, C.; Barbero, N.; Quagliotto, P.; Barolo, C.; Viscardi, G. Terpyridine and quaterpyridine complexes as sensitizers for photovoltaic applications. Materials 2016, 9, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monro, S.; Colon, K.L.; Yin, H.; Roque, J.; Konda, P.; Gujar, S.; Thummel, R.P.; Lilge, L.; Cameron, C.G.; McFarland, S.A. Transition metal complexes and photodynamic therapy from a tumor-centered approach: Challenges, opportunities, and highlights from the development of TLD1433. Chem. Rev. 2019, 119, 797–828. [Google Scholar] [CrossRef] [PubMed]
- Beller, G.; Lente, G.; Fabian, I. Kinetics and mechanism of the autocatalytic oxidation of bis(terpyridine)iron(II) by peroxomonosulfate ion (oxone) in acidic medium. Inorg. Chem. 2017, 56, 8270–8277. [Google Scholar] [CrossRef]
- Delgado, G.Y.S.; Paschoal, D.; de Oliveira, M.A.L.; Dos Santos, H.F. Structure and redox stability of [Au(III)(X^N^X)PR3] complexes (X=C or N) in aqueous solution: The role of phosphine auxiliary ligand. J. Inorg. Biochem. 2019, 200, 110804. [Google Scholar] [CrossRef] [PubMed]
- Grau, J.; Caubet, A.; Roubeau, O.; Montpeyo, D.; Lorenzo, J.; Gamez, P. Time-dependent cytotoxic properties of terpyridine-based copper complexes. Chembiochem 2020, 21, 2348–2355. [Google Scholar] [CrossRef]
- Miller, C.J.; Rose, A.L.; Waite, T.D. Importance of iron complexation for fenton-mediated hydroxyl radical production at circumneutral pH. Front. Mar. Sci. 2016, 3, 134. [Google Scholar] [CrossRef] [Green Version]
- Malarz, K.; Zych, D.; Gawecki, R.; Kuczak, M.; Musiol, R.; Mrozek-Wilczkiewicz, A. New derivatives of 4’-phenyl-2,2’:6’,2’’-terpyridine as promising anticancer agents. Eur. J. Med. Chem. 2021, 212, 113032. [Google Scholar] [CrossRef]
- Malarz, K.; Zych, D.; Kuczak, M.; Musiol, R.; Mrozek-Wilczkiewicz, A. Anticancer activity of 4’-phenyl-2,2’:6’,2’’-terpyridines—Behind the metal complexation. Eur. J. Med. Chem. 2020, 189, 112039. [Google Scholar] [CrossRef]
- Zych, D.; Slodek, A.; Krompiec, S.; Malarz, K.; Mrozek-Wilczkiewicz, A.; Musiol, R. 4′-Phenyl-2,2′:6′,2″-terpyridine Derivatives Containing 1-Substituted-2,3-Triazole Ring: Synthesis, Characterization and Anticancer Activity. ChemistrySelect 2018, 3, 7009–7017. [Google Scholar] [CrossRef]
- Rosenberg, B.; Van Camp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef]
- Anthony, E.J.; Bolitho, E.M.; Bridgewater, H.E.; Carter, O.W.L.; Donnelly, J.M.; Imberti, C.; Lant, E.C.; Lermyte, F.; Needham, R.J.; Palau, M.; et al. Metallodrugs are unique: Opportunities and challenges of discovery and development. Chem. Sci. 2020, 11, 12888–12917. [Google Scholar] [CrossRef]
- Berger, M.R.; Garzon, F.T.; Keppler, B.K.; Schmahl, D. Efficacy of new ruthenium complexes against chemically induced autochthonous colorectal carcinoma in rats. Anticancer Res. 1989, 9, 761–765. [Google Scholar]
- Štarha, P.; Trávníček, Z. Non-platinum complexes containing releasable biologically active ligands. Coord. Chem. Rev. 2019, 395, 130–145. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Popovic-Nikolic, M.R.; Nikolic, K.; Uliassi, E.; Bolognesi, M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Štarha, P.; Trávníček, Z.; Vančo, J.; Dvořák, Z. Half-sandwich Ru(II) and Os(II) bathophenanthroline complexes containing a releasable dichloroacetato ligand. Molecules 2018, 23, 420. [Google Scholar] [CrossRef] [Green Version]
- Madhok, B.M.; Yeluri, S.; Perry, S.L.; Hughes, T.A.; Jayne, D.G. Dichloroacetate induces apoptosis and cell-cycle arrest in colorectal cancer cells. Br. J. Cancer 2010, 102, 1746–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pracharova, J.; Novohradsky, V.; Kostrhunova, H.; Štarha, P.; Trávníček, Z.; Kasparkova, J.; Brabec, V. Half-sandwich Os(II) and Ru(II) bathophenanthroline complexes: Anticancer drug candidates with unusual potency and a cellular activity profile in highly invasive triple-negative breast cancer cells. Dalton Trans. 2018, 47, 12197–12208. [Google Scholar] [CrossRef]
- Novohradsky, V.; Markova, L.; Kostrhunova, H.; Trávníček, Z.; Brabec, V.; Kasparkova, J. An anticancer Os(II) bathophenanthroline complex as a human breast cancer stem cell-selective, mammosphere potent agent that kills cells by necroptosis. Sci. Rep. 2019, 9, 13327. [Google Scholar] [CrossRef] [Green Version]
- Masaryk, L.; Nemec, I.; Kašpárková, J.; Brabec, V.; Štarha, P. Unexpected solution behaviour of ester-functionalized half-sandwich Ru(II) and Ir(III) complexes. Dalton Trans. 2021, 50, 8017–8028. [Google Scholar] [CrossRef] [PubMed]
- Masaryk, L.; Muthná, D.; Halaš, P.; Zoufalý, P.; Peterová, E.; Havelek, R.; Drahoš, B.; Milde, D.; Mrkvicová, A.; Štarha, P. Stability of a half-sandwich Os(II) complex with indomethacin-functionalized ligand in the presence of carboxypeptidase A. Dalton Trans. 2022, 51, 9213–9217. [Google Scholar] [CrossRef]
- Masaryk, L.; Orvoš, J.; Słoczyńska, K.; Herchel, R.; Moncol, J.; Milde, D.; Halaš, P.; Křikavová, R.; Koczurkiewicz-Adamczyk, P.; Pękala, E.; et al. Anticancer half-sandwich Ir(III) complex and its interaction with various biomolecules and their mixtures—A case study with ascorbic acid. Inorg. Chem. Front. 2022, 9, 3758–3770. [Google Scholar] [CrossRef]
- Štarha, P.; Trávníček, Z.; Dvořák, Z. A cytotoxic tantalum(V) half-sandwich complex: A new challenge for metal-based anticancer agents. Chem. Commun. 2018, 54, 9533–9536. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.P.; Kong, X.; Jenekhe, S.A. High-performance organic light-emitting diodes based on intramolecular charge-transfer emission from donor–acceptor molecules: Significance of electron- donor strength and molecular geometry. Adv. Funct. Mater. 2006, 16, 1057–1066. [Google Scholar] [CrossRef]
- Tacca, A.; Po, R.; Caldararo, M.; Chiaberge, S.; Gila, L.; Longo, L.; Mussini, P.R.; Pellegrino, A.; Perin, N.; Salvalaggio, M.; et al. Ternary thiophene-X-thiophene semiconductor building blocks (X = fluorene, carbazole, phenothiazine): Modulating electronic properties and electropolymerization ability by tuning the X core. Electrochim. Acta 2011, 56, 6638–6653. [Google Scholar] [CrossRef]
- Slodek, A.; Zych, D.; Kotowicz, S.; Szafraniec-Gorol, G.; Zimosz, S.; Schab-Balcerzak, E.; Siwy, M.; Grzelak, J.; Maćkowski, S. “Small in size but mighty in force”—The first principle study of the impact of A/D units in A/D-phenyl-π-phenothiazine-π-dicyanovinyl systems on photophysical and optoelectronic properties. Dye. Pigment. 2021, 189, 109248. [Google Scholar] [CrossRef]
- Slodek, A.; Zych, D.; Golba, S.; Zimosz, S.; Gnida, P.; Schab-Balcerzak, E. Dyes based on the D/A-acetylene linker-phenothiazine system for developing efficient dye-sensitized solar cells. J. Mat. Chem. C 2019, 7, 5830–5840. [Google Scholar] [CrossRef]
- Slodek, A.; Zych, D.; Szafraniec-Gorol, G.; Gnida, P.; Vasylieva, M.; Schab-Balcerzak, E. Investigations of new phenothiazine-based compounds for dye-sensitized solar cells with theoretical insight. Materials 2020, 13, 2292. [Google Scholar] [CrossRef] [PubMed]
- Zimosz, S.; Slodek, A.; Gnida, P.; Glinka, A.; Ziółek, M.; Zych, D.; Pająk, A.K.; Vasylieva, M.; Schab-Balcerzak, E. New D−π–D−π–A systems based on phenothiazine derivatives with imidazole structures for photovoltaics. J. Phys. Chem. C 2022, 126, 8986–8999. [Google Scholar] [CrossRef]
- Pluta, K.; Morak-Mlodawska, B.; Jelen, M. Recent progress in biological activities of synthesized phenothiazines. Eur. J. Med. Chem. 2011, 46, 3179–3189. [Google Scholar] [CrossRef]
- Matada, M.N.; Jathi, K.; Rangappa, M.M.; Geoffry, K.; Kumar, S.R.; Nagarajappa, R.B.; Zahara, F.N. A new sulphur containing heterocycles having azo linkage: Synthesis, structural characterization and biological evaluation. J. King Saud Univ. Sci. 2020, 32, 3313–3320. [Google Scholar] [CrossRef]
- Zimosz, S.; Zych, D.; Szafraniec-Gorol, G.; Kotowicz, S.; Malarz, K.; Musioł, R.; Slodek, A. Does the change in the length of the alkyl chain bring us closer to the compounds with the expected photophysical and biological properties?—Studies based on D-π-D-A imidazole-phenothiazine system. J. Mol. Liq. 2022, 365, 120076. [Google Scholar] [CrossRef]
- Slodek, A.; Zych, D.; Maroń, A.; Gawecki, R.; Mrozek-Wilczkiewicz, A.; Malarz, K.; Musioł, R. Phenothiazine derivatives—Synthesis, characterization, and theoretical studies with an emphasis on the solvatochromic properties. J. Mol. Liq. 2019, 285, 515–525. [Google Scholar] [CrossRef]
- Kraemer, C.S.; Zeitler, K.; Mueller, T.J.J. Synthesis of functionalized ethynylphenothiazine fluorophores. Org. Lett. 2000, 2, 3723–3726. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Lu, R.; Zhou, H.; Zhang, X.; Xu, T.; Liu, X.; Zhao, Y. Synthesis of linear monodisperse vinylene-linked phenothiazine oligomers. Tetrahedron Lett. 2007, 48, 7582–7585. [Google Scholar] [CrossRef]
- Zhou, N.; Wang, L.; Thompson, D.W.; Zhao, Y. OPE/OPV H-mers: Synthesis, electronic properties, and spectroscopic responses to binding with transition metal ions. Tetrahedron 2011, 67, 125–143. [Google Scholar] [CrossRef]
- Wan, W.; Wang, H.; Lin, H.; Wang, J.; Jiang, Y.; Jiang, H.; Zhu, S.; Wang, Z.; Hao, J. Synthesis, electrochemical, photophysical, and electroluminescent properties of organic dyes containing pyrazolo [3,4-b]quinoline chromophores. Dyes Pigment. 2015, 121, 138–146. [Google Scholar] [CrossRef]
- Kraemer, C.S.; Mueller, T.J.J. Synthesis and electronic properties of alkynylated phenothiazines. Eur. J. Org. Chem. 2003, 18, 3534–3548. [Google Scholar] [CrossRef]
- Chen, F.M.; Liu, X. Advancing biomaterials of human origin for tissue engineering. Prog. Polym. Sci. 2016, 53, 86–168. [Google Scholar]
- Othman, Z.; Cillero Pastor, B.; van Rijt, S.; Habibovic, P. Understanding interactions between biomaterials and biological systems using proteomics. Biomaterials 2018, 167, 191–204. [Google Scholar] [CrossRef]
- Jurczyk, M.; Jakubowicz, J. Bionanomateriały; Wydawnictwo Politechniki Poznańskiej: Poznań, Poland, 2008. [Google Scholar]
- Mosas, K.K.A.; Chandrasekar, A.R.; Dasan, A.; Pakseresht, A.; Galusek, D. Recent advancements in materials and coatings for biomedical implants. Gels 2022, 8, 323. [Google Scholar] [CrossRef]
- Gloria, A.; De Santis, R.; Ambrosio, L. Polymer-based composite scaffolds for tissue engineering. J. Appl. Biomater. Biomech. 2010, 8, 57–67. [Google Scholar]
- Świeczko-Żurek, B. Biomateriały; Wydawnicwo Politech Gdańskiej: Gdańsk, Poland, 2009; pp. 32–45. [Google Scholar]
- Boanini, E.; Silingardi, F.; Gazzano, M.; Bigi, A. Synthesis and hydrolysis of brushite (DCPD): The role of ionic substitution. Cryst. Growth Des. 2021, 21, 1689–1697. [Google Scholar] [CrossRef]
- Singh, S.; Singh, V.; Aggarwal, S.; Mandal, U.K. Synthesis of brushite nanoparticles at different temperatures. Chem. Pap. 2010, 64, 491–498. [Google Scholar] [CrossRef]
- Grover, L.M.; Knowles, J.C.; Fleming, G.J.P.; Barralet, J.E. In vitro ageing of brushite calcium phosphate cement. Biomaterials 2003, 24, 4133–4141. [Google Scholar] [CrossRef] [PubMed]
- Penel, G.; Leroy, N.; Van Landuyt, P.; Flautre, B.; Hardouin, P.; Lemaître, J.; Leroy, G. Raman microspectrometry studies of brushite cement: In vivo evolution in a sheep model. Bone 1999, 25 (Suppl. S1), 81–84. [Google Scholar] [CrossRef]
- Pina, S.; Ferreira, J.M.F. Brushite-forming Mg-, Zn- and Sr-substituted bone cements for clinical applications. Materials 2010, 3, 519–535. [Google Scholar] [CrossRef] [Green Version]
- Tamimi, F.; Kumarasami, B.; Doillon, C.; Gbureck, U.; Le Nihouannen, D.; Cabarcos, E.L.; Barralet, J.E. Brushite-collagen composites for bone regeneration. Acta Biomater. 2008, 4, 1315–1321. [Google Scholar] [CrossRef]
- Altundal, S.; Gross, K.A. Production of a brushite/silk composite powder for coatings. In Key Engineering Materials; Trans Tech Publications Ltd.: Wallerau, Switzerland, 2019; Volume 800, pp. 75–79. [Google Scholar]
- Słota, D.; Florkiewicz, W.; Sobczak-Kupiec, A. Ceramic-polymer coatings on Ti-6Al-4V alloy modified with L-cysteine in biomedical applications. Mater Today Commun. 2020, 25, 101301. [Google Scholar] [CrossRef]
- Cateni, F.; Zacchigna, M.; Procida, G. Synthesis and Controlled Drug Delivery Studies Of A Novel Ubiquinol-Polyethylene Glycol-Vitamin E adduct. Bioorg. Chem. 2020, 105, 104329. [Google Scholar] [CrossRef]
- Tyliszczak, B.; Pielichowski, K. Charakterystyka matryc hydrożelowych—Zastosowania biomedyczne superabsorbentów polimerowych. Czas Tech. 2007, 1, 160–167. [Google Scholar]
- Zhang, X.; Qiao, J.; Zhao, H.; Huang, Z.; Liu, Y.; Fang, M.; Wu, X.; Mina, X. Preparation and performance of novel polyvinylpyrrolidone/polyethylene glycol phase change materials composite fibers by centrifugal spinning. Chem. Phys. Lett. 2018, 691, 314–318. [Google Scholar] [CrossRef]
- Arora, A.; Sharma, P.; Katti, D.S. Pullulan-based composite scaffolds for bone tissue engineering: Improved osteoconductivity by pore wall mineralization. Carbohydr. Polym. 2015, 123, 180–189. [Google Scholar]
- Cheng, K.C.; Demirci, A.; Catchmark, J.M. Pullulan: Biosynthesis, production, and applications. Appl. Microbiol. Biotechnol. 2011, 92, 29–44. [Google Scholar] [CrossRef]
- Ritz, U.; Kögler, P.; Höfer, I.; Frank, P.; Klees, S.; Gebhard, S.; Brendel, C.; Kaufmann, K.; Hofmann, A.; Rommens, P.M.; et al. Photocrosslinkable polysaccharide hydrogel composites based on dextran or pullulan-amylose blends with cytokines for a human co-culture model of human osteoblasts and endothelial cells. J. Mater. Chem. B 2016, 4, 6552–6564. [Google Scholar] [CrossRef]

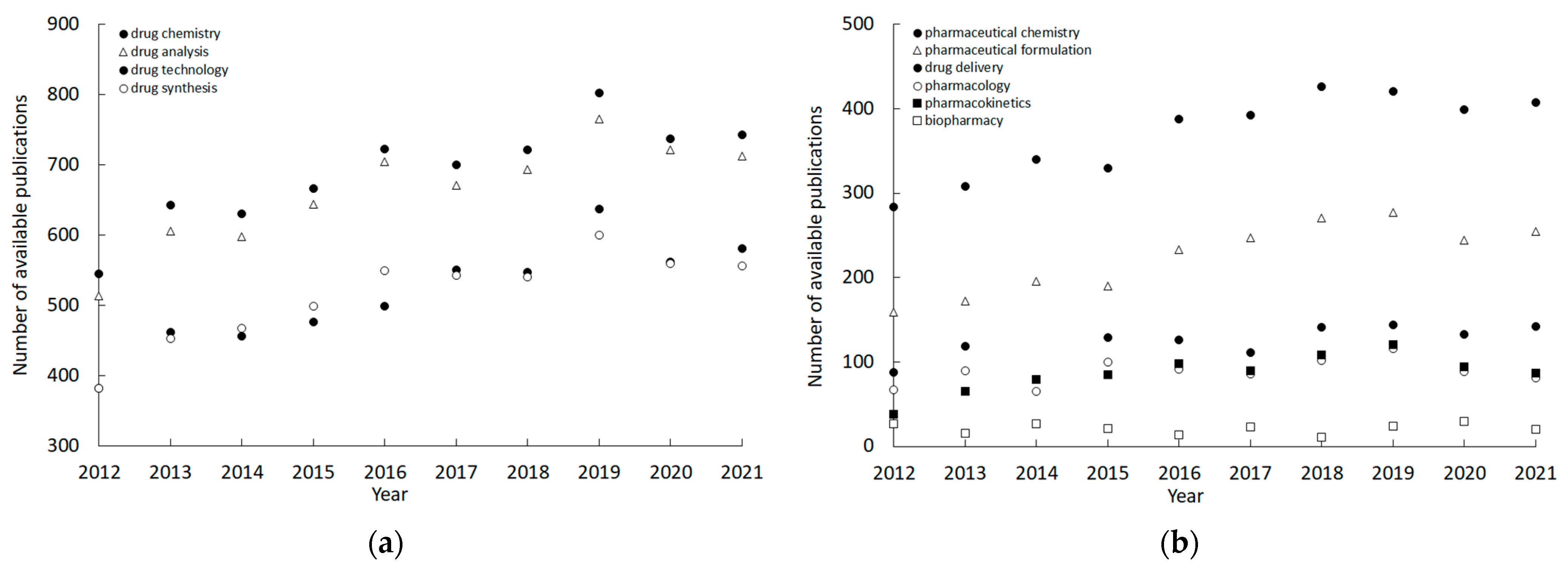

| Application Field | Number of Papers [%] |
|---|---|
| Drug chemistry | 20.97 |
| Drug analysis | 20.11 |
| Drug technology | 15.63 |
| Drug synthesis | 15.62 |
| Pharmaceutical chemistry | 11.21 |
| Pharmaceutical formulation | 6.81 |
| Drug delivery | 3.68 |
| Pharmacology | 2.69 |
| Pharmacokinetics | 2.62 |
| Biopharmacy | 0.65 |
| Rutin Derivative | Abbreviation | Corresponding Acyl | IC50 [µM] 1 | Score |
|---|---|---|---|---|
| rutin palmitate | R16 | Palmitoyl | 64 ± 12 | −8.3 |
| rutin stearate | R18 | Stearoyl | 35 ± 6.5 | −9.7 |
| rutin oleate | R18:1 | Oleoyl | 50 ± 8.5 | −9.8 |
| rutin linoleate | R18:2 | Linoleoyl | 25 ± 5.5 | −9.6 |
| rutin linolenate | R18:3 | a-linolenoyl | 62 ± 9 | −9.5 |
| rutin arachidonate | R20:4 | Arachidonoyl | 23 ± 6.5 | −11.0 |
| rutin erucate | R22:1 | Erucoyl | 50 ± 8 | −9.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hricovíni, M.; Owens, R.J.; Bak, A.; Kozik, V.; Musiał, W.; Pierattelli, R.; Májeková, M.; Rodríguez, Y.; Musioł, R.; Slodek, A.; et al. Chemistry towards Biology—Instruct: Snapshot. Int. J. Mol. Sci. 2022, 23, 14815. https://doi.org/10.3390/ijms232314815
Hricovíni M, Owens RJ, Bak A, Kozik V, Musiał W, Pierattelli R, Májeková M, Rodríguez Y, Musioł R, Slodek A, et al. Chemistry towards Biology—Instruct: Snapshot. International Journal of Molecular Sciences. 2022; 23(23):14815. https://doi.org/10.3390/ijms232314815
Chicago/Turabian StyleHricovíni, Miloš, Raymond J. Owens, Andrzej Bak, Violetta Kozik, Witold Musiał, Roberta Pierattelli, Magdaléna Májeková, Yoel Rodríguez, Robert Musioł, Aneta Slodek, and et al. 2022. "Chemistry towards Biology—Instruct: Snapshot" International Journal of Molecular Sciences 23, no. 23: 14815. https://doi.org/10.3390/ijms232314815
APA StyleHricovíni, M., Owens, R. J., Bak, A., Kozik, V., Musiał, W., Pierattelli, R., Májeková, M., Rodríguez, Y., Musioł, R., Slodek, A., Štarha, P., Piętak, K., Słota, D., Florkiewicz, W., Sobczak-Kupiec, A., & Jampílek, J. (2022). Chemistry towards Biology—Instruct: Snapshot. International Journal of Molecular Sciences, 23(23), 14815. https://doi.org/10.3390/ijms232314815


_Kim.png)








