First Insights into the Venom Composition of Two Ecuadorian Coral Snakes
Abstract
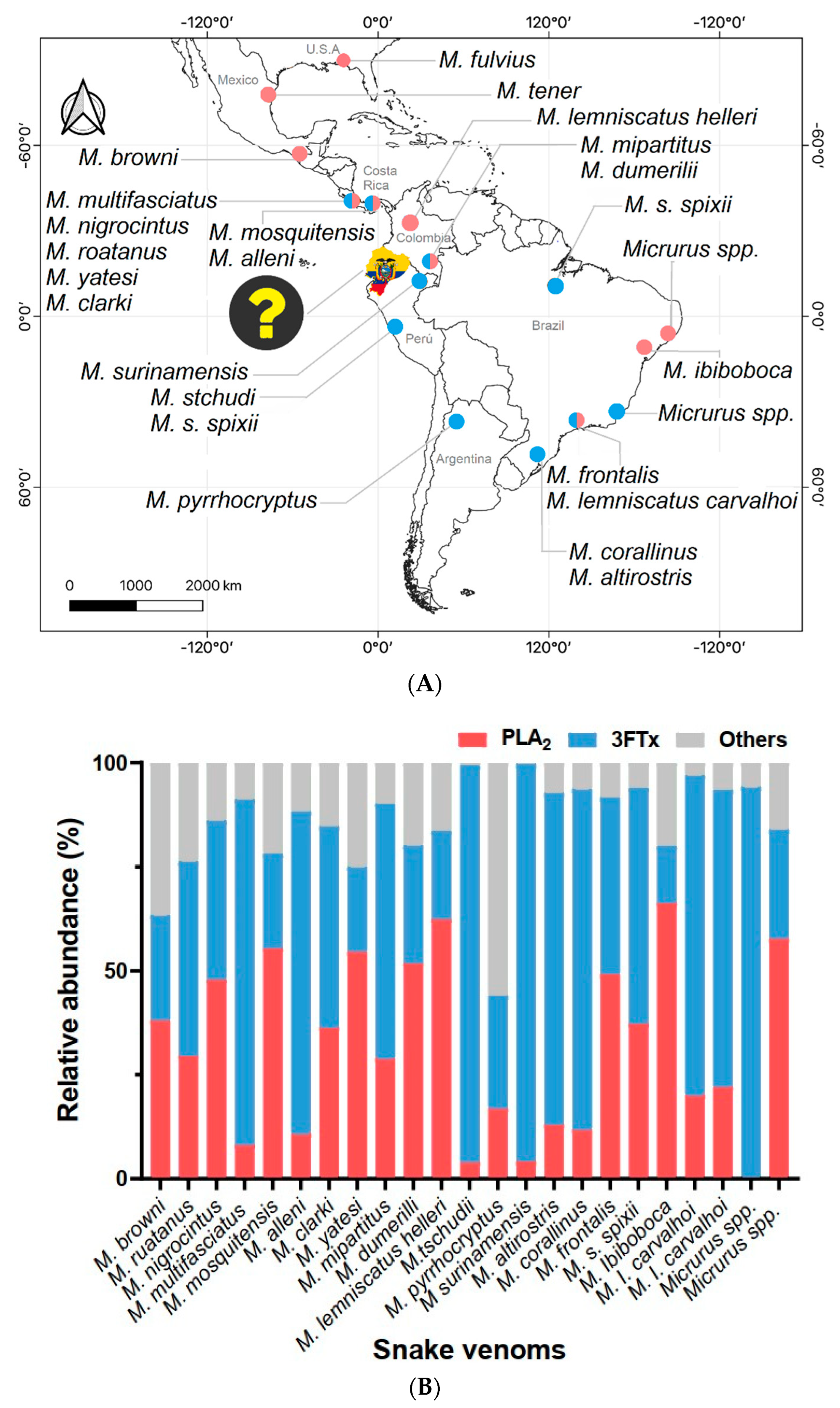
1. Introduction
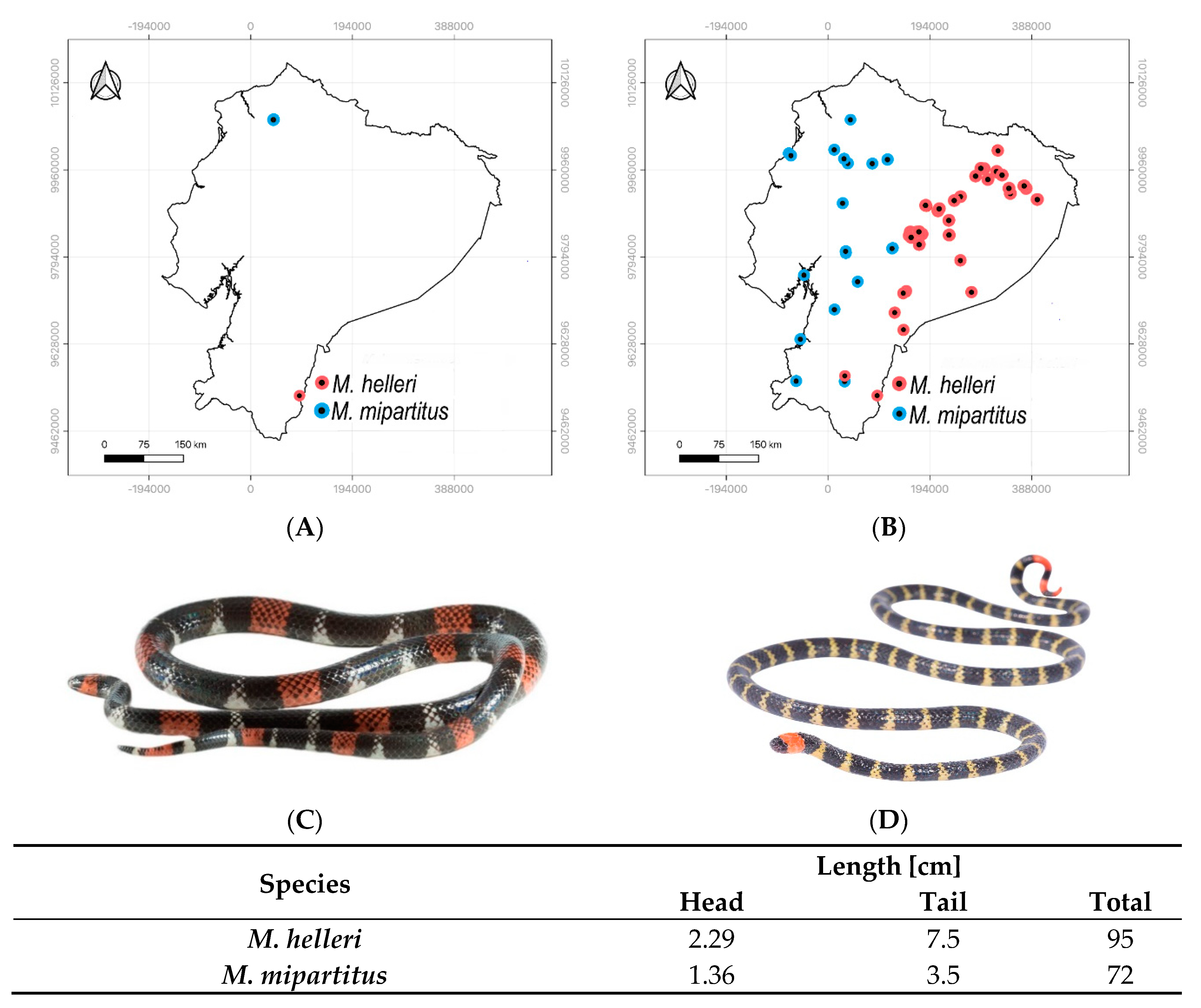
2. Results
2.1. Collection of Venoms and Their Fractionation
2.2. Biochemical Characterisation of Toxins
2.3. Protein Family Assignment by Mass Spectrometry
3. Discussion
4. Materials and Methods
4.1. Venoms
4.2. Venom Fractionation by RP-HPLC
4.3. SDS-PAGE
4.4. LC-MS/MS Analyses
4.5. MS Data Processing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munawar, A.; Ali, S.A.; Akrem, A.; Betzel, C. Snake venom peptides: Tools of biodiscovery. Toxins 2018, 10, 474. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017, 3, 17063. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Tan, N.H.; Tan, C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018, 8, 8545. [Google Scholar] [CrossRef]
- Fry, B.G.; Winkel, K.D.; Wickramaratna, J.C.; Hodgson, W.C.; Wüster, W. Effectiveness of snake antivenom: Species and regional venom variation and its clinical impact. J. Toxicol. Toxin Rev. 2003, 22, 23–34. [Google Scholar] [CrossRef]
- Zancolli, G.; Casewell, N.R. Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol. Biol. Evol. 2020, 37, 2777–2790. [Google Scholar] [CrossRef]
- Almeida, J.R.; Resende, L.M.; Watanabe, R.K.; Carregari, V.C.; Huancahuire-Vega, S.; Caldeira, C.A.d.S.; Coutinho-Neto, A.; Soares, A.M.; Vale, N.; Gomes, P.A.d.C.; et al. Snake venom peptides and low mass proteins: Molecular tools and therapeutic agents. Curr. Med. Chem. 2017, 24, 3254–3282. [Google Scholar] [CrossRef]
- Peña-Carrillo, M.S.; Pinos-Tamayo, E.A.; Mendes, B.; Domínguez-Borbor, C.; Proaño-Bolaños, C.; Miguel, D.C.; Almeida, J.R. Dissection of phospholipases A2 reveals multifaceted peptides targeting cancer cells, Leishmania and bacteria. Bioorg. Chem. 2021, 114, 105041. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Anderluh, G.; Antunes, A.; Ayvazyan, N.; Beis, D.; Caliskan, F.; Crnković, A.; Damm, M.; Dutertre, S.; Ellgaard, L.; et al. Modern venomics—Current insights, novel methods, and future perspectives in biological and applied animal venom research. GigaScience 2022, 11, 048. [Google Scholar] [CrossRef]
- Calvete, J.J.; Sanz, L.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M. Venoms, venomics, antivenomics. FEBS Lett. 2009, 583, 1736–1743. [Google Scholar] [CrossRef]
- Lomonte, B.; Calvete, J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Ledsgaard, L.; Jenkins, T.P.; Davidsen, K.; Krause, K.E.; Martos-Esteban, A.; Engmark, M.; Rørdam Andersen, M.; Lund, O.; Laustsen, A.H. Antibody cross-reactivity in antivenom research. Toxins 2018, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Pukala, T.L.; Isbister, G.K. Investigating toxin diversity and abundance in snake venom proteomes. Front. Pharmacol. 2021, 12, 768015. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A Review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Torres-Carvajal, O.; Pazmiño-Otamendi, G.; Salazar-Valenzuela, D. Reptiles of Ecuador: A resource-rich portal, with a dynamic checklist and photographic guides. Amphib. Reptile Conserv. 2019, 13, 209–229. [Google Scholar]
- Salazar-Valenzuela, D.; Mora-Obando, D.; Fernández, M.L.; Loaiza-Lange, A.; Gibbs, H.L.; Lomonte, B. Proteomic and toxicological profiling of the venom of Bothrocophias campbelli, a pitviper species from Ecuador and Colombia. Toxicon 2014, 90, 15–25. [Google Scholar] [CrossRef]
- Mora-Obando, D.; Salazar-Valenzuela, D.; Pla, D.; Lomonte, B.; Guerrero-Vargas, J.A.; Ayerbe, S.; Gibbs, H.L.; Calvete, J.J. Venom variation in Bothrops asper lineages from North-Western South America. J. Proteom. 2020, 229, 103945. [Google Scholar] [CrossRef]
- Patiño, R.S.P.; Salazar-Valenzuela, D.; Medina-Villamizar, E.; Mendes, B.; Proaño-Bolaños, C.; da Silva, S.L.; Almeida, J.R. Bothrops atrox from Ecuadorian Amazon: Initial analyses of venoms from individuals. Toxicon 2021, 193, 63–72. [Google Scholar] [CrossRef]
- Almeida, J.R.; Mendes, B.; Patiño, R.S.P.; Pico, J.; Laines, J.; Terán, M.; Mogollón, N.G.S.; Zaruma-Torres, F.; Caldeira, C.A.d.S.; da Silva, S.L. Assessing the stability of historical and desiccated snake venoms from a medically important Ecuadorian collection. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 230, 108702. [Google Scholar] [CrossRef]
- Castillo-Beltrán, M.C.; Hurtado-Gómez, J.P.; Corredor-Espinel, V.; Ruiz-Gómez, F.J. A polyvalent coral snake antivenom with broad neutralization capacity. PLoS Negl. Trop. Dis. 2019, 13, e0007250. [Google Scholar] [CrossRef]
- Mackessy, S.P. Venom production and secretion in reptiles. J. Exp. Biol. 2022, 225, 227348. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Lomonte, B.; Saviola, A.J.; Bonilla, F.; Sasa, M.; Williams, D.J.; Undheim, E.A.B.; Sunagar, K.; Jackson, T.N.W. Mutual enlightenment: A toolbox of concepts and methods for integrating evolutionary and clinical toxinology via snake venomics and the contextual stance. Toxicon X 2021, 9–10, 100070. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Pla, D.; Pérez, A.; Rodríguez, Y.; Zavaleta, A.; Salas, M.; Lomonte, B.; Calvete, J.J. Venomic analysis of the poorly studied desert coral snake, Micrurus tschudii tschudii, supports the 3FTx/PLA₂ dichotomy across Micrurus venoms. Toxins 2016, 8, 178. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Vargas-Vargas, N.; Pla, D.; Sasa, M.; Rey-Suárez, P.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in New World elapids. Toxicon 2015, 107, 217–233. [Google Scholar] [CrossRef]
- Sanz, L.; de Freitas-Lima, L.N.; Quesada-Bernat, S.; Graça-de-Souza, V.K.; Soares, A.M.; Calderón, L.A.; Calvete, J.J.; Caldeira, C.A.S. Comparative venomics of Brazilian coral snakes: Micrurus frontalis, Micrurus spixii spixii, and Micrurus surinamensis. Toxicon 2019, 166, 39–45. [Google Scholar] [CrossRef]
- Rey-Suárez, P.; Núñez, V.; Gutiérrez, J.M.; Lomonte, B. Proteomic and biological characterization of the venom of the redtail coral snake, Micrurus mipartitus (Elapidae), from Colombia and Costa Rica. J. Proteom. 2011, 75, 655–667. [Google Scholar] [CrossRef]
- Bénard-Valle, M.; Neri-Castro, E.; Yañez-Mendoza, M.F.; Lomonte, B.; Olvera, A.; Zamudio, F.; Restano-Cassulini, R.; Possani, L.D.; Jiménez-Ferrer, E.; Alagón, A. Functional, proteomic and transcriptomic characterization of the venom from Micrurus browni browni: Identification of the first lethal multimeric neurotoxin in coral snake venom. J. Proteom. 2020, 225, 103863. [Google Scholar] [CrossRef]
- Sanz, L.; Quesada-Bernat, S.; Ramos, T.; Casais-e-Silva, L.L.; Corrêa-Netto, C.; Silva-Haad, J.J.; Sasa, M.; Lomonte, B.; Calvete, J.J. New insights into the phylogeographic distribution of the 3FTx/PLA2 venom dichotomy across genus Micrurus in South America. J. Proteom. 2019, 200, 90–101. [Google Scholar] [CrossRef]
- Lomonte, B.; Rey-Suárez, P.; Fernández, J.; Sasa, M.; Pla, D.; Vargas, N.; Bénard-Valle, M.; Sanz, L.; Corrêa-Netto, C.; Núñez, V.; et al. Venoms of Micrurus coral snakes: Evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon 2016, 122, 7–25. [Google Scholar] [CrossRef]
- Mena, G.; Chaves-Araya, S.; Chacón, J.; Török, E.; Török, F.; Bonilla, F.; Sasa, M.; Gutiérrez, J.M.; Lomonte, B.; Fernández, J. Proteomic and toxicological analysis of the venom of Micrurus yatesi and its neutralization by an antivenom. Toxicon X 2022, 13, 100097. [Google Scholar] [CrossRef]
- Patiño, R.S.P.; Salazar-Valenzuela, D.; Robles-Loaiza, A.A.; Santacruz-Ortega, P.; Almeida, J.R. A retrospective study of clinical and epidemiological characteristics of snakebite in Napo Province, Ecuadorian Amazon. Trans. R. Soc. Trop. Med. Hyg. 2022, 071. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Prado, E.; Yeager, J.; Andrade, F.; Schiavi-Guzman, C.; Abedrabbo-Figueroa, P.; Terán, E.; Gómez-Barreno, L.; Simbaña-Rivera, K.; Izquierdo-Condoy, J.S. Snake antivenom production in Ecuador: Poor implementation, and an unplanned cessation leads to a call for a renaissance. Toxicon 2021, 202, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Manock, S.R.; Suarez, G.; Graham, D.; Avila-Aguero, M.L.; Warrell, D.A. Neurotoxic envenoming by South American coral snake (Micrurus lemniscatus helleri): Case report from eastern Ecuador and review. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Marques, O.A.; Pizzatto, L.; Santos, S.M.A. Reproductive strategies of new world coral snakes, genus Micrurus. Herpetologica 2013, 69, 58–66. [Google Scholar] [CrossRef]
- de Castro, K.L.P.; Lopes-de-Souza, L.; de Oliveira, D.; Machado-de-Ávila, R.A.; Paiva, A.L.B.; de Freitas, C.F.; Ho, P.L.; Chávez-Olórtegui, C.; Guerra-Duarte, C. A Combined strategy to improve the development of a coral antivenom against Micrurus spp. Front. Immunol. 2019, 10, 02422. [Google Scholar] [CrossRef]
- Olamendi-Portugal, T.; Batista, C.V.; Restano-Cassulini, R.; Pando, V.; Villa-Hernandez, O.; Zavaleta-Martínez-Vargas, A.; Salas-Arruz, M.C.; Rodríguez de la Vega, R.C.; Becerril, B.; Possani, L.D. Proteomic analysis of the venom from the fish eating coral snake Micrurus surinamensis: Novel toxins, their function and phylogeny. Proteomics 2008, 8, 1919–1932. [Google Scholar] [CrossRef]
- Calvete, J.J. Snake venomics at the crossroads between ecological and clinical toxinology. Biochemist 2019, 41, 28–33. [Google Scholar] [CrossRef]
- Laines, J.; Segura, Á.; Villalta, M.; Herrera, M.; Vargas, M.; Alvarez, G.; Gutiérrez, J.M.; León, G. Toxicity of Bothrops sp snake venoms from Ecuador and preclinical assessment of the neutralizing efficacy of a polyspecific antivenom from Costa Rica. Toxicon 2014, 88, 34–37. [Google Scholar] [CrossRef]
- Fernández, J.; Alape-Girón, A.; Angulo, Y.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Venomic and antivenomic analyses of the Central American coral snake, Micrurus nigrocinctus (Elapidae). J. Proteome Res. 2011, 10, 1816–1827. [Google Scholar] [CrossRef]
- Vergara, I.; Pedraza-Escalona, M.; Paniagua, D.; Restano-Cassulini, R.; Zamudio, F.; Batista, C.V.; Possani, L.D.; Alagón, A. Eastern coral snake Micrurus fulvius venom toxicity in mice is mainly determined by neurotoxic phospholipases A2. J Proteom. 2014, 105, 295–306. [Google Scholar] [CrossRef]
- Olamendi-Portugal, T.; Batista, C.V.F.; Pedraza-Escalona, M.; Restano-Cassulini, R.; Zamudio, F.Z.; Benard-Valle, M.; de Roodt, A.R.; Possani, L.D. New insights into the proteomic characterization of the coral snake Micrurus pyrrhocryptus venom. Toxicon 2018, 153, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Sasa, M.; Rey-Suárez, P.; Bryan, W.; Gutiérrez, J.M. Venom of the coral snake Micrurus clarki: Proteomic profile, toxicity, immunological cross-neutralization, and characterization of a three-finger toxin. Toxins 2016, 8, 138. [Google Scholar] [CrossRef]
- Rey-Suárez, P.; Núñez, V.; Fernández, J.; Lomonte, B. Integrative characterization of the venom of the coral snake Micrurus dumerilii (Elapidae) from Colombia: Proteome, toxicity, and cross-neutralization by antivenom. J. Proteom. 2016, 136, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, F.D.R.; Noronha, M.; Lozano, J.L.L. Biological and molecular properties of yellow venom of the Amazonian coral snake Micrurus surinamensis. Rev. Da Soc. Bras. De Med. Trop. 2017, 50, 365–373. [Google Scholar] [CrossRef]
- Yang, D.C.; Dobson, J.; Cochran, C.; Dashevsky, D.; Arbuckle, K.; Benard, M.; Boyer, L.; Alagón, A.; Hendrikx, I.; Hodgson, W.C.; et al. The bold and the beautiful: A neurotoxicity comparison of New World coral snakes in the Micruroides and Micrurus genera and relative neutralization by antivenom. Neurotox. Res. 2017, 32, 487–495. [Google Scholar] [CrossRef]
- Gómez, J.P.H.; Ramírez, M.V.; Gómez, F.J.R.; Fouquet, A.; Fritz, U. Multilocus phylogeny clarifies relationships and diversity within the Micrurus lemniscatus complex (Serpentes: Elapidae). SALAMANDRA 2021, 57, 229–239. [Google Scholar]
- Terribile, L.C.; Feitosa, D.T.; Pires, M.G.; de Almeida, P.C.R.; de Oliveira, G.; Diniz-Filho, J.A.F.; Silva, N.J.D., Jr. Reducing Wallacean shortfalls for the coralsnakes of the Micrurus lemniscatus species complex: Present and future distributions under a changing climate. PLoS ONE 2018, 13, e0205164. [Google Scholar] [CrossRef]
- Margres, M.J.; Aronow, K.; Loyacano, J.; Rokyta, D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013, 14, 531. [Google Scholar] [CrossRef]
- Cañas, C.A.; Castro-Herrera, F.; Castaño-Valencia, S. Envenomation by the red-tailed coral snake (Micrurus mipartitus) in Colombia. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 9. [Google Scholar] [CrossRef] [PubMed]
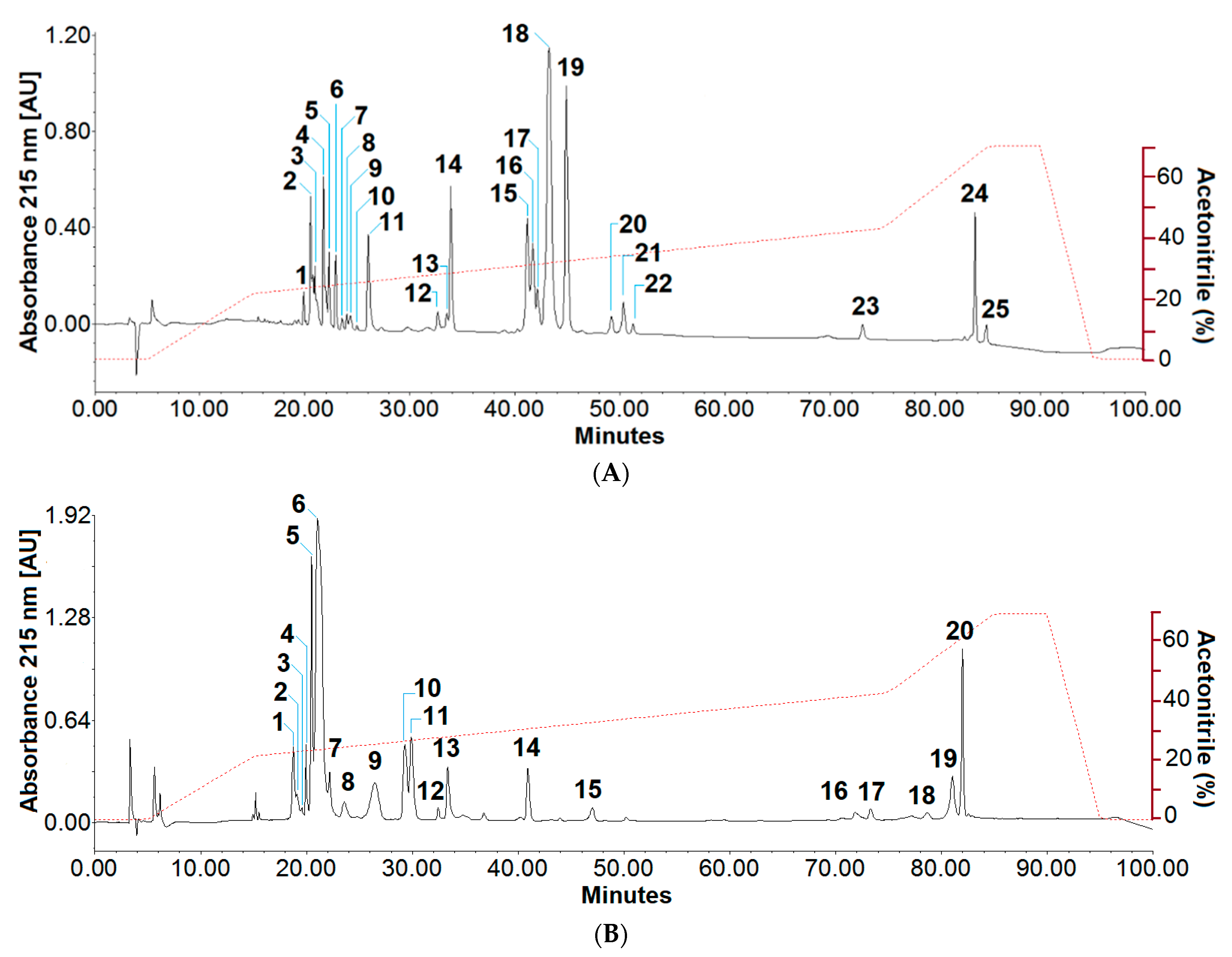


| Fractions | Relative Abundance | Peptide Ion | Peptide Sequences | Related Proteins | |||
|---|---|---|---|---|---|---|---|
| m/z | Z | Family | Species | Database | |||
| 1 | 0.65 | Unknown/unidentified | |||||
| 2 | 3.79 | 414.80 | 3+ | TRCDGFCGNR | 3FTx | Naja atra | E2IU01 |
| 461.23 | 2+ | TIDECQR | SP Kunitz inhibitor | Bungarus fasciatus | P25660 | ||
| 532.28 | 2+ | TPPAGPDVGPR | Bradykinin inhibitor peptide | Agkistrodon bilineatus | P85025 | ||
| 3 | 0.78 | 414.80 | 3+ | TRCDGFCGNR | 3FTx | Naja atra | E2IU01 |
| 4 | 5.34 | 414.80 | 3+ | TRCDGFCGNR | 3FTx | Naja atra | E2IU01 |
| 5 | 1.83 | 520.73 | 2+ | YNKISFIR | 3FTx | Micrurus altirostris | F5CPD4 |
| 6 | 2.42 | 532.28 | 2+ | TPPAGPDVGPR | Bradykinin inhibitor peptide | Agkistrodon bilineatus | P85025 |
| 7 | 0.46 | 484.27 | 2+ | IICCRSC | 3FTx | Micrurus frontralis | P86420 |
| 8 | 0.40 | 484.27 | 2+ | IICCRSC | 3FTx | Micrurus frontralis | P86420 |
| 9 | 0.51 | 484.27 | 2+ | IICCRSC | 3FTx | Micrurus frontralis | P86420 |
| 10 | 0.16 | 457.33 | 3+ | FCELPADSGSCK | SP Kunitz inhibitor | Phesudechis rossignolii | E7FL13 |
| 464.69 | 3+ | GCASSCPKNGLIK | 3FTx | Micrurus diastema | A0A0H4BKJ5 | ||
| 11 | 5.43 | 464.69 | 3+ | GCASSCPKNGLIK | 3FTx | Micrurus diastema | A0A0H4BKJ5 |
| 12 | 0.94 | 447.24 | 2+ | LAALCFAK | PLA2 | Micrurus mipartitus | C0HKB9 |
| 13 | 0.41 | 447.24 | 2+ | LAALCFAK | PLA2 | Micrurus mipartitus | C0HKB9 |
| 14 | 6.88 | 447.24 | 2+ | LAALCFAK | PLA2 | Micrurus mipartitus | C0HKB9 |
| 15 | 4.94 | 582.32 | 2+ | NLVQFGNMIK | PLA2 | Micrurus lemniscatus carvalhoi | A0A2H6NAU5 |
| 594.78 | 2+ | GGSGTPVDDLDR | PLA2 | Micrurus spixii | A0A2D4NMB1 | ||
| 16 | 3.34 | 594.78 | 2+ | GGSGTPVDDLDR | PLA2 | Micrurus spixii | A0A2D4NMB2 |
| 17 | 1.25 | 594.78 | 2+ | GGSGTPVDDLDR | PLA2 | Micrurus spixii | A0A2D4NMB3 |
| 18 | 33.72 | 594.78 | 2+ | GGSGTPVDDLDR | PLA2 | Micrurus spixii | A0A2D4NMB4 |
| 687.26 | 2+ | CQDFVCNCDR | PLA2 | Micrurus lemniscatus carvalhoi | A0A2H6NAU5 | ||
| 440.72 | 2+ | VAANCFAK | PLA2 | Micrurus lemniscatus carvalhoi | A0A2H6NAU6 | ||
| 590.31 | 2+ | NLVQFGNMOXIK | PLA2 | Micrurus lemniscatus carvalhoi | A0A2H6NAU5 | ||
| 566.27 | 2+ | GSGTPVDDLDR | PLA2 | Micrurus lemniscatus carvalhoi | A0A2H6NFP9 | ||
| 503.74 | 2+ | MIECANIR | PLA2 | Micrurus lemniscatus carvalhoi | A0A2H6NFP10 | ||
| 687.26 | 2+ | CKDFVCNCDR | PLA2 | Micrurus dumerilii | C0HKB8 | ||
| 19 | 17.41 | 594.77 | 2+ | GGSGTPVDDLDR | PLA2 | Micrurus spixii | A0A2D4NMB1 |
| 440.73 | 2+ | VAANCFAK | PLA2 | Micrurus surinamensis | A0A2D4PZ69 | ||
| 20 | 1.04 | 521.21 | 2+ | AFVCNCDR | PLA2 | Micrurus mipartitus | C0HKB9 |
| 21 | 1.82 | 521.21 | 2+ | AFVCNCDR | PLA2 | Micrurus mipartitus | C0HKB9 |
| 22 | 0.38 | 521.21 | 2+ | AFVCNCDR | PLA2 | Micrurus mipartitus | C0HKB9 |
| 23 | 0.96 | 721.69 | 3+ | ISFMOXTAHDYSLPVFVYTR | HYA | Micrurus corallinus | A0A2D4H401 |
| 24 | 4.22 | 742.85 | 2+ | EADYEEFLEIAR | LAO | Micrurus tener | A0A194ARE6 |
| 577.78 | 2+ | FDEIVGGFDR | LAO | Micrurus tener | A0A194ARE7 | ||
| 742.85 | 2+ | EADYEEFLEIAR | LAO | Micrurus mipartitus | A0A2U8QNR2 | ||
| 497.24 | 2+ | FWEADGIR | LAO | Micrurus mipartitus | A0A2U8QNR3 | ||
| 746.88 | 2+ | FDEIVGGFDRLPK | LAO | Micrurus paraensis | A0A2D4K1Y6 | ||
| 586.29 | 2+ | DHGWIDSTIK | LAO | Micrurus paraensis | A0A2D4KMW9 | ||
| 627.32 | 2+ | SASQLYQESLK | LAO | Micrurus paraensis | A0A2D4KYN2 | ||
| 25 | 0.93 | 742.85 | 2+ | EADYEEFLEIAR | LAO | Micrurus mipartitus | A0A2U8QNR2 |
| Fractions | Relative Abundance | Peptide Ion | Peptide Sequences | Related Proteins | |||
|---|---|---|---|---|---|---|---|
| m/z | z | Family | Species | Database | |||
| 1 | 3.07 | Unknown/unidentified | |||||
| 2 | 0.30 | 559.28 | 2+ | GCAVTCPKPK | 3FTx | Micrurus mipartitus | A0A2P1BSS8 |
| 3 | 0.17 | 559.28 | 2+ | GCAVTCPKPK | 3FTx | Micrurus mipartitus | A0A2P1BSS8 |
| 4 | 2.06 | 733.34 | 2+ | KGIEINCCTTDR | 3FTx | Naja sputatrix | O57327 |
| 676.02 | 3+ | VDLGCAATCPKVKPGVNIK | 3FTx | Naja nivea | P01390 | ||
| 733.34 | 2+ | KGIELNCCTTDR | 3FTx | Naja mossambica | P01431 | ||
| 5 | 7.61 | 733.34 | 2+ | KGIELNCCTTDR | 3FTx | Naja mossambica | P01431 |
| 6 | 40.26 | 733.34 | 2+ | KGIELNCCTTDR | 3FTx | Naja mossambica | P01431 |
| 722.34 | 2+ | LVPLFSKTCPPGK | 3FTx | Naja atra | P60307 | ||
| 7 | 1.07 | 733.34 | 2+ | KGIEINCCTTDR | 3FTx | Naja sputatrix | O57327 |
| 8 | 1.73 | 733.34 | 2+ | KGIEINCCTTDR | 3FTx | Naja sputatrix | O57327 |
| 9 | 7.89 | 447.25 | 2+ | LAALCFAK | PLA2 | Micrurus mipartitus | C0HKB9 |
| 521.22 | 2+ | AFVCNCDR | PLA2 | Micrurus mipartitus | C0HKB9 | ||
| 10 | 5.54 | 440.78 | 2+ | VAANCFAK | PLA2 | Micrurus surinamensis | A0A2D4PZ69 |
| 447.73 | 2+ | VAAKCFAK | PLA2 | Micrurus surinamensis | A0A2D4PRR8 | ||
| 11 | 5.16 | 448.73 | 2+ | VAAKCFAK | PLA2 | Micrurus surinamensis | A0A2D4PRR8 |
| 12 | 0.51 | 447.73 | 2+ | VAAKCFAK | PLA2 | Micrurus surinamensis | A0A2D4PRR8 |
| 13 | 3.89 | 841.43 | 3+ | KTLLLNLVVVTIVCLDFGYTIK | 3FTx | Bungarus flaviceps | D5J9P5 |
| 14 | 4.06 | 841.43 | 3+ | KTLLLNLVVVTIVCLDFGYTIK | 3FTx | Bungarus flaviceps | D5J9P5 |
| 15 | 1.26 | 841.43 | 3+ | KTLLLNLVVVTIVCLDFGYTIK | 3FTx | Bungarus flaviceps | D5J9P5 |
| 16 | 0.99 | 775.91 | 2+ | CVINATGPFTDTVR | 3FTx | Micrurus lemniscatus lemniscatus | A0A2D4IKM1 |
| 17 | 0.97 | 694.86 | 2+ | YIEFYVAVDNR | SVMP | Bungarus multicinctus | A8QL49 |
| 18 | 0.72 | 705.36 | 3+ | IDFNGNTLGLAHIGSLCSPK | SVMP | Micrurus fulvius | U3EPC7 |
| 632.84 | 2+ | SNVAVTLDLFGK | SVMP | Micrurus fulvius | U3EPC7 | ||
| 708.86 | 2+ | YIEFYVVVDNR | SVMP | Micrurus fulvius | U3EPC7 | ||
| 19 | 4.30 | 660.34 | 2+ | KMNDNAQLLTR | SVMP | Micrurus fulvius | A0A0F7YYV1 |
| 503.95 | 3+ | RPECILNKPLNR | SVMP | Micrurus fulvius | A0A0F7YYV1 | ||
| 20 | 8.45 | 934.97 | 2+ | TLPSVTADYVIVCSTSR | LAO | Micrurus mipartitus | A0A2U8QNR6 |
| 624.37 | 2+ | KVIVTYQTPAK | LAO | Micrurus mipartitus | A0A2U8QNR6 | ||
| 649.02 | 3+ | HVVVVGAGMOXSGLSAAYVLAK | LAO | Micrurus mipartitus | A0A2U8QNR6 | ||
| 742.85 | 2+ | EADYEEFLEIAR | LAO | Micrurus mipartitus | A0A2U8QNR6 | ||
| 519.79 | 2+ | IFLTCTKR | LAO | Micrurus mipartitus | A0A2U8QNR6 | ||
| 568.79 | 2+ | IHFAGEYTAK | LAO | Micrurus mipartitus | A0A2U8QNR6 | ||
| 742.85 | 2+ | EADYEEFLEIAR | LAO | Micrurus tener | A0A194ARE6 | ||
| 627.32 | 2+ | SASQLYQESLK | LAO | Micrurus paraensis | A0A2D4KYN2 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Altamirano, J.A.; Salazar-Valenzuela, D.; Medina-Villamizar, E.J.; Quirola, D.R.; Patel, K.; Vaiyapuri, S.; Lomonte, B.; Almeida, J.R. First Insights into the Venom Composition of Two Ecuadorian Coral Snakes. Int. J. Mol. Sci. 2022, 23, 14686. https://doi.org/10.3390/ijms232314686
Hernández-Altamirano JA, Salazar-Valenzuela D, Medina-Villamizar EJ, Quirola DR, Patel K, Vaiyapuri S, Lomonte B, Almeida JR. First Insights into the Venom Composition of Two Ecuadorian Coral Snakes. International Journal of Molecular Sciences. 2022; 23(23):14686. https://doi.org/10.3390/ijms232314686
Chicago/Turabian StyleHernández-Altamirano, Josselin A., David Salazar-Valenzuela, Evencio J. Medina-Villamizar, Diego R. Quirola, Ketan Patel, Sakthivel Vaiyapuri, Bruno Lomonte, and José R. Almeida. 2022. "First Insights into the Venom Composition of Two Ecuadorian Coral Snakes" International Journal of Molecular Sciences 23, no. 23: 14686. https://doi.org/10.3390/ijms232314686
APA StyleHernández-Altamirano, J. A., Salazar-Valenzuela, D., Medina-Villamizar, E. J., Quirola, D. R., Patel, K., Vaiyapuri, S., Lomonte, B., & Almeida, J. R. (2022). First Insights into the Venom Composition of Two Ecuadorian Coral Snakes. International Journal of Molecular Sciences, 23(23), 14686. https://doi.org/10.3390/ijms232314686












