Glutathione Plays a Positive Role in the Proliferation of Pinus koraiensis Embryogenic Cells
Abstract
:1. Introduction
2. Results
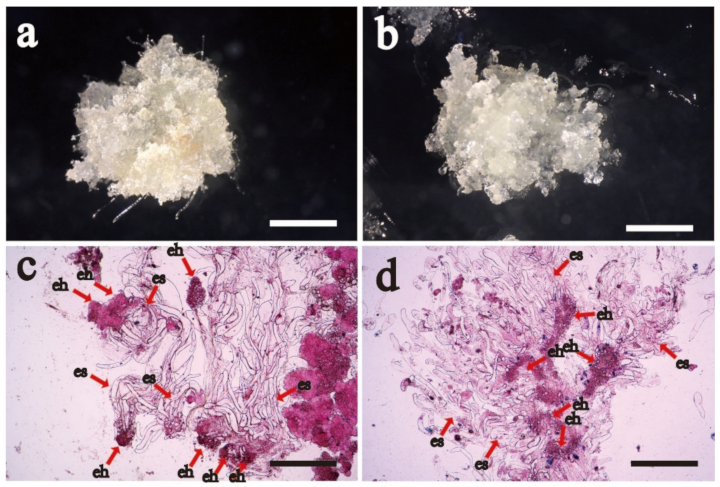
2.1. Proliferative and Cytological Observation of Cell Lines with Different Proliferative Potential
2.2. Transcriptome Analysis
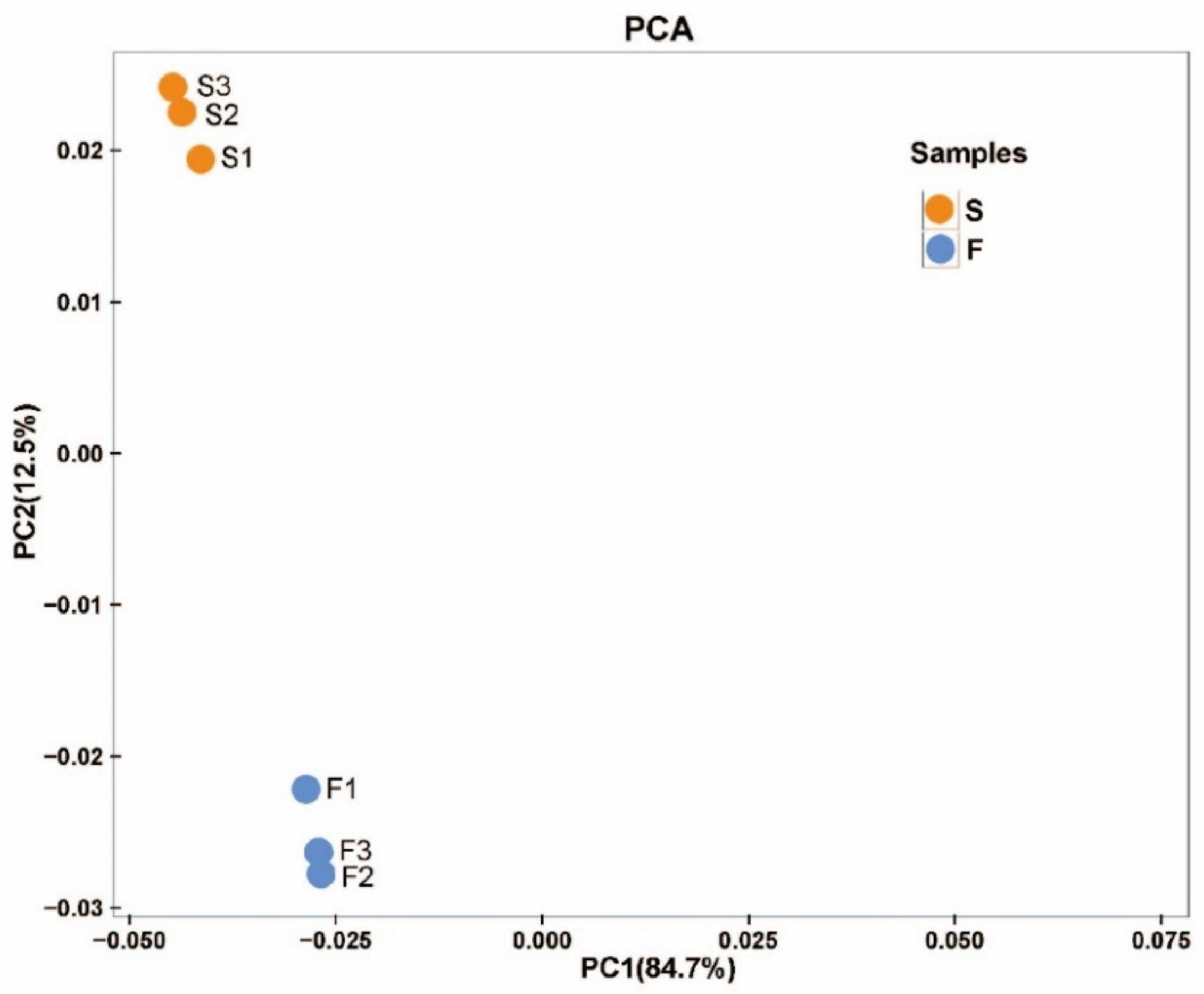
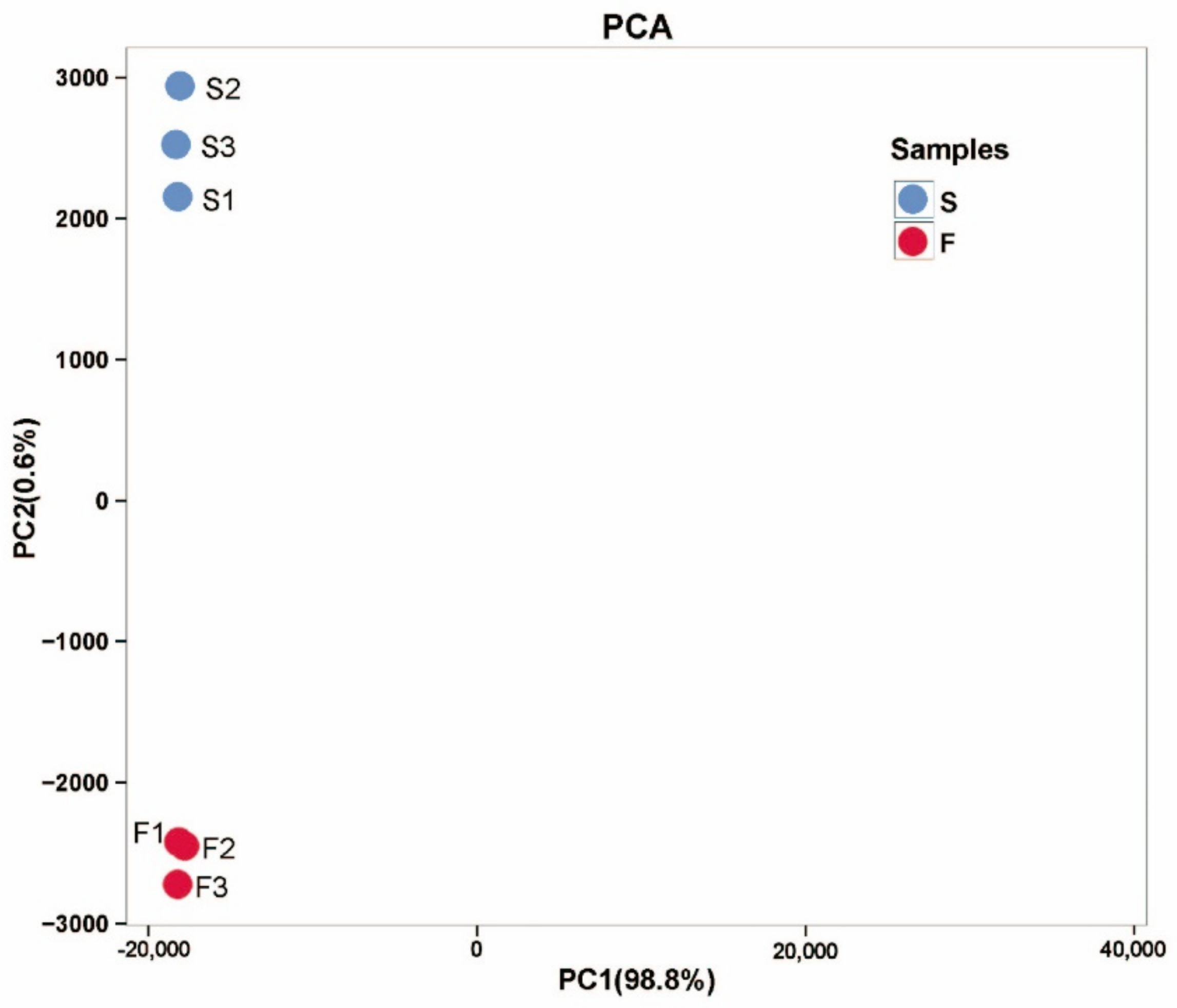
2.2.1. RNA Sequencing and Transcriptome De Novo Assembly
2.2.2. Screening of Differentially Expressed Genes and Functional Annotation
2.2.3. Screening of Differentially Expressed Genes and Functional Annotation
2.2.4. COG Classification of Differentially Expressed Genes
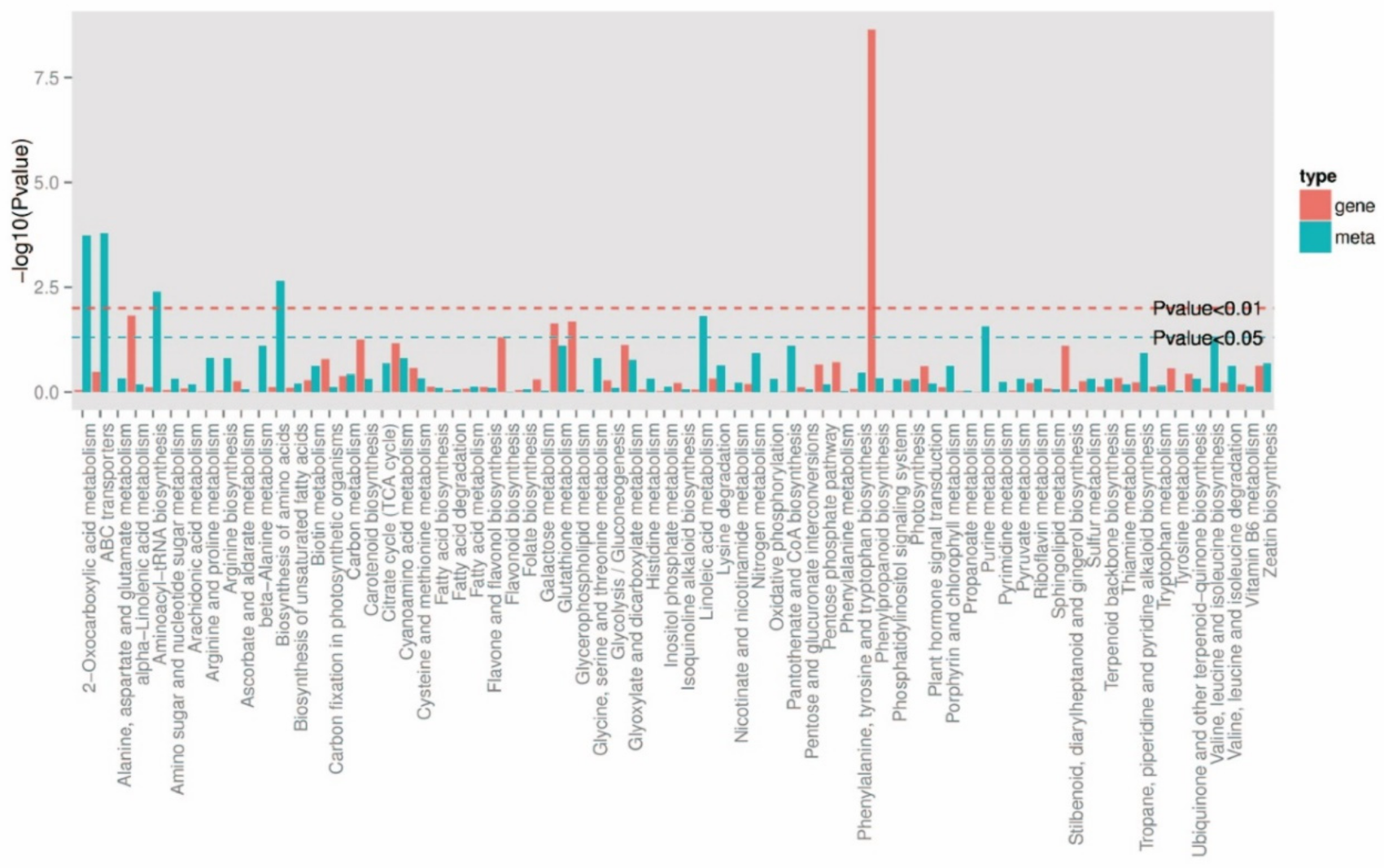
2.2.5. KEGG Enrichment Analysis in Differentially Expressed Genes
KEGG Enrichment Analysis in Differentially Expressed Genes
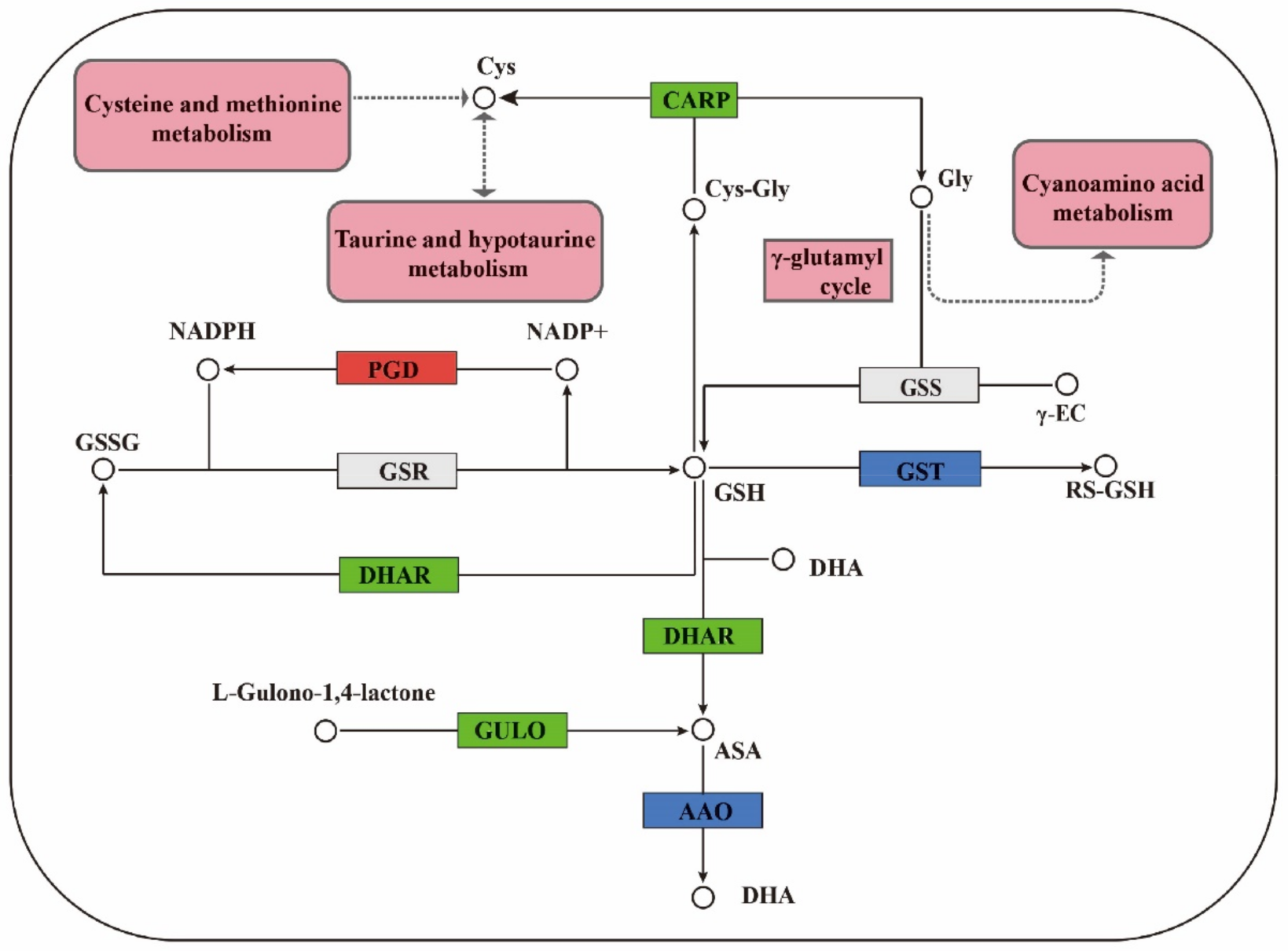
Analysis of Hormone Metabolism-Related Genes
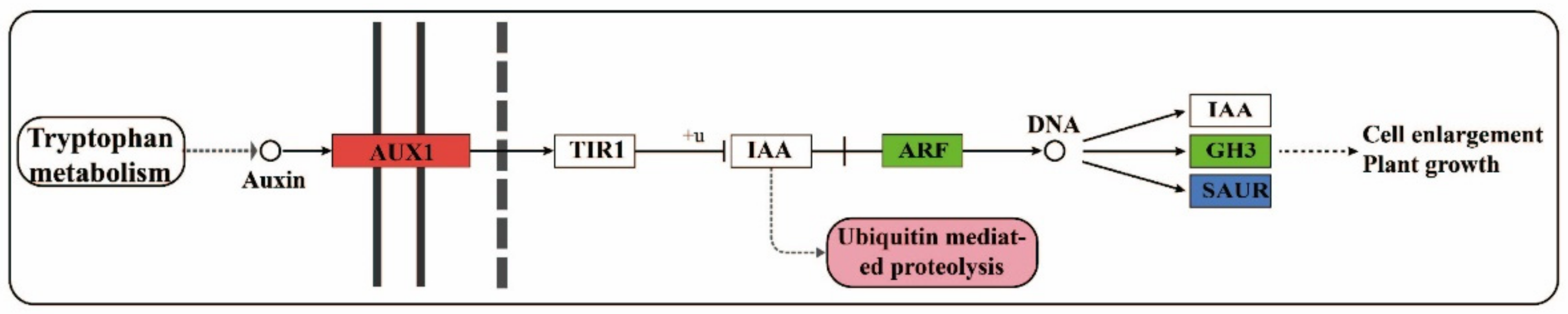
Specific Analysis of Auxin-Related Genes
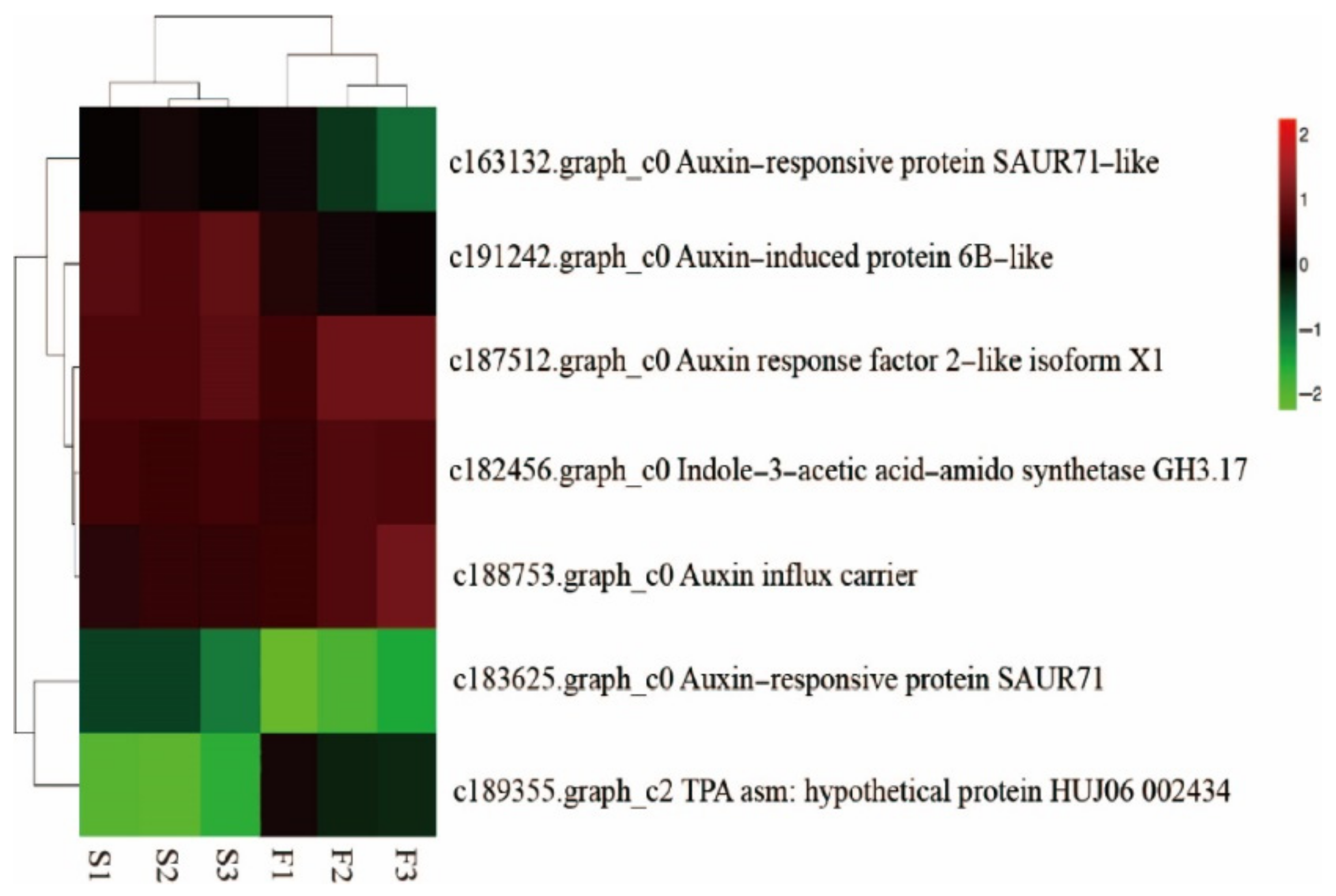
Analysis of Cytokinin-Related Genes
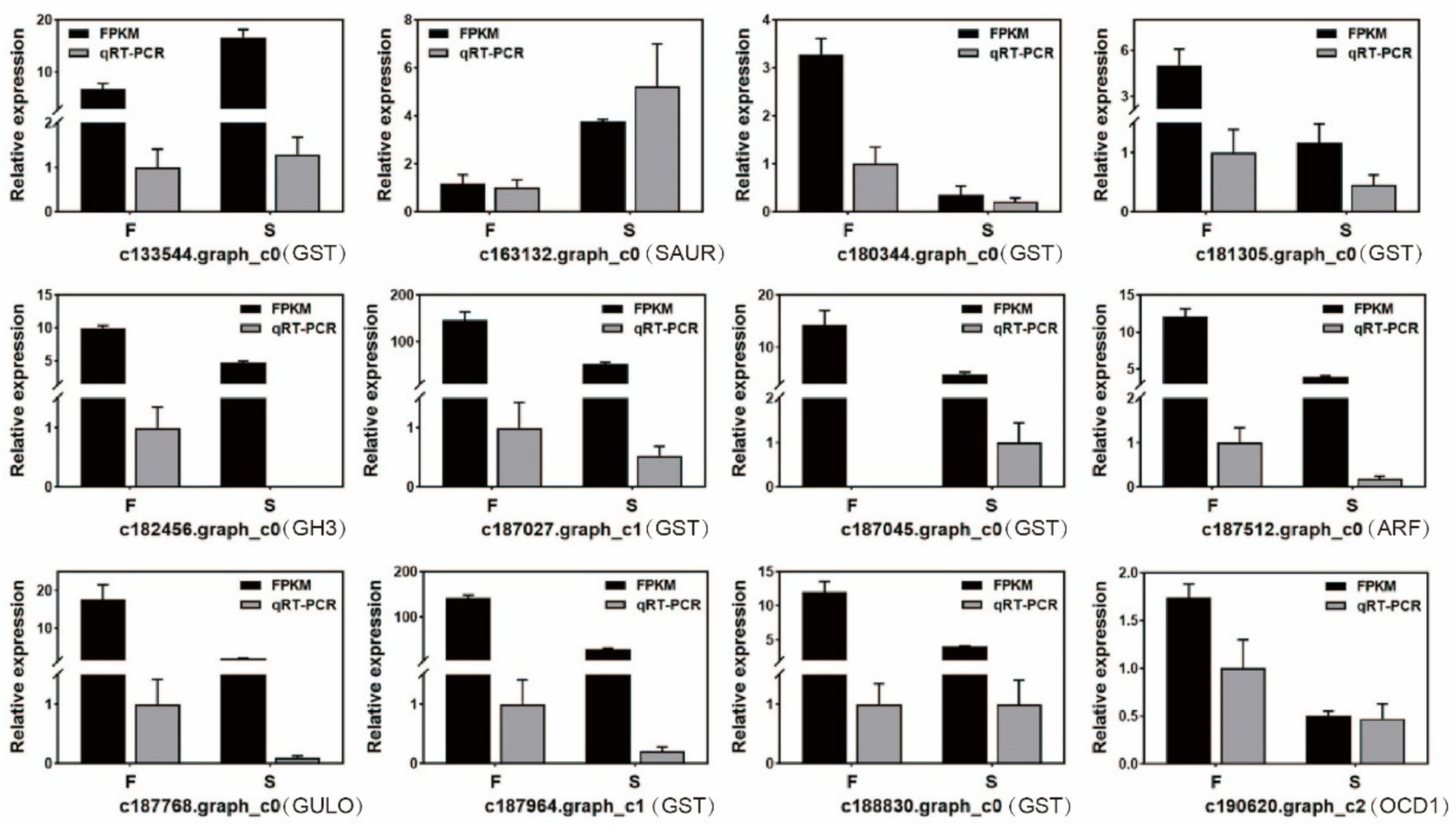
2.3. qRT-PCR
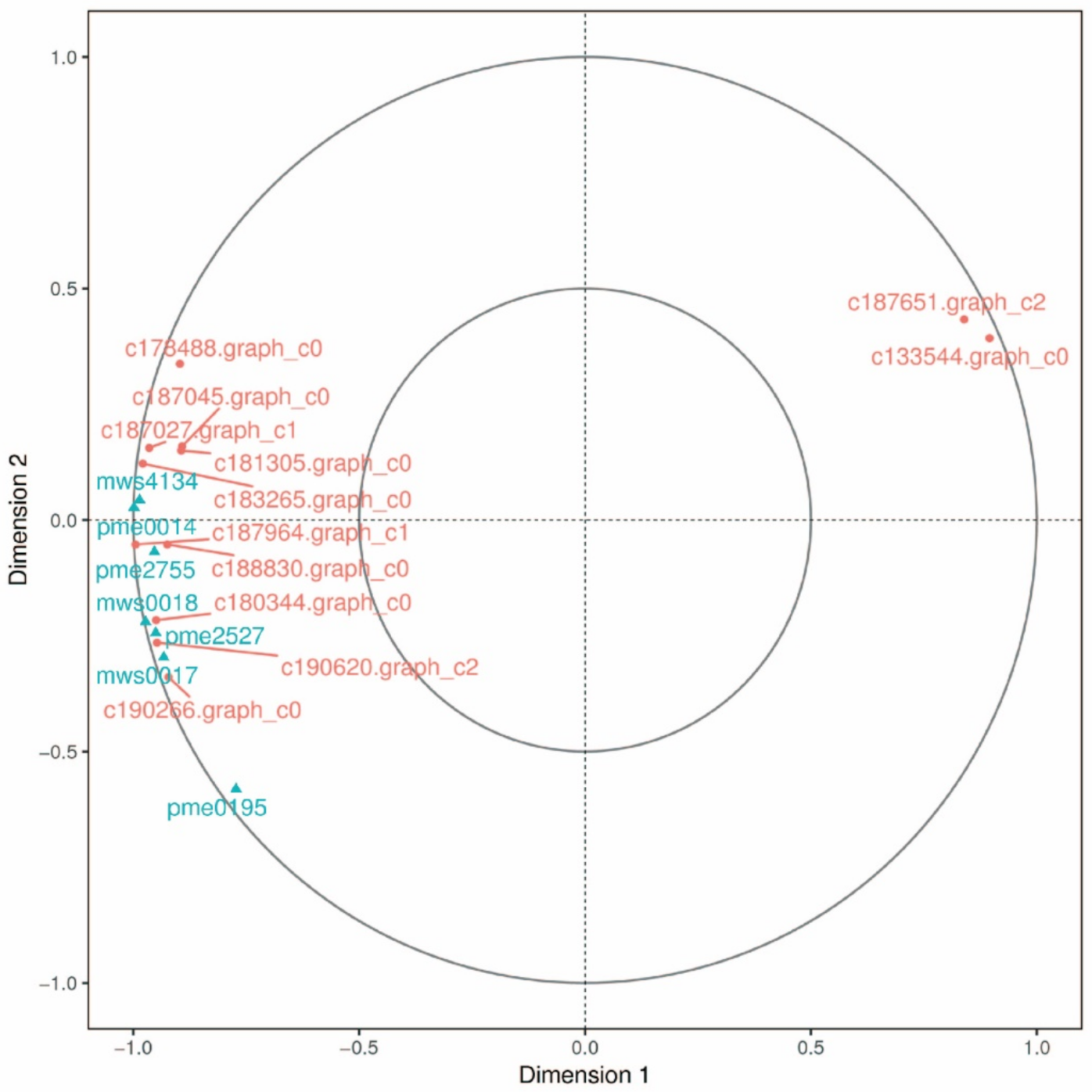
2.4. Metabolomics Analysis in Cell Lines of Korean Pine with Different Proliferative Potentials
2.4.1. Differential Metabolite Screening
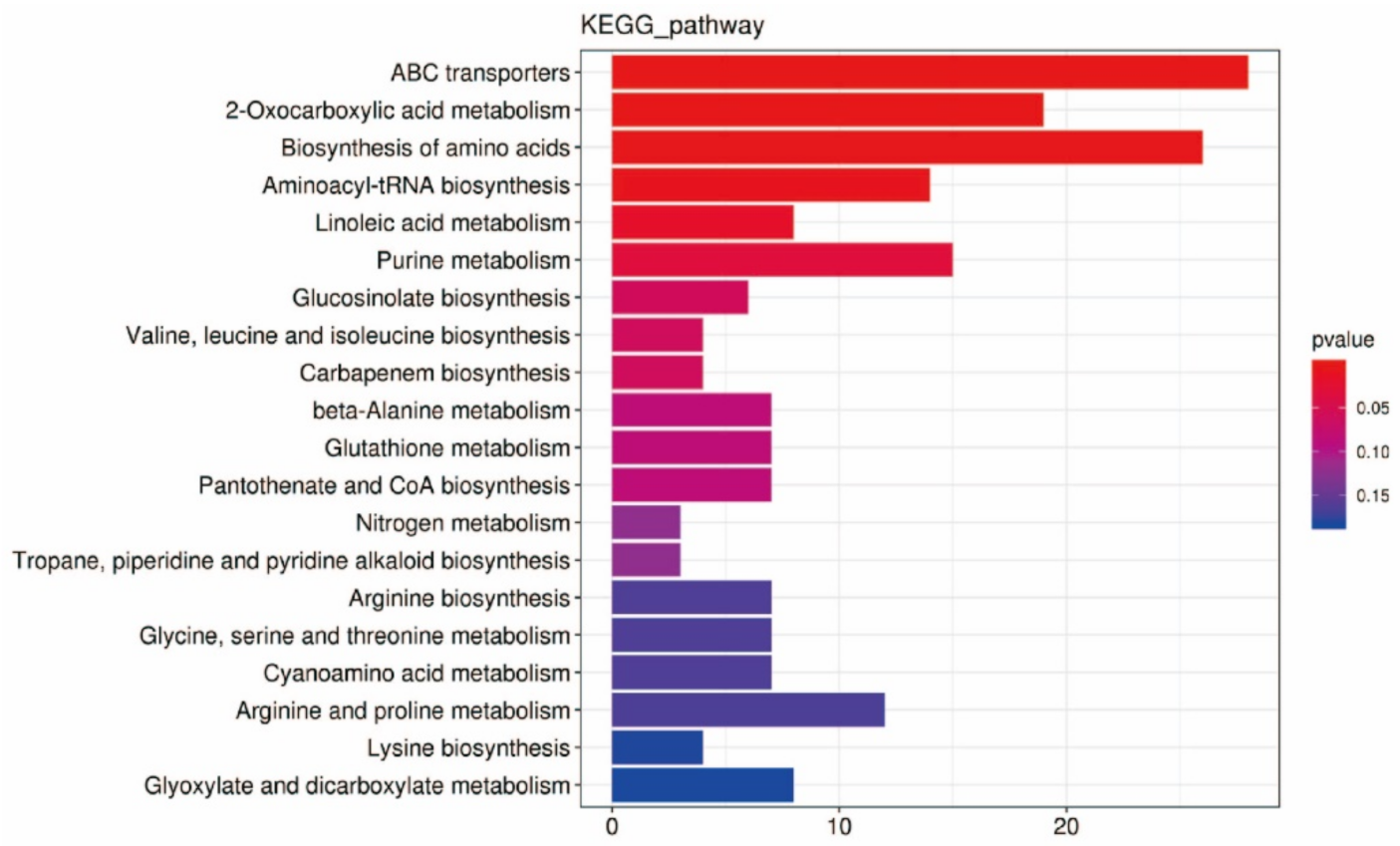
2.4.2. KEGG Functional Annotation and Enrichment Analysis of DEMs

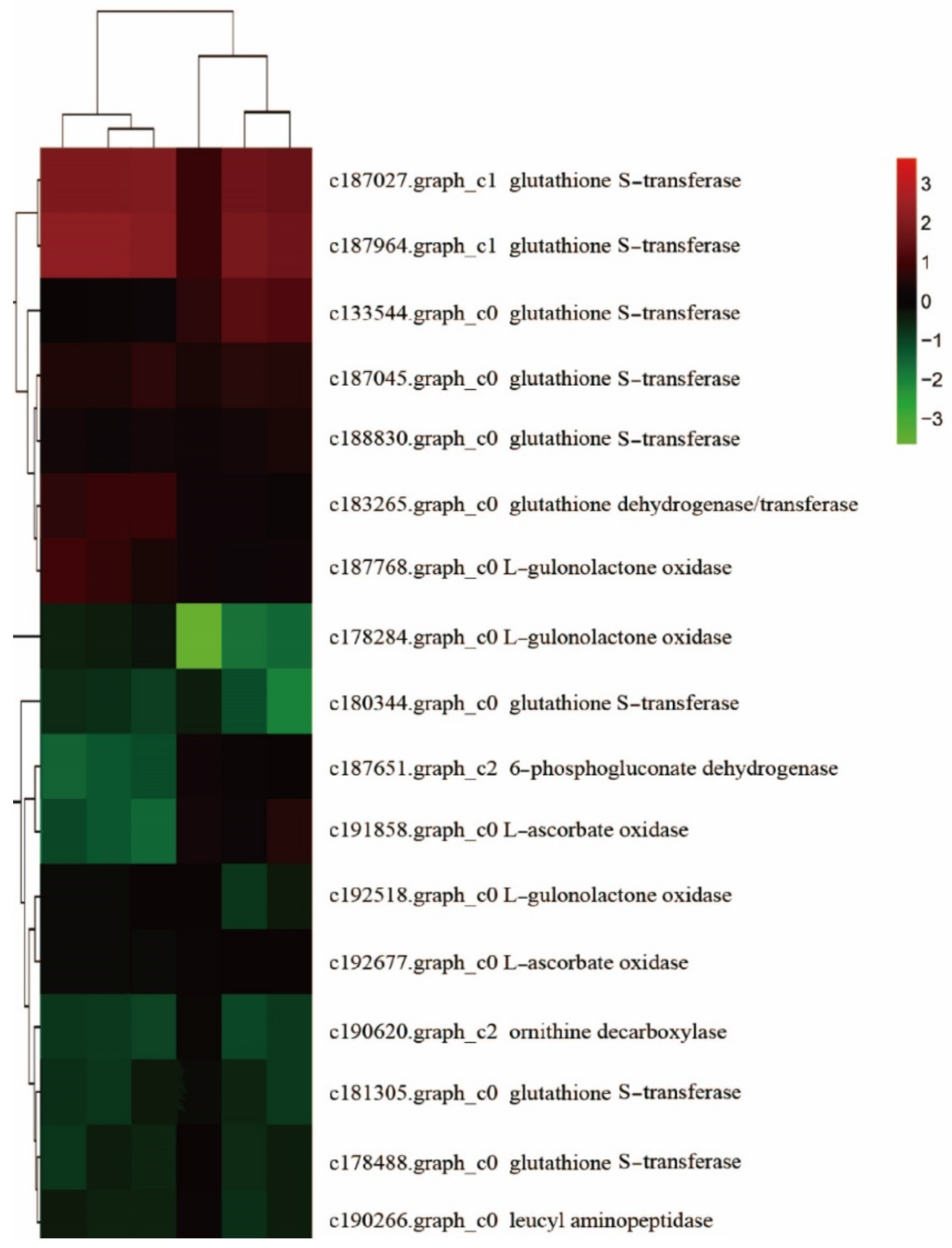
2.4.3. Combined Analysis of Metabolomics and Transcriptomics
2.4.4. Canonical Correlation Analysis in GSH-Related Genes and Metabolites
2.5. Verification
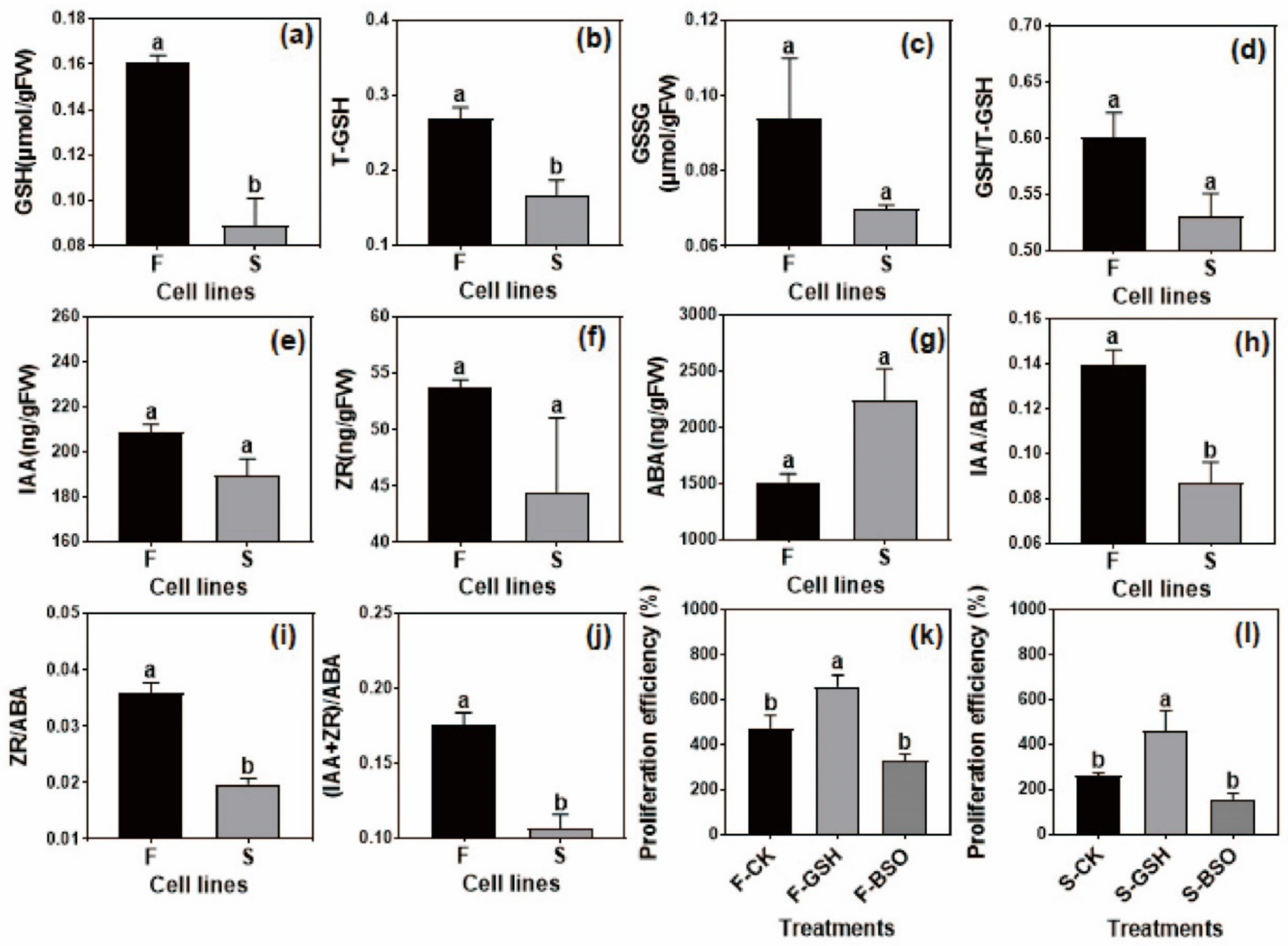
2.5.1. Determination of Intracellular GSH in F and S Cell Lines
2.5.2. Determination of Intracellular Hormone in F and S Cell Lines
2.5.3. Validation of the Effect of Exogenous GSH and L-Buthionine-Sulfoximine (BSO) on the EC Proliferation Rate of F and S Cell Lines
3. Discussion
3.1. Regulation of GSH Related Genes and Their Metabolites on Cell Proliferation
3.2. Regulation of Auxin-Related Genes and Their Metabolites on Cell Proliferation
4. Materials and Methods
4.1. Collection and Disinfection of Plant Materials
4.2. Embryogenic Callus Induction and Proliferation
4.3. Proliferative Rate Calculation and Cell Observation of EC
4.4. Transcriptome Sequencing
4.5. qRT-PCR Validation on DEGs
4.6. Metabonomic Analysis
4.6.1. Sample Extraction and Analysis
4.6.2. Association Analysis of Metabolomic and Transcriptomic Data
4.7. Validation of Important Key Substances Related to the Proliferation of Korean Pine Embryogenic Cells
4.7.1. A Determination of Intracellular Glutathione (GSH) Content in the F and S Cell Lines
4.7.2. Determination of Intracellular Hormone Content in the F and S Cell Lines
4.7.3. Validation of the Effects of Exogenous GSH and BSO on EC Proliferative Rates in the F and S Cell Lines
4.8. Data Collection and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lelu-Walter, M.A.; Gautier, F.; Eliášová, K.; Sanchez, L.; Teyssier, C.; Lomenech, A.M.; Metté, C.L.; Hargreaves, C.; Trontin, J.F.; Reeves, C. High gellan gum concentration and secondary somatic embryogenesis: Two key factors to improve somatic embryo development in Pseudotsuga menziesii [Mirb.]. Plant Cell Tissue Org. 2018, 132, 137–155. [Google Scholar] [CrossRef]
- Pullman, G.S.; Zeng, X.; Copeland-Kamp, B.; Crockett, J.; Lucrezi, J.; May, S.W.; Bucalo, K. Conifer somatic embryogenesis: Improvements by supplementation of medium with oxidation—reduction agents. Tree Physiol. 2015, 35, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Tretyakova, I.N.; Mineev, V.V. Reproductive potential of conifers, somatic embryogenesis and apomixis. Russ. J. Dev. Biol. 2021, 52, 75–86. [Google Scholar] [CrossRef]
- Uma, S.; Kumaravel, M.; Backiyarani, S.; Saraswathi, S.M.; Dura, P.; Karthic, R. Somatic embryogenesis as a tool for reproduction of genetically stable plants in banana and confirmatory field trials. Plant Cell Tissue Org. 2021, 147, 181–188. [Google Scholar] [CrossRef]
- Heringer, A.S.; Santa-Catarina, C.; Silveira, V. Insights from proteomic studies into plant somatic embryogenesis. Proteomics 2014, 18, e1700265. [Google Scholar] [CrossRef]
- Elas, G.C.; Geovanny, I.; Rosa, M.G.; Víctor, M.L. Elaboration of transcriptome during the induction of somatic embryogenesis. Methods Mol. Biol. 2018, 1815, 411–427. [Google Scholar] [CrossRef]
- Borges Araujo, A.J.; Cerruti, G.V.; Zuccarelli, R. Proteomic analysis of S-nitrosation sites during somatic embryogenesis in Brazilian pine, Araucaria angustifolia (Bertol.) Kuntze. Front. Plant Sci. 2022, 13, 902068. [Google Scholar] [CrossRef]
- Guan, Y.; Li, S.G.; Fan, X.F. Application of Somatic Embryogenesis in Woody Plants. Front. Plant Sci. 2016, 7, 938. [Google Scholar] [CrossRef] [Green Version]
- Ahn, C.H.; Choi, Y.E. In vitro clonal propagation and stable cryopreservation system for Platycladus orientalis via somatic embryogenesis. Plant Cell Tissue Org. 2011, 131, 513–523. [Google Scholar] [CrossRef]
- Ávila, C.; Llebrés, M.T.; Castro-Rodríguez, V.; Lobato-Fernández, C.; Reymond, I.; Harvengt, L.; Trontin, J.-F.; Cánovas, F.M. Identification of metabolic pathways differentially regulated in somatic and zygotic embryos of Maritime Pine. Front. Plant Sci. 2022, 13, 877960. [Google Scholar] [CrossRef]
- Li, F.Q.; Yao, J.B.; Hu, L.F.; Chen, J.H.; Shi, J. Multiple methods synergistically promote the synchronization of somatic embryogenesis through suspension culture in the new hybrid between Pinus elliottii and Pinus caribaea. Front. Plant Sci. 2022, 13, 857972. [Google Scholar] [CrossRef]
- Belmonte, M.; Stasolla, C.; Loukanina, N.; Yeung, E.; Thorpe, T. Glutathione modulation of purine metabolism in cultured white spruce embryogenic tissue. Plant Sci. 2003, 165, 1377–1385. [Google Scholar] [CrossRef]
- Silvia, D.S.; Emanuele, D.P.; Chiara, P. Stress responses and epigenomic instability mark the loss of somatic embryogenesis competence in grapevine. Plant Physiol. 2021, 188, 490–508. [Google Scholar] [CrossRef]
- Zhang, L.F.; Fan, Y.R.; Lan, Q.; Qi, L.W.; Han, S.Y. Expression of the SPL-like gene LaSPL9 in Japanese larch (Larix leptolepis) is regulated by miR156 during somatic embryogenesis. Trees 2021, 35, 1727–1737. [Google Scholar] [CrossRef]
- Ci, H.T.; Li, C.Y.; Aung, T.T.; Wang, S.L.; Yun, C.; Wang, F.; Ren, X.X.; Zhang, X.X. A Comparative transcriptome analysis reveals the molecular mechanisms that underlie somatic embryogenesis in Peaonia ostii ‘Fengdan’. Int. J. Mol. Sci. 2022, 23, 10595. [Google Scholar] [CrossRef]
- Yuan, J.B.; Chao, Y.H.; Han, L.B. Uncovering a phenomenon of active hormone transcriptional regulation during early somatic embryogenesis in Medicago sativa. Int. J. Mol. Sci. 2022, 23, 8633. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, W.Y.; Kim, Y.W.; Moon, H.K. Characterization of metabolic differences between embryogenic and non-embryogenic cells in forest trees. BMC Proc. 2011, 5, P146. [Google Scholar] [CrossRef] [Green Version]
- Izabela, D.; Edward, B.; Abreu, I.N.; Thomas, M.; Ulrika, E. Metabolome and transcriptome profiling reveal new insights into somatic embryo germination in Norway spruce (Picea abies). Tree Physiol. 2017, 37, 1752–1766. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.F.; Deng, C.; Zhu, T.; Ling, J.; Zhang, H.; Kong, L.S.; Zhang, S.G.; Wang, J.H.; Chen, X.Y. Dynamics of Physiological and miRNA Changes after Long-Term Proliferation in Somatic Embryogenesis of Picea balfouriana; Springer: Berlin/Heidelberg, Germany, 2019; Volume 33, pp. 469–480. [Google Scholar] [CrossRef]
- Businge, E.; Brackmann, K.; Moritz, T.; Egertsdotter, U. Metabolite profiling reveals clear metabolic changes during somatic embryo development of Norway spruce (Picea abies). Tree Physiol. 2012, 32, 232–244. [Google Scholar] [CrossRef]
- Gao, F.; Peng, C.; Wang, H.; Shen, H.L.; Yang, L. Selection of culture conditions for callus induction and proliferation by somatic embryogenesis of Pinus koraiensis. J. For. Res. 2021, 32, 9. [Google Scholar] [CrossRef]
- Gao, F.; Peng, C.; Wang, H.; Tretyakova, I.N.; Nosov, A.M.; Shen, H.L.; Yang, L. Key Techniques for somatic embryogenesis and plant regeneration of Pinus koraiensis. Forests 2020, 11, 912. [Google Scholar] [CrossRef]
- Peng, C.; Gao, F.; Wang, H.; Shen, H.L.; Yang, L. Optimization of maturation process for somatic embryo production and cryopreservation of embryogenic tissue in Pinus koraiensis. Plant Cell Tissue Org. 2020, 144, 185–194. [Google Scholar] [CrossRef]
- Peng, C.; Gao, F.; Wang, H.; Shen, H.L.; Yang, L. Physiological and biochemical traits in Korean Pine somatic embryogenesis. Forests 2020, 11, 577. [Google Scholar] [CrossRef]
- Belmonte, M.F.; Ambrose, S.J.; Ross, A.; Abrams, S.; Stasolla, C. Improved development of microspore-derived embryo cultures of Brassica napus cv Topaz following changes in glutathione metabolism. Physiol. Plantarum. 2006, 127, 690–700. [Google Scholar] [CrossRef]
- Dixon, R.A. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Claudio, S.; Belmonte, M.F.; Craig, D.L.; Liu, W.; Yeung, E.C.; Sederoff, R.R. The effect of reduced glutathione on morphology and gene expression of white spruce (Picea glauca) somatic embryos. J. Exp. Bot. 2004, 55, 695–709. [Google Scholar] [CrossRef]
- Belmonte, M.F.; Gillian, D.; Reid, D.M.; Yeung, E.C.; Stasolla, C. Alterations of the glutathione redox state improve apical meristem structure and somatic embryo quality in white spruce (Picea glauca). J. Exp. Bot. 2005, 56, 2355–2364. [Google Scholar] [CrossRef] [Green Version]
- Hacham, Y.; Koussevitzky, S.; Kirma, M.; Amir, R. Glutathione application affects the transcript profile of genes in Arabidopsis seedling. J. Plant Physiol. 2014, 171, 1444–1451. [Google Scholar] [CrossRef]
- Noshi, M.; Yamada, H.; Hatanaka, R.; Tanabe, N.; Tamoi, M.; Shigeoka, S. Arabidopsis dehydroascorbate reductase 1 and 2 modulate redox states of ascorbate-glutathione cycle in the cytosol in response to photooxidative stress. Biosci. Biotechnol. Biochem. 2017, 81, 523–533. [Google Scholar] [CrossRef]
- Shohael, A.M.; Ali, M.B.; Hahn, E.J.; Paek, K.Y. Glutathione metabolism and antioxidant responses during Eleutherococcus senticosus somatic embryo development in a bioreactor. Plant Cell Tissue Org. 2007, 89, 121–129. [Google Scholar] [CrossRef]
- Guilfoyle, T.J. Chapter 19 Auxin-regulated genes and promoters. New Compr. Biochem. 1999, 33, 423–459. [Google Scholar] [CrossRef]
- Bhosale, M.; Pande, S.; Kumar, A.; Kairamkonda, S.; Nandi, D. Characterization of two M17 family members in Escherichia coli, Peptidase A and Peptidase B. Biochem. Biophys. Res. Commun. 2010, 395, 76–81. [Google Scholar] [CrossRef]
- Reijns, M.; Lu, Y.; Leach, S.; Colloms, S.D. Mutagenesis of PepA suggests a new model for the Xer/cer synaptic complex. Mol. Microbiol. 2010, 57, 927–941. [Google Scholar] [CrossRef]
- Gali, R.R.; Board, P.G. Sequencing and expression of a cDNA for human glutathione synthetase. Biochem. J. 1995, 310, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.J.; Alan, M.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A. Arabidopsis AUX1 gene: A permease-like regulator of root gravitopism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef]
- Grieneisen, V.A.; Jian, X.; Maree, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [Green Version]
- Mateo-Bonmati, E.; Casanova-Saez, R.; Simura, J.; Ljung, K. Redefining the roles of UDP-glycosyltransferases in auxin metabolism and homeostasis during plant development. New Phytol. 2021, 26, 427012. [Google Scholar] [CrossRef]
- Qiu, T.; Chen, Y.; Li, M.M.; Kong, Y.Y.; Zhu, Y.B.; Han, N.; Bian, H.W.; Zhu, M.Y.; Wang, J.H. The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily of SMALL AUXIN UP RNA genes: Potential implications. Plant Signal. Behav. 2013, 8, e25283. [Google Scholar] [CrossRef] [Green Version]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inze, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Mockaitis, K.; Estelle, M. Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef] [Green Version]
- Litvay, J.D.; Verma, D.C.; Johnson, M.A. Influence of a loblolly pine (Pinus taeda L.). Culture medium and its components on growth and somatic embryogenesis of the wild carrot (Daucus carota L.). Plant Cell Rep. 1985, 4, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Chao, X.; Daniel, H. Fast and sensitive protein alignment using diamond. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.; Chen, Y.; Fuchu, H.E. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–72. [Google Scholar] [CrossRef]
- Rolf, A.; Amos, B.; Wu, C.H.; Barker, W.C.; Brigitte, B.; Serenella, F.; Elisabeth, G.; Huang, H.; Rodrigo, L.; Michele, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [Green Version]
- Tatusov, L.R.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Jaime, H.C.; Damian, S.; Kristoffer, F.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, 286–293. [Google Scholar] [CrossRef] [Green Version]
- Minoru, K.; Susumu, G.; Shuichi, K.; Yasushi, O. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, F.; Peng, C.; Zhang, Y.; Wang, H.; Shen, H.; Yang, L. Glutathione Plays a Positive Role in the Proliferation of Pinus koraiensis Embryogenic Cells. Int. J. Mol. Sci. 2022, 23, 14679. https://doi.org/10.3390/ijms232314679
Gao F, Peng C, Zhang Y, Wang H, Shen H, Yang L. Glutathione Plays a Positive Role in the Proliferation of Pinus koraiensis Embryogenic Cells. International Journal of Molecular Sciences. 2022; 23(23):14679. https://doi.org/10.3390/ijms232314679
Chicago/Turabian StyleGao, Fang, Chunxue Peng, Yue Zhang, Hao Wang, Hailong Shen, and Ling Yang. 2022. "Glutathione Plays a Positive Role in the Proliferation of Pinus koraiensis Embryogenic Cells" International Journal of Molecular Sciences 23, no. 23: 14679. https://doi.org/10.3390/ijms232314679




