Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation
Abstract
1. Introduction
2. Results
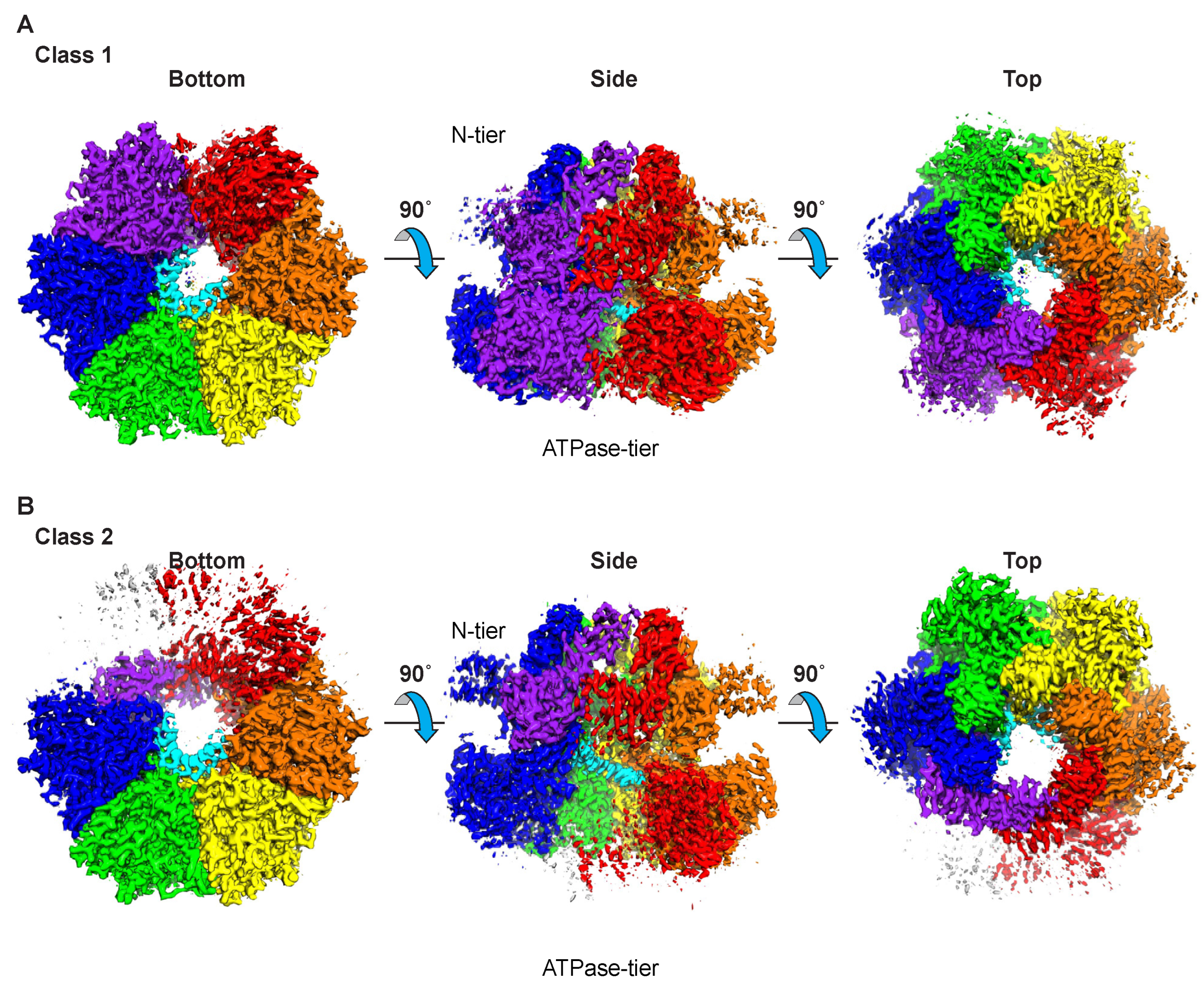
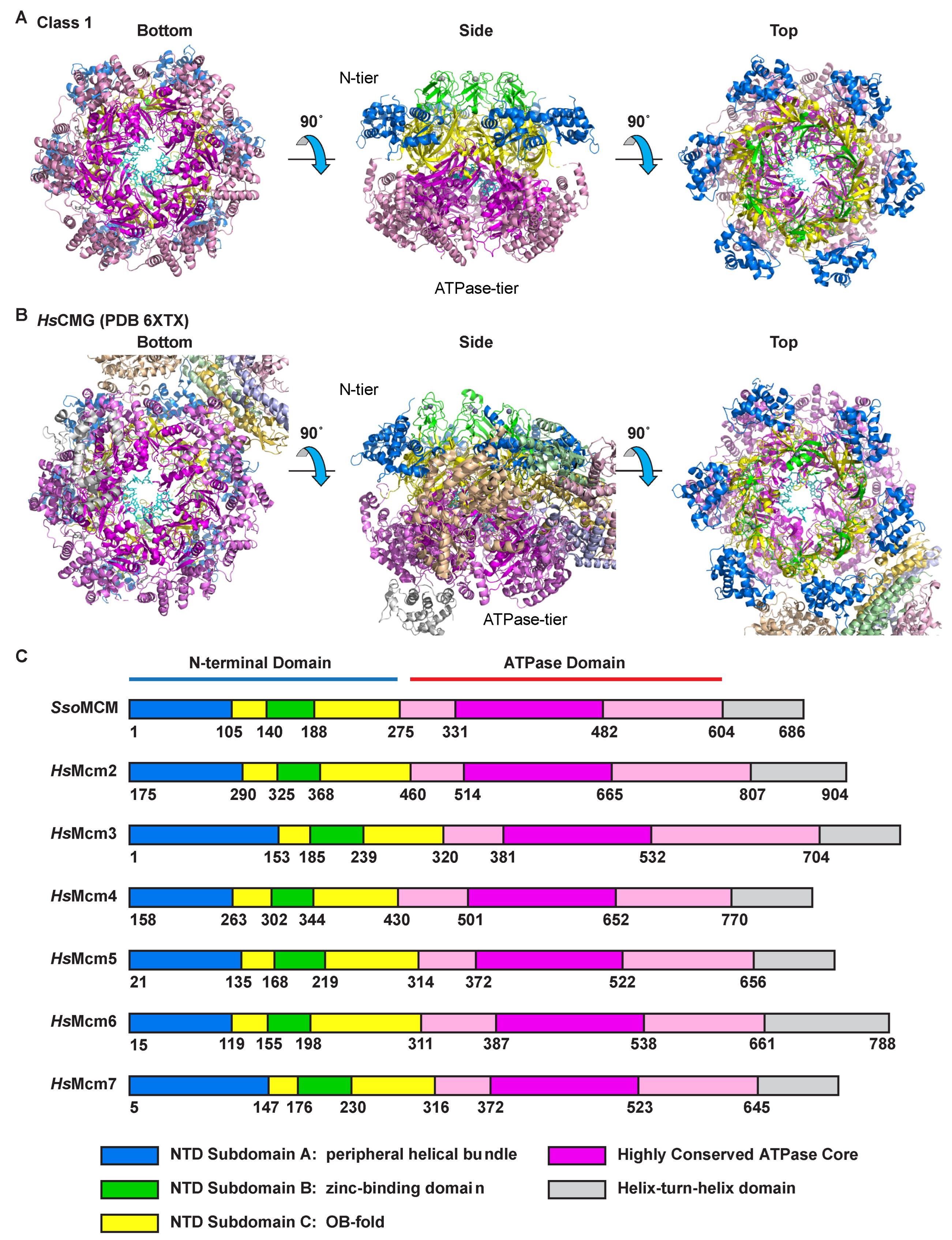
2.1. Overall Architecture
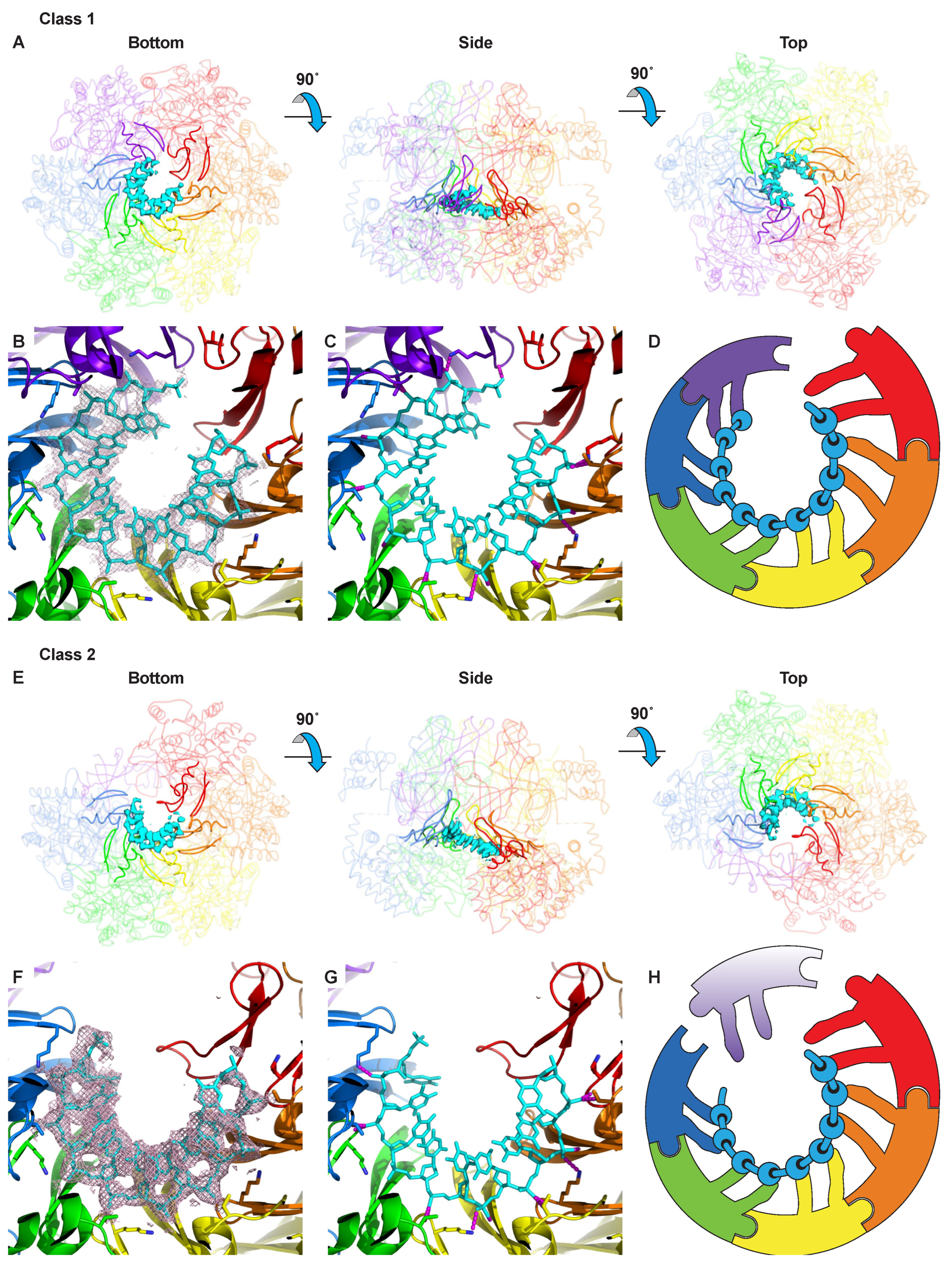
2.2. Class 1: Six Subunits Bound to ssDNA
2.3. Class 2: Five Subunits Bound to ssDNA
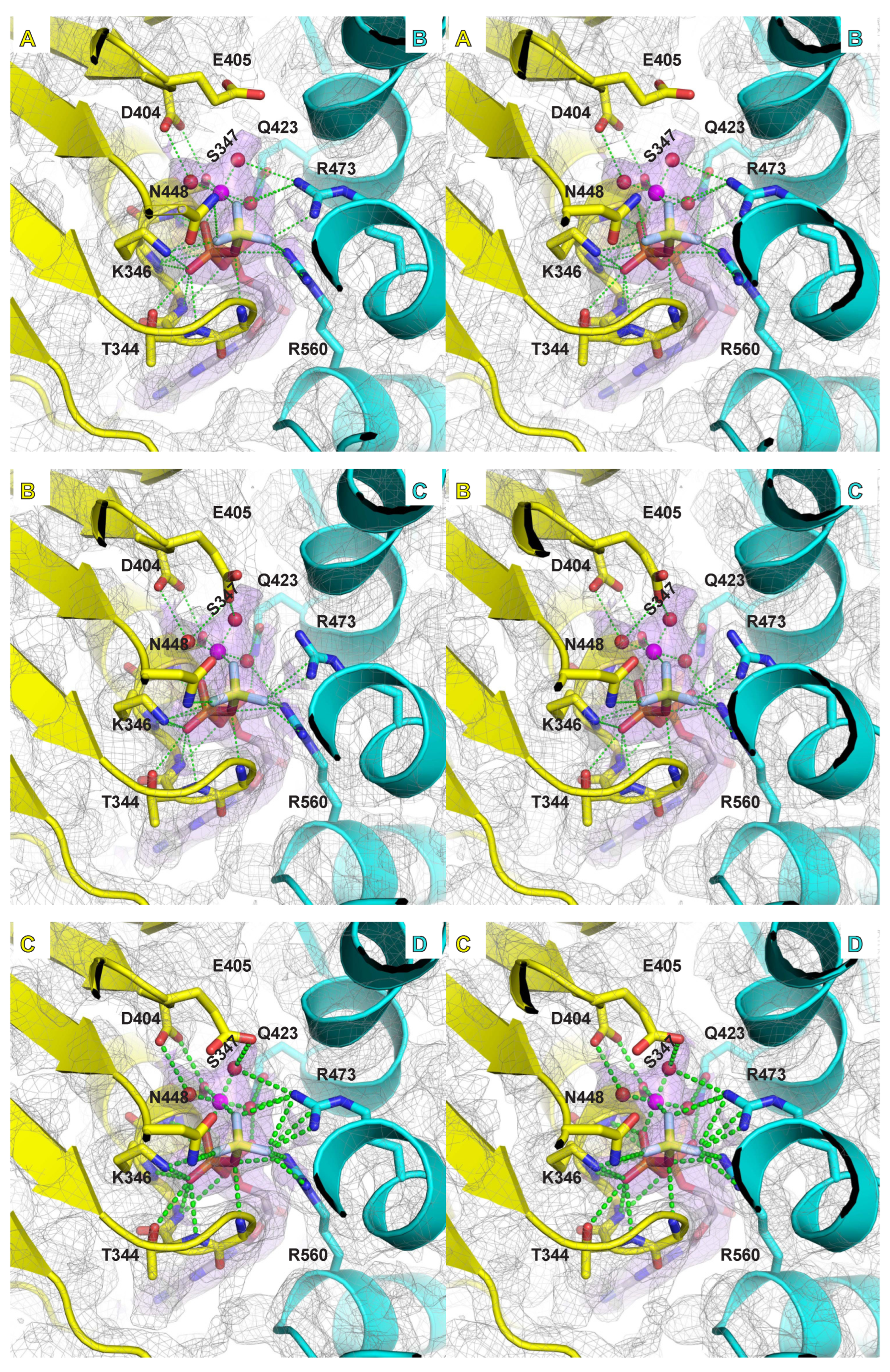
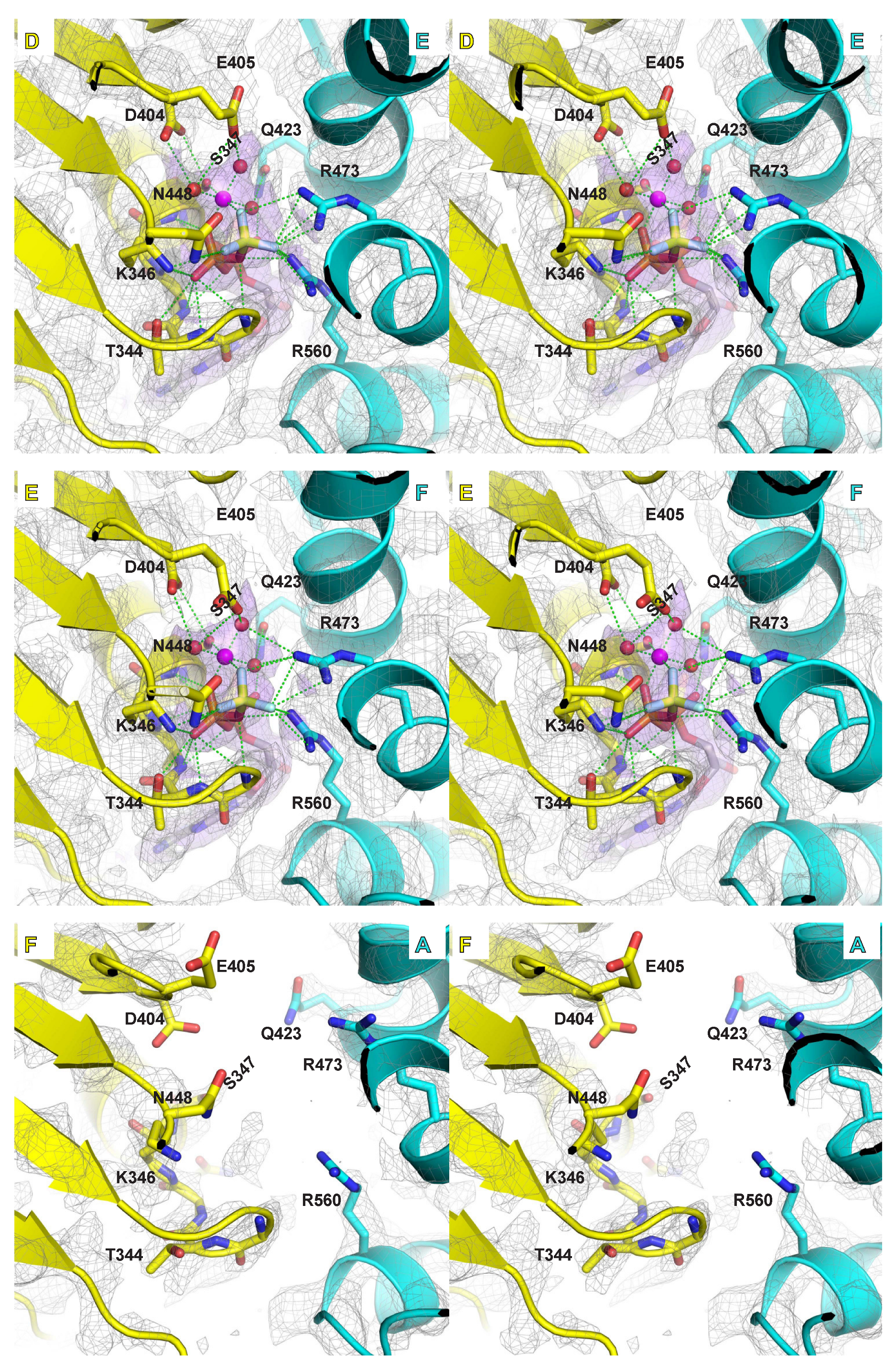
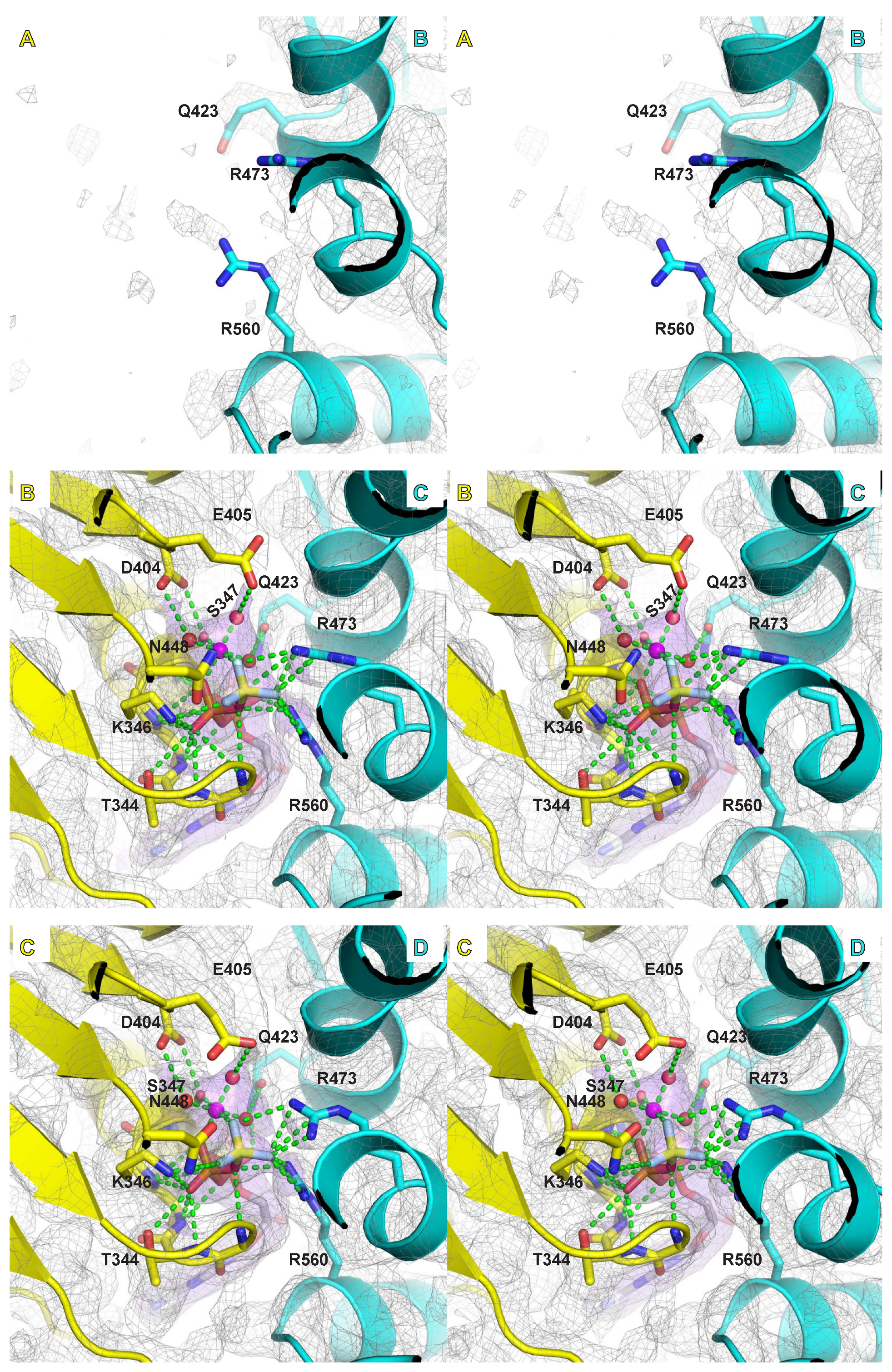
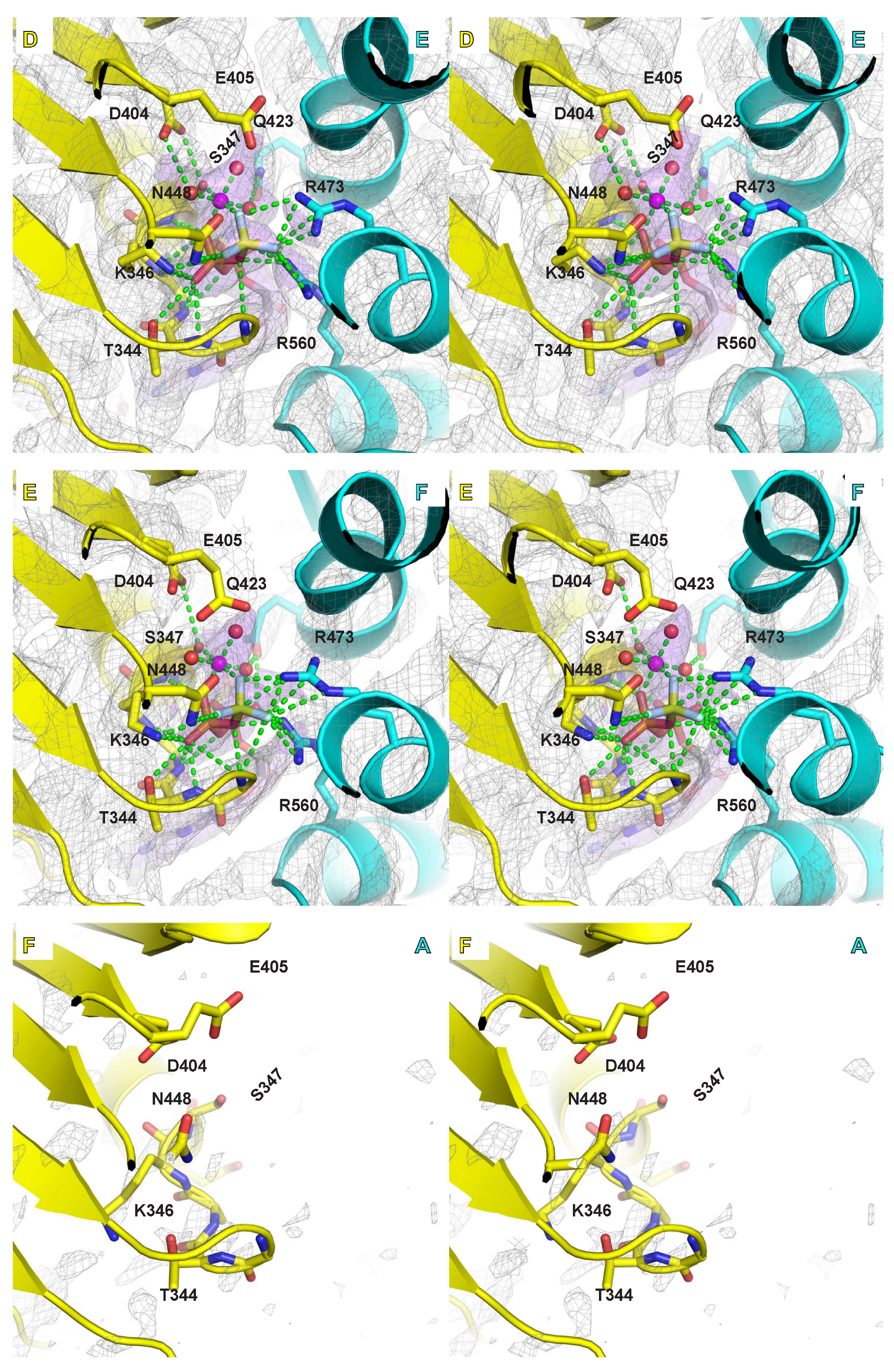
2.4. Mg/ATPase Sites
3. Discussion
Translocation Mechanism
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Cryo-EM Sample Preparation
4.3. Data Collection, Structure Determination, and Structure Refinement
4.4. SsoMCM–T16–MgADP-BeF3
4.5. SsoMCM–T20-CTATAG-T20–MgADP-BeF3
4.6. SsoMCM–T12–MgADP-BeF3
4.7. Merged Structures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
| SsoMCM–T12–MgADP-BeF3 class 1: | 8EAF, EMD-27974 |
| SsoMCM–T12–MgADP-BeF3 class 2: | 8EAG, EMD-27975 |
| SsoMCM–T16–MgADP-BeF3 class 1: | 8EAH, EMD-27976 |
| SsoMCM–T16–MgADP-BeF3 class 2: | 8EAI, EMD-27977 |
| SsoMCM–T20-CTATAG-T20–MgADP-BeF3 class 1: | 8EAJ, EMD-27978 |
| SsoMCM–T20-CTATAG-T20–MgADP-BeF3 class 2: | 8EAK, EMD-27979 |
| Merged class 1 particles: | 8EAL, EMD-27980 |
| Merged class 2 particles: | 8EAM, EMD-27981 |
Acknowledgments
Conflicts of Interest
References
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef]
- Patel, S.S.; Picha, K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000, 69, 651–697. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, W. Different mechanisms for translocation by monomeric and hexameric helicases. Curr. Opin. Struct. Biol. 2020, 61, 25–32. [Google Scholar] [CrossRef]
- Yu, X.; Hingorani, M.M.; Patel, S.S.; Egelman, E.H. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 1996, 3, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Enemark, E.J.; Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 2006, 442, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.V.; Yardimci, H.; Long, D.T.; Ho, T.V.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Scharer, O.D.; Walter, J.C. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Itsathitphaisarn, O.; Wing, R.A.; Eliason, W.K.; Wang, J.; Steitz, T.A. The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation. Cell 2012, 151, 267–277. [Google Scholar] [CrossRef]
- Lee, S.J.; Syed, S.; Enemark, E.J.; Schuck, S.; Stenlund, A.; Ha, T.; Joshua-Tor, L. Dynamic look at DNA unwinding by a replicative helicase. Proc. Natl. Acad. Sci. USA 2014, 111, E827–E835. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [CrossRef]
- Chong, J.P.; Hayashi, M.K.; Simon, M.N.; Xu, R.M.; Stillman, B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 2000, 97, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Koonin, E.V.; Wolf, Y.I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990, 262, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.T. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 2000, 55, 75–134. [Google Scholar] [PubMed]
- Stenlund, A. Initiation of DNA replication: Lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 2003, 4, 777–785. [Google Scholar] [CrossRef]
- Oakley, A.J. A structural view of bacterial DNA replication. Protein Sci. 2019, 28, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.; Falkenberg, M. TWINKLE and Other Human Mitochondrial DNA Helicases: Structure, Function and Disease. Genes 2020, 11, 408. [Google Scholar] [CrossRef]
- Kulczyk, A.W.; Richardson, C.C. The Replication System of Bacteriophage T7. Enzymes 2016, 39, 89–136. [Google Scholar]
- Brennan, C.A.; Dombroski, A.J.; Platt, T. Transcription termination factor rho is an RNA-DNA helicase. Cell 1987, 48, 945–952. [Google Scholar] [CrossRef]
- Tye, B.K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999, 68, 649–686. [Google Scholar] [CrossRef]
- Davey, M.J.; Indiani, C.; O’Donnell, M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 2003, 278, 4491–4499. [Google Scholar] [CrossRef]
- Li, N.; Zhai, Y.; Zhang, Y.; Li, W.; Yang, M.; Lei, J.; Tye, B.K.; Gao, N. Structure of the eukaryotic MCM complex at 3.8 A. Nature 2015, 524, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Rzechorzek, N.J.; Hardwick, S.W.; Jatikusumo, V.A.; Chirgadze, D.Y.; Pellegrini, L. CryoEM structures of human CMG-ATPgammaS-DNA and CMG-AND-1 complexes. Nucleic Acids Res. 2020, 48, 6980–6995. [Google Scholar] [CrossRef] [PubMed]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gristwood, T.; Hodgson, B.; Trinidad, J.C.; Albers, S.V.; Bell, S.D. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc. Natl. Acad. Sci. USA 2016, 113, 13390–13395. [Google Scholar] [CrossRef]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef]
- Bochman, M.L.; Schwacha, A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 2008, 31, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.D. The ATP synthase—A splendid molecular machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Boyer, P.D. The binding change mechanism for ATP synthase—Some probabilities and possibilities. Biochim. Biophys. Acta 1993, 1140, 215–250. [Google Scholar] [CrossRef]
- Stock, D.; Gibbons, C.; Arechaga, I.; Leslie, A.G.; Walker, J.E. The rotary mechanism of ATP synthase. Curr. Opin. Struct. Biol. 2000, 10, 672–679. [Google Scholar] [CrossRef]
- Sobti, M.; Ueno, H.; Noji, H.; Stewart, A.G. The six steps of the complete F1-ATPase rotary catalytic cycle. Nat. Commun. 2021, 12, 4690. [Google Scholar] [CrossRef]
- Egelman, E.H.; Yu, X.; Wild, R.; Hingorani, M.M.; Patel, S.S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl. Acad. Sci. USA 1995, 92, 3869–3873. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Sawaya, M.R.; Ellenberger, T.; Wigley, D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 2000, 101, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, J.P.; Leslie, A.G.; Lutter, R.; Walker, J.E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 1994, 370, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cui, Y.; Fox, T.; Lin, S.; Wang, H.; de Val, N.; Zhou, Z.H.; Yang, W. Structures and operating principles of the replisome. Science 2019, 363, eaav7003. [Google Scholar] [CrossRef]
- Thomsen, N.D.; Berger, J.M. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell 2009, 139, 523–534. [Google Scholar] [CrossRef]
- Meagher, M.; Epling, L.B.; Enemark, E.J. DNA translocation mechanism of the MCM complex and implications for replication initiation. Nat. Commun. 2019, 10, 3117. [Google Scholar] [CrossRef]
- Pape, T.; Meka, H.; Chen, S.; Vicentini, G.; van Heel, M.; Onesti, S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003, 4, 1079–1083. [Google Scholar] [CrossRef]
- Costa, A.; Pape, T.; van Heel, M.; Brick, P.; Patwardhan, A.; Onesti, S. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006, 34, 5829–5838. [Google Scholar] [CrossRef]
- Bochman, M.L.; Schwacha, A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 2007, 282, 33795–33804. [Google Scholar] [CrossRef]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef]
- Costa, A.; Ilves, I.; Tamberg, N.; Petojevic, T.; Nogales, E.; Botchan, M.R.; Berger, J.M. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011, 18, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Bai, L.; Sun, J.; Georgescu, R.; Liu, J.; O’Donnell, M.E.; Li, H. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 2016, 23, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Yuan, Z.; Bai, L.; Schneider, S.; Zhao, G.; Stillman, B.; Speck, C.; Li, H. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc. Natl. Acad. Sci. USA 2017, 114, E9529–E9538. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.J.; Bishop, B.E.; Leon, R.P.; Sclafani, R.A.; Ogata, C.M.; Chen, X.S. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003, 10, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.M.; Enemark, E.J. Archaeal MCM Proteins as an Analog for the Eukaryotic Mcm2-7 Helicase to Reveal Essential Features of Structure and Function. Archaea 2015, 2015, 305497. [Google Scholar] [CrossRef] [PubMed]
- Croll, T.I. ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018, 74 Pt 6, 519–530. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, version 1.3r1; Schrodinger Inc.: New York, NY, USA, 2010.
- Kagawa, R.; Montgomery, M.G.; Braig, K.; Leslie, A.G.; Walker, J.E. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004, 23, 2734–2744. [Google Scholar] [CrossRef]
- Gai, D.; Zhao, R.; Li, D.; Finkielstein, C.V.; Chen, X.S. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 2004, 119, 47–60. [Google Scholar] [CrossRef]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [CrossRef]
- Bochman, M.L.; Bell, S.P.; Schwacha, A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell Biol. 2008, 28, 5865–5873. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003, 333, 781–815. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, P.; Kose, H.B.; Martino, F.; Petojevic, T.; Abid Ali, F.; Locke, J.; Tamberg, N.; Nans, A.; Berger, J.M.; Botchan, M.R.; et al. Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome. Cell Rep. 2019, 28, 2673–2688.e8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Georgescu, R.; Bai, L.; Zhang, D.; Li, H.; O’Donnell, M.E. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun. 2020, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Baretic, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef]
- Mossessova, E.; Lima, C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 2000, 5, 865–876. [Google Scholar] [CrossRef]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Zivanov, J.; Nakane, T.; Forsberg, B.O.; Kimanius, D.; Hagen, W.J.; Lindahl, E.; Scheres, S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018, 7, e42166. [Google Scholar] [CrossRef] [PubMed]

| Dataset | SsoMCM–T12– MgADP-BeF3 | SsoMCM–T16– MgADP-BeF3 | SsoMCM– T20-CTATAG-T20– MgADP-BeF3 | Merged Particles | ||||
|---|---|---|---|---|---|---|---|---|
| Microscope | Titan Krios | Titan Krios | Titan Krios | N/A | ||||
| kEV | 300 | 300 | 300 | N/A | ||||
| Micrographs | 1928 | 5171 | 2718 | N/A | ||||
| Detector | K3 | K3 | K3 | N/A | ||||
| Magnification | 81,000 | 81,000 | 81,000 | N/A | ||||
| Pixel size (Å) | 1.08 | 1.08 | 1.08 | N/A | ||||
| Particles after 2D classification | 1,067,534 | 2,839,559 | 1,568,736 | N/A | ||||
| Class | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 |
| Particles | 645,002 | 268,300 | 1,649,356 | 776,823 | 832,313 | 448,473 | 3,126,671 | 1,493,596 |
| Resolution * (Å) | 2.62 | 3.01 | 2.48 | 2.76 | 2.45 | 2.67 | 2.34 | 2.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meagher, M.; Myasnikov, A.; Enemark, E.J. Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation. Int. J. Mol. Sci. 2022, 23, 14678. https://doi.org/10.3390/ijms232314678
Meagher M, Myasnikov A, Enemark EJ. Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation. International Journal of Molecular Sciences. 2022; 23(23):14678. https://doi.org/10.3390/ijms232314678
Chicago/Turabian StyleMeagher, Martin, Alexander Myasnikov, and Eric J. Enemark. 2022. "Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation" International Journal of Molecular Sciences 23, no. 23: 14678. https://doi.org/10.3390/ijms232314678
APA StyleMeagher, M., Myasnikov, A., & Enemark, E. J. (2022). Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation. International Journal of Molecular Sciences, 23(23), 14678. https://doi.org/10.3390/ijms232314678






