Transcriptomic Dysregulation of Inflammation-Related Genes in Leukocytes of Patients with Gestational Diabetes Mellitus (GDM) during and after Pregnancy: Identifying Potential Biomarkers Relevant to Glycemic Abnormality
Abstract
:1. Introduction
2. Results
2.1. Clinical Data for the Groups Studied
2.2. Gene Expression Alterations in Inflammation-Related Genes
2.3. Correlations between Gene Expression and Clinical Variables
2.4. Gene–Gene Expression Correlations
2.5. Diagnostic Potential of Inflammation-Related Genes
2.6. Prediction of Postpartum AGT Incidence
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Clinical and Biochemical Measurements
4.3. Leukocytes’ Separation and Total RNA Extraction
4.4. cDNA Synthesis and Nested Quantitative Polymerase Chain Reaction (qPCR)
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paulo, M.S.; Abdo, N.M.; Bettencourt-Silva, R.; Al-Rifai, R.H. Gestational Diabetes Mellitus in Europe: A Systematic Review and Meta-Analysis of Prevalence Studies. Front. Endocrinol. (Lausanne) 2021, 12, 691033. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37 (Suppl. S1), S81–S90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wierzba, W.; Śliwczyński, A.; Karnafel, W.; Bojar, I.; Pinkas, J. Gestational diabetes mellitus/hyperglycaemia during pregnancy in Poland in the years 2010–2012 based on the data from the National Health Fund. Ginekol. Pol. 2017, 88, 244–248. [Google Scholar] [CrossRef] [Green Version]
- Gilmartin, A.B.; Ural, S.H.; Repke, J.T. Gestational diabetes mellitus. Rev. Obstet. Gynecol. 2008, 1, 129–134. [Google Scholar] [PubMed]
- Li, Z.; Cheng, Y.; Wang, D.; Chen, H.; Chen, H.; Ming, W.K.; Wang, Z. Incidence Rate of Type 2 Diabetes Mellitus after Gestational Diabetes Mellitus: A Systematic Review and Meta-Analysis of 170,139 Women. J. Diabetes Res. 2020, 2020, 3076463. [Google Scholar] [CrossRef]
- Kramer, C.K.; Campbell, S.; Retnakaran, R. Gestational diabetes and the risk of cardiovascular disease in women: A systematic review and meta-analysis. Diabetologia 2019, 62, 905–914. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, S.D.; Umans, J.G.; Ratner, R. Gestational diabetes: Implications for cardiovascular health. Curr. Diabetes Rep. 2012, 12, 43–52. [Google Scholar] [CrossRef]
- Kim, C.; Newton, K.M.; Knopp, R.H. Gestational diabetes and the incidence of type 2 diabetes: A systematic review. Diabetes Care 2002, 25, 1862–1868. [Google Scholar] [CrossRef] [Green Version]
- Noctor, E.; Dunne, F.P. Type 2 diabetes after gestational diabetes: The influence of changing diagnostic criteria. World J. Diabetes 2015, 6, 234–244. [Google Scholar] [CrossRef]
- Vounzoulaki, E.; Khunti, K.; Abner, S.C.; Tan, B.K.; Davies, M.J.; Gillies, C.L. Progression to type 2 diabetes in women with a known history of gestational diabetes: Systematic review and meta-analysis. BMJ 2020, 369, m1361. [Google Scholar] [CrossRef]
- Unwin, N.; Shaw, J.; Zimmet, P.; Alberti, K.G. Impaired glucose tolerance and impaired fasting glycaemia: The current status on definition and intervention. Diabet. Med. 2002, 19, 708–723. [Google Scholar] [CrossRef]
- Tobias, D.K.; Hu, F.B.; Chavarro, J.; Rosner, B.; Mozaffarian, D.; Zhang, C. Healthful dietary patterns and type 2 diabetes mellitus risk among women with a history of gestational diabetes mellitus. Arch. Intern. Med. 2012, 172, 1566–1572. [Google Scholar] [CrossRef] [Green Version]
- Bao, W.; Tobias, D.K.; Bowers, K.; Chavarro, J.; Vaag, A.; Grunnet, L.G.; Strøm, M.; Mills, J.; Liu, A.; Kiely, M.; et al. Physical activity and sedentary behaviors associated with risk of progression from gestational diabetes mellitus to type 2 diabetes mellitus: A prospective cohort study. JAMA Intern. Med. 2014, 174, 1047–1055. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.H.; Wang, D.P.; Zhang, L.L.; Zhang, F.; Wang, D.M.; Zhang, W.Y. Genomic expression profiles of blood and placenta reveal significant immune-related pathways and categories in Chinese women with gestational diabetes mellitus. Diabet. Med. 2011, 28, 237–246. [Google Scholar] [CrossRef]
- McElwain, C.J.; McCarthy, F.P.; McCarthy, C.M. Gestational Diabetes Mellitus and Maternal Immune Dysregulation: What We Know So Far. Int. J. Mol. Sci. 2021, 22, 4261. [Google Scholar] [CrossRef]
- Gomes, C.P.; Torloni, M.R.; Gueuvoghlanian-Silva, B.Y.; Alexandre, S.M.; Mattar, R.; Daher, S. Cytokine levels in gestational diabetes mellitus: A systematic review of the literature. Am. J. Reprod. Immunol. 2013, 69, 545–557. [Google Scholar] [CrossRef]
- Skórzyńska-Dziduszko, K.E.; Kimber-Trojnar, Ż.; Patro-Małysza, J.; Olszewska, A.; Zaborowski, T.; Małecka-Massalska, T. An Interplay between Obesity and Inflammation in Gestational Diabetes Mellitus. Curr. Pharm. Biotechnol. 2016, 17, 603–613. [Google Scholar] [CrossRef]
- Liew, C.C.; Ma, J.; Tang, H.C.; Zheng, R.; Dempsey, A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef]
- Mac-Marcjanek, K.; Zieleniak, A.; Zurawska-Klis, M.; Cypryk, K.; Wozniak, L.; Wojcik, M. Expression Profile of Diabetes-Related Genes Associated with Leukocyte Sirtuin 1 Overexpression in Gestational Diabetes. Int. J. Mol. Sci. 2018, 19, 3826. [Google Scholar] [CrossRef] [Green Version]
- Bartáková, V.; Malúšková, D.; Mužík, J.; Bělobrádková, J.; Kaňková, K. Possibility to predict early postpartum glucose abnormality following gestational diabetes mellitus based on the results of routine mid-gestational screening. Biochem. Med. 2015, 25, 460–468. [Google Scholar] [CrossRef]
- Miao, Z.; Wu, H.; Ren, L.; Bu, N.; Jiang, L.; Yang, H.; Zhang, J.; Guo, X. Long-Term Postpartum Outcomes of Insulin Resistance and β-cell Function in Women with Previous Gestational Diabetes Mellitus. Int. J. Endocrinol. 2020, 2020, 7417356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Zhang, C. Prevalence of Gestational Diabetes and Risk of Progression to Type 2 Diabetes: A Global Perspective. Curr. Diabetes Rep. 2016, 16, 7. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.; Kucuk, M.; Ilgin, A.; Dagdelen, M. Assessment of insulin sensitivity/resistance and their relations with leptin concentrations and anthropometric measures in a pregnant population with and without gestational diabetes mellitus. J. Diabetes Complicat. 2010, 24, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.A.; Kasim, M.M.; Noor Aizuddin, A.; Umar, N.A. Homeostatic indices of insulin resistance among gestational diabetics in anticipating pregnancy complications. Gynecol. Endocrinol. 2013, 29, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Lekva, T.; Bollerslev, J.; Godang, K.; Roland, M.C.; Friis, C.M.; Voldner, N.; Henriksen, T.; Ueland, T. β-cell dysfunction in women with previous gestational diabetes is associated with visceral adipose tissue distribution. Eur. J. Endocrinol. 2015, 173, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Szymański, F.M.; Barylski, M.; Cybulska, B.; Wożakowska-Kapłon, B.; Krasiński, Z.; Mamcarz, A.; Widecka, K.; Płatek, A.E.; Dudek, D.; Mickiewicz, A.; et al. Recommendation for the management of dyslipidemia in Poland—Third Declaration of Sopot. Interdisciplinary Expert Position Statement endorsed by the Polish Cardiac Society Working Group on Cardiovascular Pharmacotherapy. Cardiol. J. 2018, 25, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Özyer, Ş.; Engin-Üstün, Y.; Uzunlar, Ö.; Katar, C.; Danışman, N. Inflammation and glycemic tolerance status in pregnancy: The role of maternal adiposity. Gynecol. Obstet. Investig. 2014, 78, 53–58. [Google Scholar] [CrossRef]
- Retnakaran, R.; Hanley, A.J.; Raif, N.; Connelly, P.W.; Sermer, M.; Zinman, B. C-reactive protein and gestational diabetes: The central role of maternal obesity. J. Clin. Endocrinol. Metab. 2003, 88, 3507–3512. [Google Scholar] [CrossRef] [Green Version]
- Morisset, A.S.; Dubé, M.C.; Côté, J.A.; Robitaille, J.; Weisnagel, S.J.; Tchernof, A. Circulating interleukin-6 concentrations during and after gestational diabetes mellitus. Acta Obstet. Gynecol. Scand. 2011, 90, 524–530. [Google Scholar] [CrossRef]
- Qazi, B.S.; Tang, K.; Qazi, A. Recent advances in underlying pathologies provide insight into interleukin-8 expression-mediated inflammation and angiogenesis. Int. J. Inflam. 2011, 2011, 908468. [Google Scholar] [CrossRef]
- Pan, X.; Kaminga, A.C.; Wen, S.W.; Liu, A. Chemokines in Prediabetes and Type 2 Diabetes: A Meta-Analysis. Front. Immunol. 2021, 12, 622438. [Google Scholar] [CrossRef]
- Lappas, M.; Permezel, M.; Rice, G.E. Release of proinflammatory cytokines and 8-isoprostane from placenta, adipose tissue, and skeletal muscle from normal pregnant women and women with gestational diabetes mellitus. J. Clin. Endocrinol. Metab. 2004, 89, 5627–5633. [Google Scholar] [CrossRef] [Green Version]
- Kleiblova, P.; Dostalova, I.; Bartlova, M.; Lacinova, Z.; Ticha, I.; Krejci, V.; Springer, D.; Kleibl, Z.; Haluzik, M. Expression of adipokines and estrogen receptors in adipose tissue and placenta of patients with gestational diabetes mellitus. Mol. Cell. Endocrinol. 2010, 314, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Kuzmicki, M.; Telejko, B.; Wawrusiewicz-Kurylonek, N.; Citko, A.; Lipinska, D.; Pliszka, J.; Wilk, J.; Kalejta, K.; Lemancewicz, A.; Grabiec, M.; et al. The expression of suppressor of cytokine signaling 1 and 3 in fat and placental tissue from women with gestational diabetes. Gynecol. Endocrinol. 2012, 28, 841–844. [Google Scholar] [CrossRef]
- Purohit, S.; Sharma, A.; Hopkins, D.; Steed, L.; Bode, B.; Anderson, S.W.; Reed, J.C.; Steed, R.D.; Yang, T.; She, J.X. Large-Scale Discovery and Validation Studies Demonstrate Significant Reductions in Circulating Levels of IL8, IL-1Ra, MCP-1, and MIP-1β in Patients With Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2015, 100, E1179–E1187. [Google Scholar] [CrossRef] [Green Version]
- Kuzmicki, M.; Telejko, B.; Zonenberg, A.; Szamatowicz, J.; Kretowski, A.; Nikolajuk, A.; Laudanski, P.; Gorska, M. Circulating pro- and anti-inflammatory cytokines in Polish women with gestational diabetes. Horm. Metab. Res. 2008, 40, 556–560. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, J.; Shen, L.; Wang, A.; Wang, R. Correlation between inflammatory markers (hs-CRP, TNF-α, IL-1β, IL-6, IL-18), glucose intolerance, and gestational diabetes mellitus in pregnant women. Int. J. Clin. Exp. Med. 2018, 11, 8310–8316. [Google Scholar]
- Amirian, A.; Mahani, M.B.; Abdi, F. Role of interleukin-6 (IL-6) in predicting gestational diabetes mellitus. Obstet. Gynecol. Sci. 2020, 63, 407–416. [Google Scholar] [CrossRef]
- Tsigos, C.; Papanicolaou, D.A.; Kyrou, I.; Defensor, R.; Mitsiadis, C.S.; Chrousos, G.P. Dose-dependent effects of recombinant human interleukin-6 on glucose regulation. J. Clin. Endocrinol. Metab. 1997, 82, 4167–4170. [Google Scholar] [CrossRef]
- Devaraj, S.; Venugopal, S.K.; Singh, U.; Jialal, I. Hyperglycemia induces monocytic release of interleukin-6 via induction of protein kinase C-α and-β. Diabetes 2005, 54, 85–91. [Google Scholar] [CrossRef]
- Kang, J.; Lee, C.N.; Li, H.Y.; Hsu, K.H.; Wang, S.H.; Lin, S.Y. Association of Interleukin-10 Methylation Levels With Gestational Diabetes in a Taiwanese Population. Front. Genet. 2018, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Sabat, R.; Grütz, G.; Warszawska, K.; Kirsch, S.; Witte, E.; Wolk, K.; Geginat, J. Biology of interleukin-10. Cytokine Growth Factor Rev. 2010, 21, 331–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, K.; Nappo, F.; Marfella, R.; Giugliano, G.; Giugliano, F.; Ciotola, M.; Quagliaro, L.; Ceriello, A.; Giugliano, D. Inflammatory cytokine concentrations are acutely increased by hyperglycemia in humans: Role of oxidative stress. Circulation 2002, 106, 2067–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynn, T.A. Type 2 cytokines: Mechanisms and therapeutic strategies. Nat. Rev. Immunol. 2015, 15, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Stanya, K.J.; Jacobi, D.; Liu, S.; Bhargava, P.; Dai, L.; Gangl, M.R.; Inouye, K.; Barlow, J.L.; Ji, Y.; Mizgerd, J.P.; et al. Direct control of hepatic glucose production by interleukin-13 in mice. J. Clin. Investig. 2013, 123, 261–271. [Google Scholar] [CrossRef]
- Jiang, L.Q.; Franck, N.; Egan., B.; Sjögren, R.J.; Katayama, M.; Duque-Guimaraes, D.; Arner, P.; Zierath, J.R.; Krook, A. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E1359–E1366. [Google Scholar] [CrossRef] [Green Version]
- Lakshmi, L.J.; Rathore, S.; Faizal, M.; Yusuf, F.N.K.; Zephy, D. Insulin resistance alter the levels of IL-4, IL-5 and IL-13 in type 2 diabetes mellitus with less than 5 years of duration. Indian J. Appl. Res. 2022, 12, 40–43. [Google Scholar] [CrossRef]
- Georgiou, H.M.; Lappas, M.; Georgiou, G.M.; Marita, A.; Bryant, V.J.; Hiscock, R.; Permezel, M.; Khalil, Z.; Rice, G.E. Screening for biomarkers predictive of gestational diabetes mellitus. Acta Diabetol. 2008, 45, 157–165. [Google Scholar] [CrossRef]
- Li, Q.; Verma, I.M. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef]
- Abbad, L.; Prakoura, N.; Michon, A.; Chalghoumi, R.; Reichelt-Wurm, S.; Banas, M.C.; Chatziantoniou, C. Role of Periostin and Nuclear Factor-κB Interplay in the Development of Diabetic Nephropathy. Cells 2022, 11, 2212. [Google Scholar] [CrossRef]
- Kuzmicki, M.; Telejko, B.; Wawrusiewicz-Kurylonek, N.; Lipinska, D.; Pliszka, J.; Wilk, J.; Zielinska, A.; Skibicka, J.; Szamatowicz, J.; Kretowski, A.; et al. The expression of genes involved in NF-κB activation in peripheral blood mononuclear cells of patients with gestational diabetes. Eur. J. Endocrinol. 2013, 168, 419–427. [Google Scholar] [CrossRef]
- Hofmann, M.A.; Schiekofer, S.; Kanitz, M.; Klevesath, M.S.; Joswig, M.; Lee, V.; Morcos, M.; Tritschler, H.; Ziegler, R.; Wahl, P.; et al. Insufficient glycemic control increases nuclear factor-kappa B binding activity in peripheral blood mononuclear cells isolated from patients with type 1 diabetes. Diabetes Care 1998, 21, 1310–1316. [Google Scholar] [CrossRef]
- Schiekofer, S.; Galasso, G.; Andrassy, M.; Aprahamian, T.; Schneider, J.; Rocnik, E. Glucose control with insulin results in reduction of NF-kappaB-binding activity in mononuclear blood cells of patients with recently manifested type 1 diabetes. Diabetes Obes. Metab. 2006, 8, 473–482. [Google Scholar] [CrossRef]
- Saucedo, R.; Zarate, A.; Basurto, L.; Hernandez, M.; Puello, E.; Galvan, R.; Campos, S. Relationship between circulating adipokines and insulin resistance during pregnancy and postpartum in women with gestational diabetes. Arch. Med. Res. 2011, 42, 318–323. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, Y.H.; Chen, Y.P.; Yuan, X.L.; Wang, J.; Zhu, H.; Lu, C.M. Maternal circulating concentrations of tumor necrosis factor-alpha, leptin, and adiponectin in gestational diabetes mellitus: A systematic review and meta-analysis. Sci. World J. 2014, 2014, 926932. [Google Scholar] [CrossRef] [Green Version]
- Moller, D.E. Potential role of TNF-alpha in the pathogenesis of insulin resistance and type 2 diabetes. Trends Endocrinol. Metab. 2000, 11, 212–217. [Google Scholar] [CrossRef]
- Wang, S.L.; Liu, P.Q.; Ding, Y.; Peng, W.; Qu, X. [Maternal serum tumor necrosis factor-alpha concentration and correlation with insulin resistance in gestational diabetes]. Zhonghua Fu Chan Ke Za Zhi 2004, 39, 737–740. (In Chinese) [Google Scholar]
- Mohammed, A.; Aliyu, I.S.; Manu, M. Correlation between circulating level of tumor necrosis factor-alpha and insulin resistance in Nigerian women with gestational diabetes mellitus. Ann. Afr. Med. 2018, 17, 168–171. [Google Scholar] [CrossRef]
- Altinova, A.E.; Toruner, F.; Bozkurt, N.; Bukan, N.; Karakoc, A.; Yetkin, I.; Ayvaz, G.; Cakir, N.; Arslan, M. Circulating concentrations of adiponectin and tumor necrosis factor-alpha in gestational diabetes mellitus. Gynecol. Endocrinol. 2007, 23, 161–165. [Google Scholar] [CrossRef]
- Coughlan, M.T.; Oliva, K.; Georgiou, H.M.; Permezel, J.M.; Rice, G.E. Glucose-induced release of tumour necrosis factor-alpha from human placental and adipose tissues in gestational diabetes mellitus. Diabet. Med. 2001, 18, 921–927. [Google Scholar] [CrossRef]
- Bogdanet, D.; O’Shea, P.; Lyons, C.; Shafat, A.; Dunne, F. The Oral Glucose Tolerance Test-Is it Time for a Change?-A Literature Review with an Emphasis on Pregnancy. J. Clin. Med. 2020, 9, 3451. [Google Scholar] [CrossRef] [PubMed]
- Cosson, E.; Carbillon, L.; Valensi, P. High Fasting Plasma Glucose during Early Pregnancy: A Review about Early Gestational Diabetes Mellitus. J. Diabetes Res. 2017, 2017, 8921712. [Google Scholar] [CrossRef] [PubMed]
- Koning, S.H.; Lutgers, H.L.; Hoogenberg, K.; Trompert, C.A.; van den Berg, P.P.; Wolffenbuttel, B.H. Postpartum glucose follow-up and lifestyle management after gestational diabetes mellitus: General practitioner and patient perspectives. J. Diabetes Metab. Disord. 2016, 15, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastore, I.; Chiefari, E.; Vero, R.; Brunetti, A. Postpartum glucose intolerance: An updated overview. Endocrine 2018, 59, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Sears, B. Anti-inflammatory Diets. J. Am. Coll. Nutr. 2015, 34 (Suppl. S1), 14–21. [Google Scholar] [CrossRef]
- Li, P.; Luo, L. Identification of Critical Genes and Signaling Pathways in Human Monocytes Following High-Intensity Exercise. Healthcare 2021, 9, 618. [Google Scholar] [CrossRef]
- Sisay, M.; Edessa, D.; Ali, T.; Mekuria, A.N.; Gebrie, A. The relationship between advanced glycation end products and gestational diabetes: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0240382. [Google Scholar] [CrossRef]
- Khalid, M.; Petroianu, G.; Adem, A. Advanced Glycation End Products and Diabetes Mellitus: Mechanisms and Perspectives. Biomolecules 2022, 12, 542. [Google Scholar] [CrossRef]
- Metzger, B.E.; Gabbe, S.G.; Persson, B.; Buchanan, T.A.; Catalano, P.A.; Damm, P.; Dyer, A.R.; Leiva, A.; Hod, M.; Kitzmiler, J.L.; et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 2010, 33, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Diagnostic criteria and classification of hyperglycaemia first detected in pregnancy: A World Health Organization Guideline. Diabetes Res. Clin. Pract. 2014, 103, 341–363. [CrossRef]
- Zalecenia kliniczne dotyczące postępowania u chorych na cukrzycę 2014. Stanowisko Polskiego Towarzystwa Diabetologicznego. Clin. Diabetol. 2014, 3, 1–81.
- Wojcik, M.; Zieleniak, A.; Mac-Marcjanek, K.; Wozniak, L.A.; Cypryk, K. The elevated gene expression level of the A(2B) adenosine receptor is associated with hyperglycemia in women with gestational diabetes mellitus. Diabetes Metab. Res. Rev. 2014, 30, 42–53. [Google Scholar] [CrossRef]
- Wojcik, M.; Zieleniak, A.; Zurawska-Klis, M.; Cypryk, K.; Wozniak, L.A. Increased expression of immune-related genes in leukocytes of patients with diagnosed gestational diabetes mellitus (GDM). Exp. Biol. Med. 2016, 241, 457–465. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Katz, A.; Nambi, S.S.; Mather, K.; Baron, A.D.; Follmann, D.A.; Sullivan, G.; Quon, M.J. Quantitative insulin sensitivity check index: A simple, accurate method for assessing insulin sensitivity in humans. J. Clin. Endocrinol. Metab. 2000, 85, 2402–2410. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Nested Polymerase Chain Reaction (PCR). Cold Spring Harb. Protoc. 2019, 2019, 175–178. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]



| Pregnancy | Postpartum | |||||
|---|---|---|---|---|---|---|
| Variable | NGT (n = 31) | GDM (n = 28) | GDM vs. NGT p | pGDM (n = 28) | pGDM vs. GDM p | pGDM vs. NGT p |
| Maternal age (years) | 30.00 (26.00–31.00) | 30.50 (28.00–34.00) | 0.2363 | - | - | - |
| Gestational age at OGTT (week) | 25.00 (24.00–26.00) | 25.00 (24.00–26.00) | 0.6309 | - | - | - |
| Pre-pregnancy BMI (kg/m2) | 23.29 (20.84–27.15) | 25.30 (22.92–28.23) | 0.1871 | - | - | - |
| Pregnancy weight (kg) | 71.35 (62.70–81.70) | 76.40 (67.80–85.00) | 0.1817 | - | - | - |
| Gestational weight gain (kg) | 8.00 (5.40–9.85) | 9.15 (6.05–11.05) | 0.3717 | - | - | - |
| Postpartum BMI (kg/m2) | - | - | - | 24.74 (21.85–30.51) | 0.1068 # | 0.2879 # |
| FPG (mg/dL) | 81.00 (74.00–85.00) | 86.50 (81.00–101.00) | 0.0024 * | 93.50 (86.00–101.50) | 0.0400 * | 0.0000 * |
| 1-h OGTT (mg/dL) | 159.00 (144.00–170.00) | 182.00 (161.00–189.00) | 0.0012 * | - | - | - |
| 2-h OGTT (mg/dL) | 137.00 (125.00–147.00) | 158.00 (147.00–170.50) | 0.0000 * | 102.00 (89.00–108.00) | 0.0000 * | 0.0000 * |
| HbA1C (%) | 5.29 (5.02–5.49) | 5.09 (4.88–5.34) | 0.2709 | 5.11 (4.96–5.29) | 0.9595 | 0.1311 |
| Fasting insulin (μIU/mL) | 8.05 (6.50–9.80) | 12.60 (7.60–16.30) | 0.0223 * | 9.05 (5.30–11.50) | 0.0012 * | 0.5038 |
| HOMA-B | 156.25 (122.40–183.00) | 187.26 (138.09–242.96) | 0.3445 | 104.21 (81.18–168.00) | 0.0019 * | 0.0398 * |
| HOMA-IR | 1.63 (1.34–1.85) | 2.66 (1.61–3.74) | 0.0065 * | 1.84 (1.13–3.07) | 0.0119 * | 0.3308 |
| QUICKI-IS | 0.15 (0.15–0.16) | 0.14 (0.14–0.15) | 0.0065 * | 0.15 (0.14–0.16) | 0.0049 * | 0.3308 |
| TC (mg/dL) | 271.35 (241.00–296.72) | 242.55 (209.20–268.15) | 0.0124 * | 197.50 (167.00–209.00) | 0.0000 * | 0.0000 * |
| HDL-C (mg/dL) | 84.35 (71.78–98.00) | 73.05 (60.75–86.70) | 0.0465 * | 58.00 (48.00–75.00) | 0.0001 * | 0.0001 * |
| LDL-C (mg/dL) | 144.50 (126.00–170.00) | 123.50 (99.00–153.50) | 0.0304 * | 105.00 (88.00–133.50) | 0.0123 * | 0.0005 * |
| TGs (mg/dL) | 212.95 (182.60–240.00) | 210.60 (169.10–246.10) | 0.5503 | 91.50 (50.85–124.95) | 0.0001 * | 0.0000 * |
| CRP (mg/dL) | 3.16 (1.80–7.33) | 3.72 (1.93–5.47) | 0.8307 | 1.41 (1.00–2.34) | 0.0062 * | 0.0071 * |
| Pregnancy | GDM vs. NGT | 1-Year Postpartum | pGDM vs. GDM | pGDM vs. NGT | |||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | NGT (n = 31) | GDM (n = 28) | FC | p | pGDM (n = 28) | FC | p | FC | p |
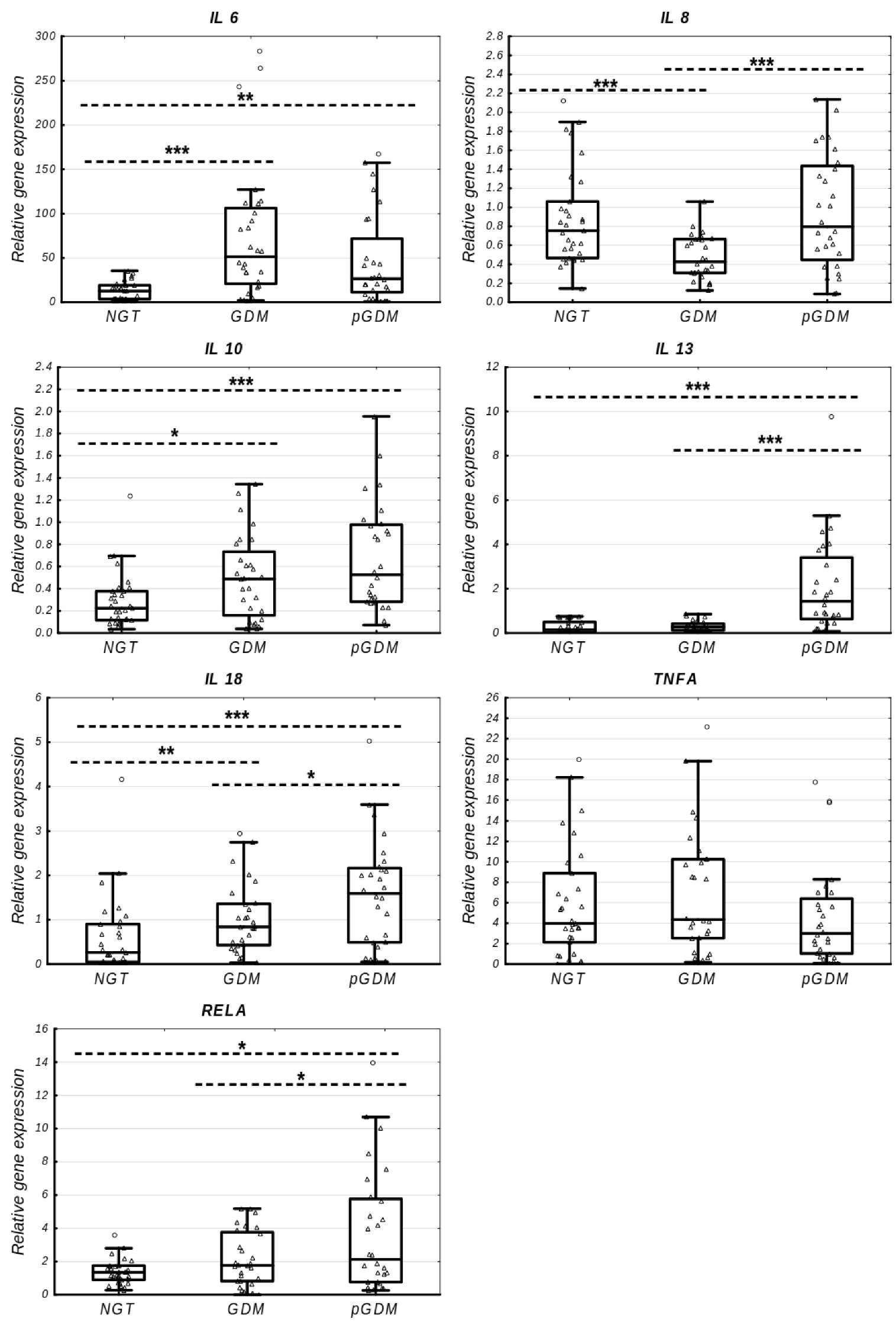
| IL6 | 12.63 (3.59–19.22) | 51.40 (20.85–106.19) | 4.07 | 0.0000 * | 26.50 (11.30–71.80) | 0.52 | 0.1514 | 2.10 | 0.0022 * |
| IL8 | 0.75 (0.47–1.06) | 0.43 (0.31–0.66) | 0.57 | 0.0004 * | 0.80 (0.45–1.44) | 1.86 | 0.0006 * | 1.06 | 0.7905 |
| IL10 | 0.22 (0.12–0.38) | 0.49 (0.16–0.73) | 2.18 | 0.0369 * | 0.53 (0.28–0.98) | 1.08 | 0.1329 | 2.35 | 0.0004 * |
| IL13 | 0.15 (0.02–0.49) | 0.27 (0.12–0.42) | 1.85 | 0.1649 | 1.43 (0.63–3.41) | 5.30 | 0.0000 * | 9.68 | 0.0000 * |
| IL18 | 0.26 (0.05–0.90) | 0.84 (0.43–1.36) | 3.21 | 0.0049 * | 1.59 (0.49–2.17) | 1.89 | 0.0382 * | 6.1 | 0.0004 * |
| TNFA | 3.97 (2.13–8.88) | 4.34 (2.52–10.24) | 1.09 | 0.4990 | 2.99 (1.03–6.39) | 0.69 | 0.0795 | 0.75 | 0.3055 |
| RELA | 1.35 (0.90–1.75) | 1.77 (0.82–3.76) | 1.31 | 0.1234 | 2.13 (0.77–5.77) | 1.20 | 0.0323 * | 1.58 | 0.0443 * |
| IL6 | IL8 | IL10 | IL13 | IL18 | TNFA | RELA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | r | p | r | p | r | p | r | p | r | p | r | p | r | p |
| Maternal age (years) | 0.18 | 0.1975 | −0.03 | 0.8335 | 0.16 | 0.2335 | −0.25 | 0.0604 | 0.24 | 0.0728 | −0.03 | 0.8003 | −0.02 | 0.9067 |
| Pre-pregnancy BMI (kg/m2) | 0.04 | 0.7870 | −0.15 | 0.2777 | −0.01 | 0.9534 | −0.05 | 0.7123 | 0.01 | 0.9410 | 0.30 | 0.0244 | 0.05 | 0.7394 |
| Pregnancy weight (kg) | 0.05 | 0.7015 | −0.08 | 0.5611 | −0.02 | 0.9065 | 0.01 | 0.9520 | 0.00 | 0.9809 | 0.30 | 0.0229 | 0.11 | 0.4242 |
| Gestational weight gain (kg) | 0.01 | 0.9358 | 0.08 | 0.5570 | −0.10 | 0.4504 | −0.12 | 0.3752 | −0.10 | 0.4727 | −0.08 | 0.5619 | 0.00 | 0.9774 |
| FPG (mg/dL) | 0.42 | 0.0010 * | −0.15 | 0.2722 | 0.22 | 0.0990 | 0.13 | 0.3264 | 0.25 | 0.0605 | 0.26 | 0.0454 | 0.22 | 0.0944 |
| 1-h OGTT (mg/dL) | 0.32 | 0.0207 | −0.06 | 0.6473 | 0.04 | 0.7639 | −0.01 | 0.9660 | 0.16 | 0.2614 | 0.14 | 0.3231 | −0.09 | 0.5322 |
| 2-h OGTT (mg/dL) | 0.39 | 0.0022 * | −0.21 | 0.1064 | 0.27 | 0.0374 | −0.10 | 0.4575 | 0.29 | 0.0275 | 0.13 | 0.3120 | 0.08 | 0.5353 |
| HbA1C (%) | −0.19 | 0.1439 | 0.18 | 0.1737 | −0.19 | 0.1587 | −0.29 | 0.0297 | −0.15 | 0.2547 | −0.04 | 0.7454 | −0.43 | 0.0007 * |
| Fasting insulin (μIU/mL) | 0.13 | 0.4047 | −0.13 | 0.3898 | −0.13 | 0.4073 | 0.04 | 0.7860 | −0.17 | 0.2624 | 0.15 | 0.3279 | 0.04 | 0.8140 |
| HOMA-B | 0.01 | 0.9261 | −0.10 | 0.4971 | −0.21 | 0.1716 | −0.06 | 0.6785 | −0.05 | 0.7205 | 0.08 | 0.6143 | −0.04 | 0.7732 |
| HOMA-IR | 0.20 | 0.1803 | −0.16 | 0.2991 | −0.05 | 0.7339 | 0.06 | 0.6755 | −0.13 | 0.3921 | 0.15 | 0.3236 | 0.08 | 0.6037 |
| QUICKI-IS | −0.20 | 0.1803 | 0.16 | 0.2991 | 0.05 | 0.7339 | −0.06 | 0.6755 | 0.13 | 0.3921 | −0.15 | 0.3236 | −0.08 | 0.6037 |
| TC (mg/dL) | −0.23 | 0.1018 | 0.28 | 0.0418 | −0.18 | 0.1844 | −0.10 | 0.4818 | −0.17 | 0.2115 | −0.13 | 0.3402 | −0.20 | 0.1562 |
| HDL-C (mg/dL) | 0.03 | 0.8357 | 0.04 | 0.7905 | −0.13 | 0.3514 | −0.15 | 0.2642 | −0.01 | 0.9575 | −0.20 | 0.1418 | −0.21 | 0.1278 |
| LDL-C (mg/dL) | −0.22 | 0.1067 | 0.31 | 0.0209 | −0.10 | 0.4924 | −0.02 | 0.9033 | −0.12 | 0.3834 | −0.07 | 0.6334 | −0.10 | 0.4530 |
| TGs (mg/dL) | −0.26 | 0.0550 | 0.12 | 0.3909 | −0.21 | 0.1361 | −0.13 | 0.3439 | −0.36 | 0.0079 | 0.05 | 0.7382 | −0.03 | 0.8402 |
| CRP (mg/dL) | −0.18 | 0.2408 | 0.00 | 0.9847 | 0.08 | 0.5882 | 0.05 | 0.7537 | 0.20 | 0.1786 | 0.16 | 0.2769 | −0.05 | 0.7593 |
| IL6 | - | - | −0.18 | 0.1745 | 0.48 | 0.0001 * | 0.10 | 0.4424 | 0.72 | 0.0000 * | 0.33 | 0.0110 | 0.26 | 0.0490 |
| IL8 | −0.18 | 0.1745 | - | - | 0.24 | 0.0685 | −0.19 | 0.1406 | −0.13 | 0.3342 | −0.04 | 0.7587 | 0.20 | 0.1272 |
| IL10 | 0.48 | 0.0001 * | 0.24 | 0.0685 | - | - | 0.14 | 0.2857 | 0.61 | 0.0000 * | 0.23 | 0.0789 | 0.58 | 0.0000 * |
| IL13 | 0.10 | 0.4424 | −0.19 | 0.1406 | 0.14 | 0.2857 | - | - | 0.22 | 0.0924 | 0.24 | 0.0729 | 0.24 | 0.0636 |
| IL18 | 0.72 | 0.0000 * | −0.13 | 0.3342 | 0.61 | 0.0000 * | 0.22 | 0.0924 | - | - | 0.45 | 0.0004 * | 0.28 | 0.0288 |
| TNFA | 0.33 | 0.0110 | −0.04 | 0.7587 | 0.23 | 0.0789 | 0.24 | 0.0729 | 0.45 | 0.0004 * | - | - | 0.10 | 0.4516 |
| RELA | 0.26 | 0.0490 | 0.20 | 0.1272 | 0.58 | 0.0000 * | 0.24 | 0.0636 | 0.28 | 0.0288 | 0.10 | 0.4516 | - | - |
| IL6 | IL8 | IL10 | IL13 | IL18 | TNFA | RELA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | r | p | r | p | r | p | r | p | r | p | r | p | r | p |
| Postpartum BMI (kg/m2) | 0.26 | 0.1920 | 0.32 | 0.1114 | 0.42 | 0.0332 | 0.31 | 0.1249 | 0.21 | 0.3010 | 0.25 | 0.2182 | 0.32 | 0.1139 |
| FPG (mg/dL) | 0.18 | 0.3578 | 0.12 | 0.5383 | 0.27 | 0.1702 | −0.19 | 0.3232 | 0.21 | 0.2754 | 0.04 | 0.8214 | 0.36 | 0.0567 |
| 2-h OGTT (mg/dL) | 0.07 | 0.7244 | 0.42 | 0.0312 | 0.62 | 0.0006 * | 0.21 | 0.2908 | 0.31 | 0.1097 | 0.10 | 0.6278 | 0.22 | 0.2638 |
| HbA1C (%) | −0.06 | 0.7737 | 0.22 | 0.2677 | 0.09 | 0.6560 | −0.07 | 0.7231 | −0.12 | 0.5458 | 0.02 | 0.9097 | −0.08 | 0.6826 |
| Fasting insulin (μIU/mL) | 0.00 | 0.9894 | −0.05 | 0.8215 | 0.12 | 0.5558 | 0.17 | 0.4161 | −0.07 | 0.7249 | 0.19 | 0.3460 | 0.14 | 0.5062 |
| HOMA-B | −0.06 | 0.7869 | −0.10 | 0.6435 | −0.14 | 0.5115 | 0.34 | 0.0963 | −0.19 | 0.3679 | 0.26 | 0.2171 | −0.08 | 0.6956 |
| HOMA-IR | 0.06 | 0.7729 | −0.03 | 0.8868 | 0.19 | 0.3551 | 0.11 | 0.6110 | −0.01 | 0.9593 | 0.21 | 0.3210 | 0.19 | 0.3730 |
| QUICKI-IS | −0.06 | 0.7729 | 0.03 | 0.8868 | −0.19 | 0.3551 | −0.11 | 0.6110 | 0.01 | 0.9593 | −0.21 | 0.3210 | −0.19 | 0.3730 |
| TC (mg/dL) | 0.04 | 0.8389 | 0.33 | 0.0835 | 0.31 | 0.1086 | −0.15 | 0.4597 | 0.18 | 0.3488 | 0.11 | 0.5754 | 0.00 | 0.9945 |
| HDL-C (mg/dL) | −0.21 | 0.2861 | −0.03 | 0.8758 | −0.29 | 0.1380 | 0.03 | 0.8758 | −0.10 | 0.6198 | −0.33 | 0.0881 | −0.09 | 0.6365 |
| LDL-C (mg/dL) | 0.04 | 0.8432 | 0.27 | 0.1638 | 0.38 | 0.0436 | 0.04 | 0.8259 | 0.17 | 0.3940 | 0.16 | 0.4027 | 0.02 | 0.9284 |
| TGs (mg/dL) | 0.03 | 0.8879 | −0.01 | 0.9427 | 0.28 | 0.1562 | −0.21 | 0.2729 | −0.04 | 0.8487 | −0.09 | 0.6476 | 0.10 | 0.6179 |
| CRP (mg/dL) | 0.41 | 0.0361 | 0.24 | 0.2304 | 0.17 | 0.3871 | 0.25 | 0.2060 | 0.34 | 0.0821 | 0.44 | 0.0208 | 0.05 | 0.8161 |
| IL6 | - | - | 0.33 | 0.0814 | 0.41 | 0.0320 | 0.09 | 0.6437 | 0.69 | 0.0001 * | 0.44 | 0.0194 | 0.27 | 0.1676 |
| IL8 | 0.33 | 0.0814 | - | - | 0.59 | 0.0009 * | 0.15 | 0.4547 | 0.61 | 0.0005 * | 0.23 | 0.2358 | 0.44 | 0.0183 |
| IL10 | 0.41 | 0.0320 | 0.59 | 0.0009 * | - | - | 0.15 | 0.4581 | 0.66 | 0.0001 * | 0.30 | 0.1154 | 0.35 | 0.0640 |
| IL13 | 0.09 | 0.6437 | 0.15 | 0.4547 | 0.15 | 0.4581 | - | - | 0.09 | 0.6557 | 0.26 | 0.1815 | 0.06 | 0.7609 |
| IL18 | 0.69 | 0.0001 * | 0.61 | 0.0005 * | 0.66 | 0.0001 * | 0.09 | 0.6557 | - | - | 0.41 | 0.0288 | 0.47 | 0.0115 |
| TNFA | 0.44 | 0.0194 | 0.23 | 0.2358 | 0.30 | 0.1154 | 0.26 | 0.1815 | 0.41 | 0.0288 | - | - | 0.03 | 0.8770 |
| RELA | 0.27 | 0.1676 | 0.44 | 0.0183 | 0.35 | 0.0640 | 0.06 | 0.7609 | 0.47 | 0.0115 | 0.03 | 0.8770 | - | - |
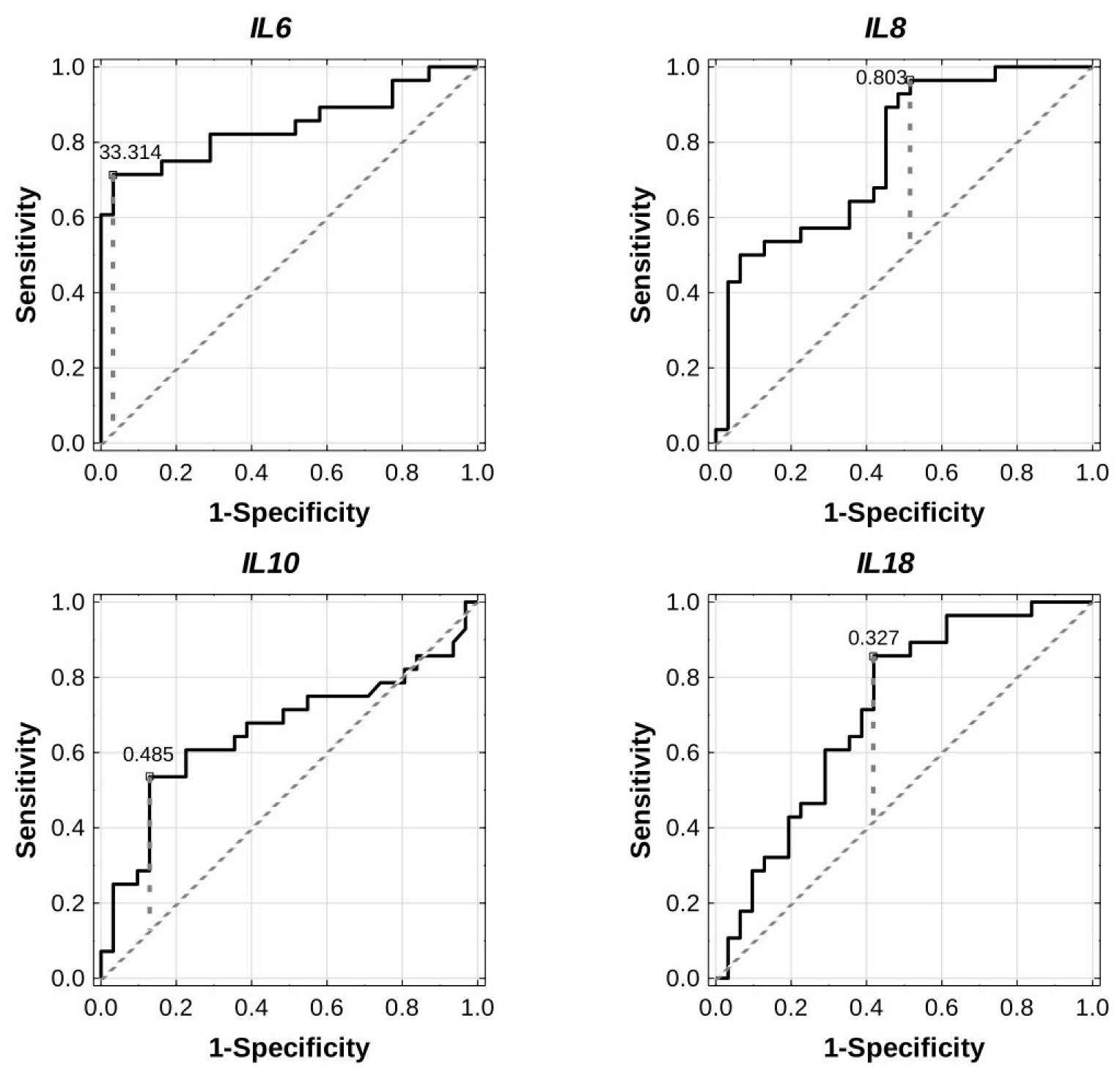
| Gene | AUC | SE | 95% CI | p | Effect | Cut-Off Value | 95% CI | YI | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| IL6 | 0.844 | 0.055 | 0.736–0.953 | 0.0000 | ↑ | 33.314 | 16.608–33.314 | 0.682 | 0.4712–0.8249 |
| IL8 | 0.771 | 0.061 | 0.651–0.890 | 0.0000 | ↓ | 0.803 | 0.546–1.060 | 0.4482 | 0.1774–0.7190 |
| IL10 | 0.658 | 0.075 | 0.511–0.805 | 0.0354 | ↑ | 0.485 | 0.302–0.536 | 0.4067 | 0.1601–0.5818 |
| IL13 | 0.605 | 0.075 | 0.459–0.752 | 0.1588 | ns | 0.068 | 0.034–0.097 | 0.2869 | 0.1071–0.4021 |
| IL18 | 0.714 | 0.067 | 0.582–0.846 | 0.0014 | ↑ | 0.327 | 0.031–0.353 | 0.4378 | 0.2477–0.6129 |
| TNFA | 0.552 | 0.076 | 0.403–0.701 | 0.4945 | ns | 8.334 | 0.179–8.437 | 0.2062 | 0.0357–0.4136 |
| RELA | 0.618 | 0.079 | 0.464–0.772 | 0.1328 | ns | 1.715 | 0.806–1.801 | 0.3134 | 0.0392–0.432 |
| Variable | AUC | SE | 95% CI | p | Effect | Cut-Off Value | 95% CI | YI | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| FPG | 0.711 | 0.099 | 0.517–0.905 | 0.0328 | ↑ | 101.0 | 81.0–106.0 | 0.333 | 0.100–0.456 |
| IL6 | 0.411 | 0.122 | 0.171–0.651 | 0.4677 | ns | 264.260 | 1.944–264.260 | 0.200 | 0.000–0.400 |
| IL8 | 0.750 | 0.112 | 0.53–0.970 | 0.0260 | ↑ | 0.667 | 0.307–0.669 | 0.544 | 0.200–0.789 |
| IL10 | 0.650 | 0.107 | 0.441–0.859 | 0.1598 | ns | 0.618 | 0.089–1.343 | 0.278 | 0.056–0.400 |
| IL13 | 0.733 | 0.1 | 0.538–0.929 | 0.0193 | ↑ | 0.492 | 0.105–0.553 | 0.444 | 0.167–0.611 |
| IL18 | 0.550 | 0.113 | 0.329–0.771 | 0.6574 | ns | 1.341 | 0.353–2.316 | 0.178 | 0.000–0.344 |
| TNFA | 0.761 | 0.1 | 0.565–0.957 | 0.0089 | ↑ | 4.442 | 0.974–4.442 | 0.467 | 0.111–0.633 |
| RELA | 0.594 | 0.12 | 0.358–0.831 | 0.4331 | ns | 1.916 | 0.092–3.865 | 0.267 | 0.000–0.511 |
| (IL8 × IL13 × TNFA) | 0.850 | 0.082 | 0.689–1.000 | 0.0000 | ↑ | 1.353 | 0.089–1.353 | 0.644 | 0.300–0.844 |
| (IL8 × IL13 × TNFA × FPG) | 0.850 | 0.082 | 0.689–1.000 | 0.0001 | ↑ | 140.713 | 7.27–140.713 | 0.644 | 0.300–0.844 |
| Variable | Estimate | SE | Wald’s Test | p | OR (95% CI) |
|---|---|---|---|---|---|
| (Intercept) | −2.230 | 0.759 | 8.636 | 0.0033 | 0.108 (0.024, 0.476) |
| (IL8 × L13 × TNFA) | 1.422 | 0.579 | 6.037 | 0.0140 | 4.147 (1.333, 12.896) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zieleniak, A.; Zurawska-Klis, M.; Cypryk, K.; Wozniak, L.; Wojcik, M. Transcriptomic Dysregulation of Inflammation-Related Genes in Leukocytes of Patients with Gestational Diabetes Mellitus (GDM) during and after Pregnancy: Identifying Potential Biomarkers Relevant to Glycemic Abnormality. Int. J. Mol. Sci. 2022, 23, 14677. https://doi.org/10.3390/ijms232314677
Zieleniak A, Zurawska-Klis M, Cypryk K, Wozniak L, Wojcik M. Transcriptomic Dysregulation of Inflammation-Related Genes in Leukocytes of Patients with Gestational Diabetes Mellitus (GDM) during and after Pregnancy: Identifying Potential Biomarkers Relevant to Glycemic Abnormality. International Journal of Molecular Sciences. 2022; 23(23):14677. https://doi.org/10.3390/ijms232314677
Chicago/Turabian StyleZieleniak, Andrzej, Monika Zurawska-Klis, Katarzyna Cypryk, Lucyna Wozniak, and Marzena Wojcik. 2022. "Transcriptomic Dysregulation of Inflammation-Related Genes in Leukocytes of Patients with Gestational Diabetes Mellitus (GDM) during and after Pregnancy: Identifying Potential Biomarkers Relevant to Glycemic Abnormality" International Journal of Molecular Sciences 23, no. 23: 14677. https://doi.org/10.3390/ijms232314677
APA StyleZieleniak, A., Zurawska-Klis, M., Cypryk, K., Wozniak, L., & Wojcik, M. (2022). Transcriptomic Dysregulation of Inflammation-Related Genes in Leukocytes of Patients with Gestational Diabetes Mellitus (GDM) during and after Pregnancy: Identifying Potential Biomarkers Relevant to Glycemic Abnormality. International Journal of Molecular Sciences, 23(23), 14677. https://doi.org/10.3390/ijms232314677






