Differential Expression of RSK4 Transcript Isoforms in Cancer and Its Clinical Relevance
Abstract
:1. Introduction
2. Results
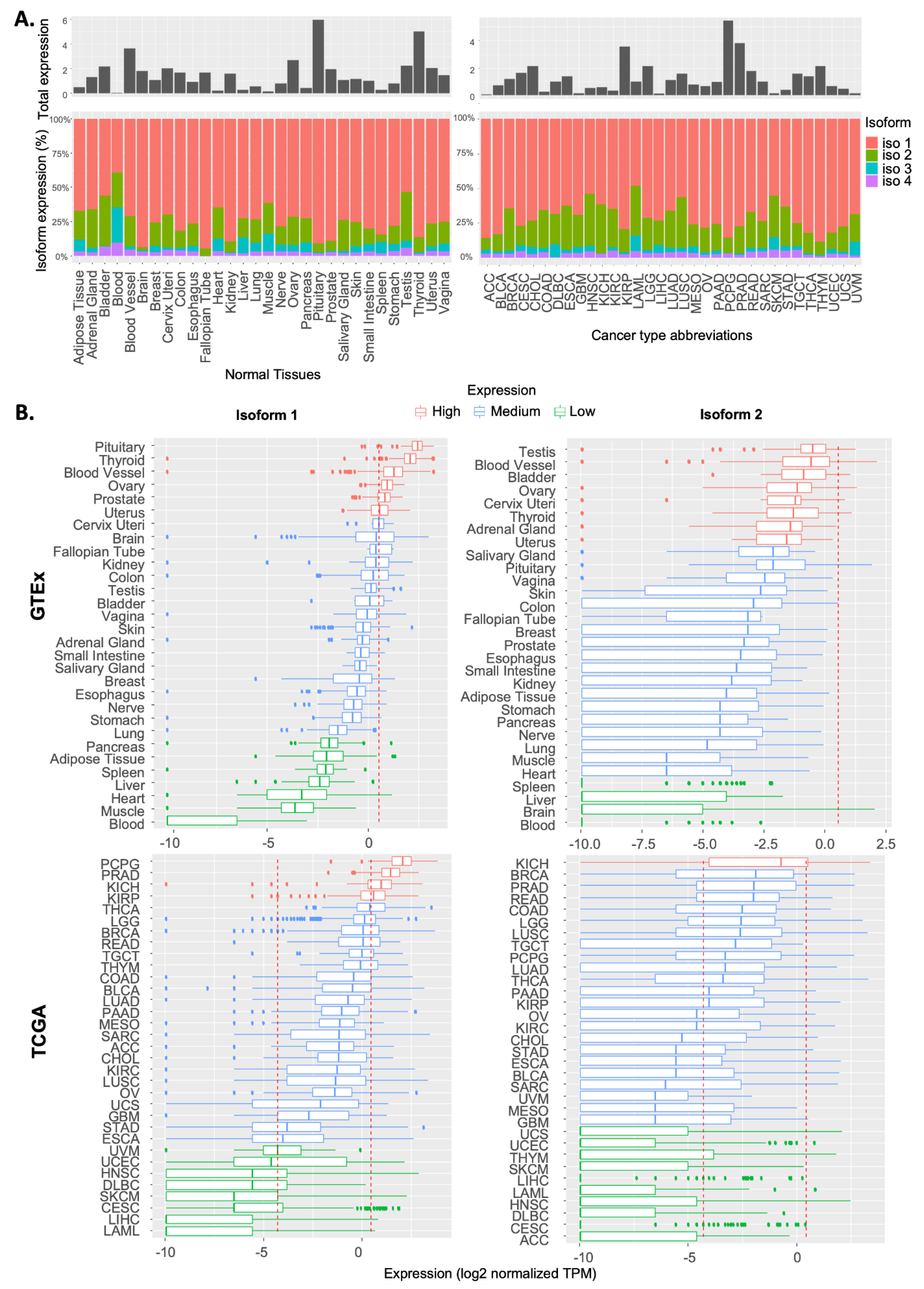
2.1. Expression Pattern of RSK4 Transcripts in Normal and Cancer Tissues
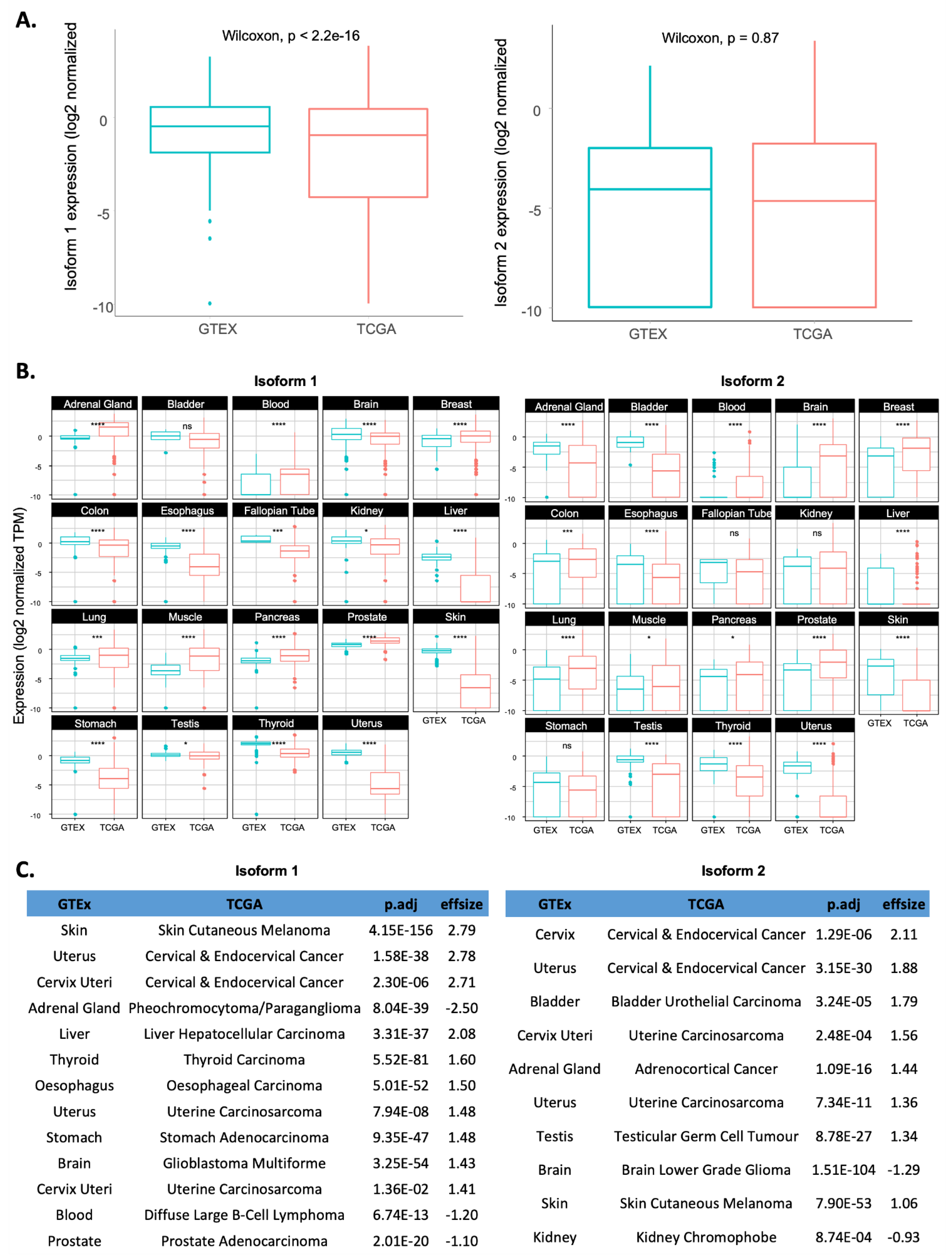
2.2. Comparison of RSK4 Isoform Expression between Tumour Samples and Their Matched Normal Counterparts
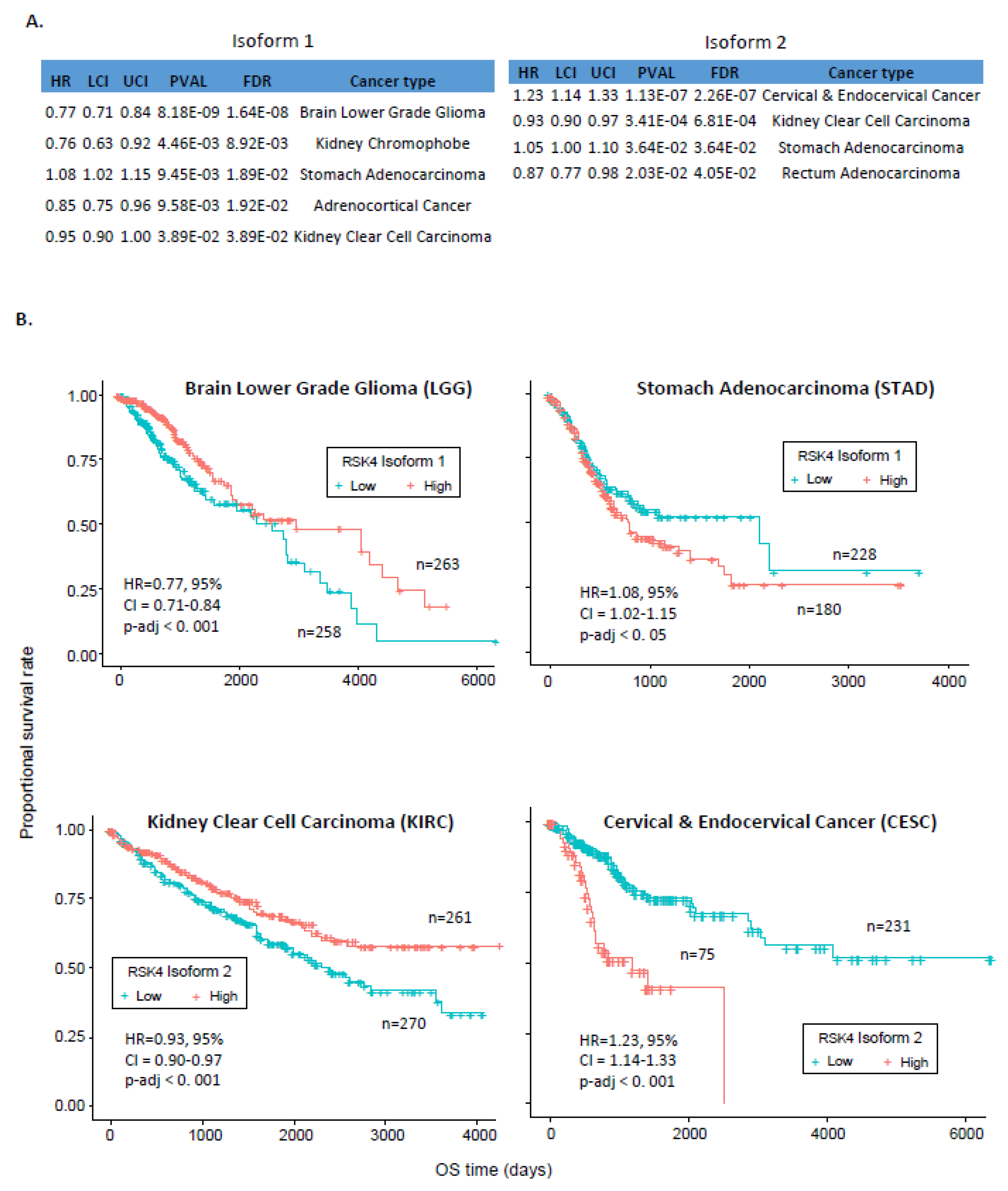
2.3. Association between RSK4 Isoform and Patients’ Survival
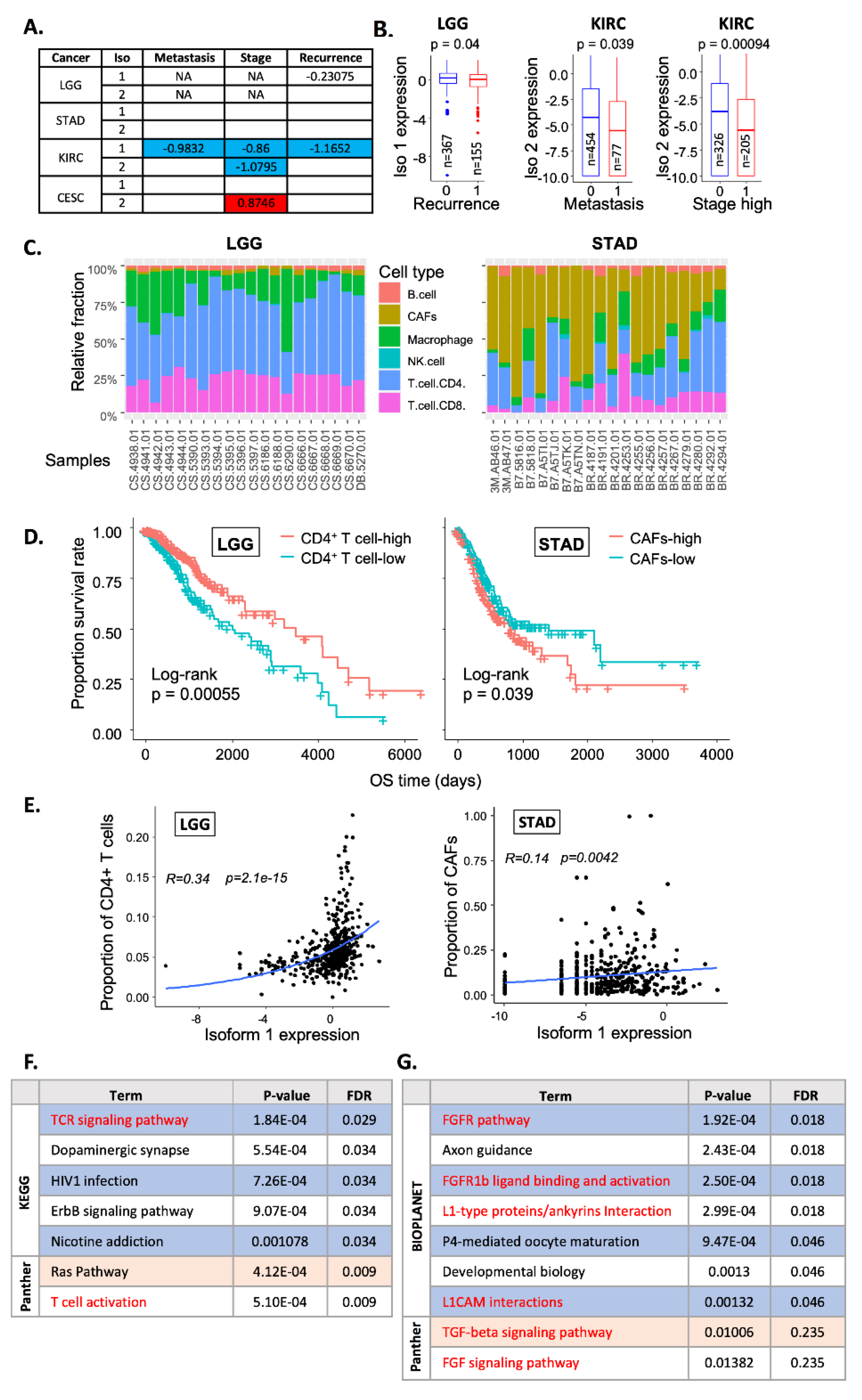
2.4. Correlation with Clinical Features
3. Discussion
4. Material and Methods
4.1. Data Sources
4.2. Statistical Tests
4.3. Survival Analysis
4.4. Cell Type Deconvolution Analysis
4.5. Gene Co-Expression and Pathway Enrichment Analysis
4.6. Data and Code Availability
4.7. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romeo, Y.; Zhang, X.; Roux, P.P. Regulation and Function of the RSK Family of Protein Kinases. Biochem. J. 2012, 441, 553–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.C.; Jivan, A.; Shao, C.; Duan, L.; Goad, D.; Zaganjor, E.; Osborne, J.; McGlynn, K.; Stippec, S.; Earnest, S.; et al. The Roles of MAPKs in Disease. Cell Res. 2008, 18, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Houles, T.; Roux, P.P. Defining the Role of the RSK Isoforms in Cancer. Semin. Cancer Biol. 2018, 48, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Carriere, A.; Ray, H.; Blenis, J.; Roux, P.P. The RSK Factors of Activating the Ras/MAPK Signaling Cascade. Front. Biosci. 2008, 13, 4258–4275. [Google Scholar] [CrossRef]
- Lara, R.; Seckl, M.J.; Pardo, O.E. The P90 RSK Family Members: Common Functions and Isoform Specificity. Cancer Res. 2013, 73, 5301–5308. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.W.; Erikson, E.; Blenis, J.; Maller, J.L.; Erikson, R.L. A Xenopus Ribosomal Protein S6 Kinase Has Two Apparent Kinase Domains That Are Each Similar to Distinct Protein Kinases. Proc. Natl. Acad. Sci. USA 1988, 85, 3377–3381. [Google Scholar] [CrossRef] [Green Version]
- Fisher, T.L.; Blenis, J. Evidence for Two Catalytically Active Kinase Domains in Pp90rsk. Mol. Cell. Biol. 1996, 16, 1212–1219. [Google Scholar] [CrossRef] [Green Version]
- Ludwik, K.A.; Lannigan, D.A. Ribosomal S6 Kinase (RSK) Modulators: A Patent Review. Expert Opin. Ther. Pat. 2016, 26, 1061–1078. [Google Scholar] [CrossRef]
- Casalvieri, K.A.; Matheson, C.J.; Backos, D.S.; Reigan, P. Selective Targeting of RSK Isoforms in Cancer. Trends Cancer 2017, 3, 302–312. [Google Scholar] [CrossRef]
- Cho, Y.-Y.; Yao, K.; Kim, H.-G.; Kang, B.S.; Zheng, D.; Bode, A.M.; Dong, Z. Ribosomal S6 Kinase 2 Is a Key Regulator in Tumor Promoter–Induced Cell Transformation. Cancer Res. 2007, 67, 8104. [Google Scholar] [CrossRef]
- Xu, J.; Jia, Q.; Zhang, Y.; Yuan, Y.; Xu, T.; Yu, K.; Chai, J.; Wang, K.; Chen, L.; Xiao, T.; et al. Prominent Roles of Ribosomal S6 Kinase 4 (RSK4) in Cancer. Pathol. Res. Pr. 2021, 219, 153374. [Google Scholar] [CrossRef] [PubMed]
- Serra, V.; Eichhorn, P.J.A.; García-García, C.; Ibrahim, Y.H.; Prudkin, L.; Sánchez, G.; Rodríguez, O.; Antón, P.; Parra, J.L.; Marlow, S.; et al. RSK3/4 Mediate Resistance to PI3K Pathway Inhibitors in Breast Cancer. J. Clin. Investig. 2013, 123, 2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Martin, T.A.; Davies, E.L.; Ruge, F.; Yu, H.; Zhang, Y.; Teng, X.; Jiang, W.G. The Clinical Implications of RSK1-3 in Human Breast Cancer. Anticancer Res. 2016, 36, 1267–1274. [Google Scholar]
- Kohn, M.; Hameister, H.; Vogel, M.; Kehrer-Sawatzki, H. Expression Pattern of the Rsk2, Rsk4 and Pdk1 Genes during Murine Embryogenesis. Gene Expr. Patterns 2003, 3, 173–177. [Google Scholar] [CrossRef]
- Chen, R.H.; Sarnecki, C.; Blenis, J. Nuclear Localization and Regulation of Erk- and Rsk-Encoded Protein Kinases. Mol. Cell. Biol. 1992, 12, 915–927. [Google Scholar] [PubMed] [Green Version]
- Lenormand, P.; Sardet, C.; Pagès, G.; L’Allemain, G.; Brunet, A.; Pouysségur, J. Growth Factors Induce Nuclear Translocation of MAP Kinases (P42mapk and P44mapk) but Not of Their Activator MAP Kinase Kinase (P45mapkk) in Fibroblasts. J. Cell. Biol. 1993, 122, 1079–1088. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Bjørbaek, C.; Weremowicz, S.; Morton, C.C.; Moller, D.E. RSK3 Encodes a Novel Pp90rsk Isoform with a Unique N-Terminal Sequence: Growth Factor-Stimulated Kinase Function and Nuclear Translocation. Mol. Cell. Biol. 1995, 15, 4353–4363. [Google Scholar] [CrossRef] [Green Version]
- Dümmler, B.A.; Hauge, C.; Silber, J.; Yntema, H.G.; Kruse, L.S.; Kofoed, B.; Hemmings, B.A.; Alessi, D.R.; Frödin, M. Functional Characterization of Human RSK4, a New 90-KDa Ribosomal S6 Kinase, Reveals Constitutive Activation in Most Cell Types. J. Biol. Chem. 2005, 280, 13304–13314. [Google Scholar] [CrossRef] [Green Version]
- Poomakkoth, N.; Issa, A.; Abdulrahman, N.; Abdelaziz, S.G.; Mraiche, F. P90 Ribosomal S6 Kinase: A Potential Therapeutic Target in Lung Cancer. J. Transl. Med. 2016, 14, 14. [Google Scholar] [CrossRef] [Green Version]
- Chrysostomou, S.; Roy, R.; Prischi, F.; Thamlikitkul, L.; Chapman, K.L.; Mufti, U.; Peach, R.; Ding, L.; Hancock, D.; Moore, C.; et al. Repurposed Floxacins Targeting RSK4 Prevent Chemoresistance and Metastasis in Lung and Bladder Cancer. Sci. Transl. Med. 2021, 13, 602. [Google Scholar] [CrossRef]
- Fang, Y.-Y.; Ma, F.-C.; Gan, X.-L.; Luo, W.-Q.; He, R.-Q.; Xie, H.-M.; Li, S.-Y.; Chen, G.; Wei, D.-M.; Hu, X.-H. Clinicopathological Significance of Ribosomal Protein S6 Kinase A6 in Lung Squamous Cell Carcinoma: An Immunohistochemical and RNA-Seq Study. Int. J. Clin. Exp. Pathol. 2018, 11, 1318–1327. [Google Scholar] [PubMed]
- He, Q.; He, R.; Luo, W.; Gan, X.; Ma, J.; Chen, G.; Li, P.; Lan, D.; Hu, X. Expression of RSK4 in Lung Adenocarcinoma Tissue and Its Clinicopathological Value: A Study Based on RNA-Seq Data and Immunohistochemistry. Int. J. Clin. Exp. Pathol. 2017, 10, 11405. [Google Scholar] [PubMed]
- Li, M.-Y.; Fan, L.-N.; Han, D.-H.; Yu, Z.; Ma, J.; Liu, Y.-X.; Li, P.-F.; Zhao, D.-H.; Chai, J.; Jiang, L.; et al. Ribosomal S6 Protein Kinase 4 Promotes Radioresistance in Esophageal Squamous Cell Carcinoma. J. Clin. Investig. 2020, 130, 4301–4319. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Dai, J.; Lin, X.; Meng, Y.; Liang, H. Effect of RSK4 on Biological Characteristics of Gastric Cancer. Cancer Manag. Res. 2020, 12, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Li, P.; Yin, Z.; Fu, G.; Liao, D.J.; Liu, Y.; Zhu, J.; Zhang, Y.; Wang, L.; Yan, Q.; et al. Ribosomal S6 Protein Kinase 4: A Prognostic Factor for Renal Cell Carcinoma. Br. J. Cancer 2013, 109, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Wang, K.; Chai, J.; Xu, T.; Wei, J.; Liu, Y.; Wang, Y.; Xu, J.; Li, M.; Fan, L. High RSK4 Expression Constitutes a Predictor of Poor Prognosis for Patients with Clear Cell Renal Carcinoma. Pathol. Res. Pr. 2021, 227, 153642. [Google Scholar] [CrossRef]
- Chen, L.; Xu, T.; Jia, Q.; Wang, X.; Li, M.; Liang, G. RSK4: A New Prognostic Factor in Glioma. Pathol. Res. Pr. 2020, 216, 153020. [Google Scholar] [CrossRef]
- Huo, H.; Ye, X.; Yang, H.; Li, Q.; Jiang, Y. RSK4 Inhibits Breast Cancer Cell Proliferation and Invasion in Vitro, and Is Correlated with Estrogen Receptor Upregulation in Breast Cancer. Oncol. Rep. 2019, 42, 2777–2787. [Google Scholar] [CrossRef]
- Mei, Y.; Liao, X.; Zhu, L.; Yang, H. Overexpression of RSK4 Reverses Doxorubicin Resistance in Human Breast Cancer Cells via PI3K/AKT Signalling Pathway. J. Biochem. 2020, 167, 603–611. [Google Scholar] [CrossRef]
- Thakur, A.; Sun, Y.; Bollig, A.; Wu, J.; Biliran, H.; Banerjee, S.; Sarkar, F.H.; Liao, D.J. Anti-Invasive and Antimetastatic Activities of Ribosomal Protein S6 Kinase 4 in Breast Cancer Cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 4427. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Rahman, K.W.; Wu, J.; Bollig, A.; Biliran, H.; Lin, X.; Nassar, H.; Grignon, D.J.; Sarkar, F.H.; Liao, J.D. Aberrant Expression of X-Linked Genes RbAp46, Rsk4, and Cldn2 in Breast Cancer. Mol. Cancer Res. 2007, 5, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiang, Y.; Wei, W.; Ji, Y.; Gao, H.; Liu, J. Frequent Epigenetic Inactivation of RSK4 by Promoter Methylation in Cancerous and Non-Cancerous Tissues of Breast Cancer. Med. Oncol. 2014, 31, 793. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, Q.Y.; Liu, J.L.; Wei, W.; Yang, H.W.; Tang, W. RSK4 Knockdown Promotes Proliferation, Migration and Metastasis of Human Breast Adenocarcinoma Cells. Oncol. Rep. 2015, 34, 3156–3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bologna, G.; Yvon, C.; Duvaud, S.; Veuthey, A.-L. N-Terminal Myristoylation Predictions by Ensembles of Neural Networks. Proteomics 2004, 4, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Program—National Cancer Institute. Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 3 April 2022).
- The Genotype-Tissue Expression (GTEx) Project|Nature Genetics. Available online: https://www.nature.com/articles/ng.2653 (accessed on 3 April 2022).
- Pagès, F.; Galon, J.; Dieu-Nosjean, M.-C.; Tartour, E.; Sautès-Fridman, C.; Fridman, W.-H. Immune Infiltration in Human Tumors: A Prognostic Factor That Should Not Be Ignored. Oncogene 2010, 29, 1093–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aran, D.; Hu, Z.; Butte, A.J. XCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [Green Version]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous Enumeration of Cancer and Immune Cell Types from Bulk Tumor Gene Expression Data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and Pharmacological Modulators of the Tumor Immune Contexture Revealed by Deconvolution of RNA-Seq Data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the Population Abundance of Tissue-Infiltrating Immune and Stromal Cell Populations Using Gene Expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.D.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive Evaluation of Transcriptome-Based Cell-Type Quantification Methods for Immuno-Oncology. Bioinformatics 2019, 35, i436. [Google Scholar] [CrossRef] [PubMed]
- Vancauwenberghe, E.; Noyer, L.; Derouiche, S.; Lemonnier, L.; Gosset, P.; Sadofsky, L.R.; Mariot, P.; Warnier, M.; Bokhobza, A.; Slomianny, C.; et al. Activation of Mutated TRPA1 Ion Channel by Resveratrol in Human Prostate Cancer Associated Fibroblasts (CAF). Mol. Carcinog 2017, 56, 1851–1867. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yi, Z.; Li, J.; Wei, Y.; Feng, R.; Liu, J.; Huang, J.; Chen, Y.; Wang, X.; Sun, J.; et al. FGFR Blockade Boosts T Cell Infiltration into Triple-Negative Breast Cancer by Regulating Cancer-Associated Fibroblasts. Theranostics 2022, 12, 4564–4580. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling Pathways in Cancer-Associated Fibroblasts and Targeted Therapy for Cancer. Signal Transduct. Target 2021, 6, 218. [Google Scholar] [CrossRef]
- Blethrow, J.; Zhang, C.; Shokat, K.M.; Weiss, E.L. Design and Use of Analog-Sensitive Protein Kinases. Curr. Protoc. Mol. Biol. 2004, 66, 18.11.1–18.11.19. [Google Scholar] [CrossRef]
- Puig, O.; Caspary, F.; Rigaut, G.; Rutz, B.; Bouveret, E.; Bragado-Nilsson, E.; Wilm, M.; Séraphin, B. The Tandem Affinity Purification (TAP) Method: A General Procedure of Protein Complex Purification. Methods 2001, 24, 218–229. [Google Scholar] [CrossRef] [Green Version]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive Analysis of Normal Adjacent to Tumor Transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Cao, S.; Yang, M.; Wu, S.; Wang, Z.; Lin, X.; Song, X.; Liao, D.J. Basic Anatomy and Tumor Biology of the RPS6KA6 Gene That Encodes the P90 Ribosomal S6 Kinase. Oncogene 2013, 32, 1794–1810. [Google Scholar] [CrossRef] [Green Version]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-Coding RNA Networks in Cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil Enables Reproducible, Open Source, Big Biomedical Data Analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Seckl, M.J.; Lorentzen, M.P.G.; Pardo, O.E. Differential Expression of RSK4 Transcript Isoforms in Cancer and Its Clinical Relevance. Int. J. Mol. Sci. 2022, 23, 14569. https://doi.org/10.3390/ijms232314569
Chen S, Seckl MJ, Lorentzen MPG, Pardo OE. Differential Expression of RSK4 Transcript Isoforms in Cancer and Its Clinical Relevance. International Journal of Molecular Sciences. 2022; 23(23):14569. https://doi.org/10.3390/ijms232314569
Chicago/Turabian StyleChen, Sisi, Michael J. Seckl, Marc P. G. Lorentzen, and Olivier E. Pardo. 2022. "Differential Expression of RSK4 Transcript Isoforms in Cancer and Its Clinical Relevance" International Journal of Molecular Sciences 23, no. 23: 14569. https://doi.org/10.3390/ijms232314569
APA StyleChen, S., Seckl, M. J., Lorentzen, M. P. G., & Pardo, O. E. (2022). Differential Expression of RSK4 Transcript Isoforms in Cancer and Its Clinical Relevance. International Journal of Molecular Sciences, 23(23), 14569. https://doi.org/10.3390/ijms232314569






