Effects of Subtoxic Concentrations of Atrazine, Cypermethrin, and Vinclozolin on microRNA-Mediated PI3K/Akt/mTOR Signaling in SH-SY5Y Cells
Abstract
:1. Introduction
2. Results
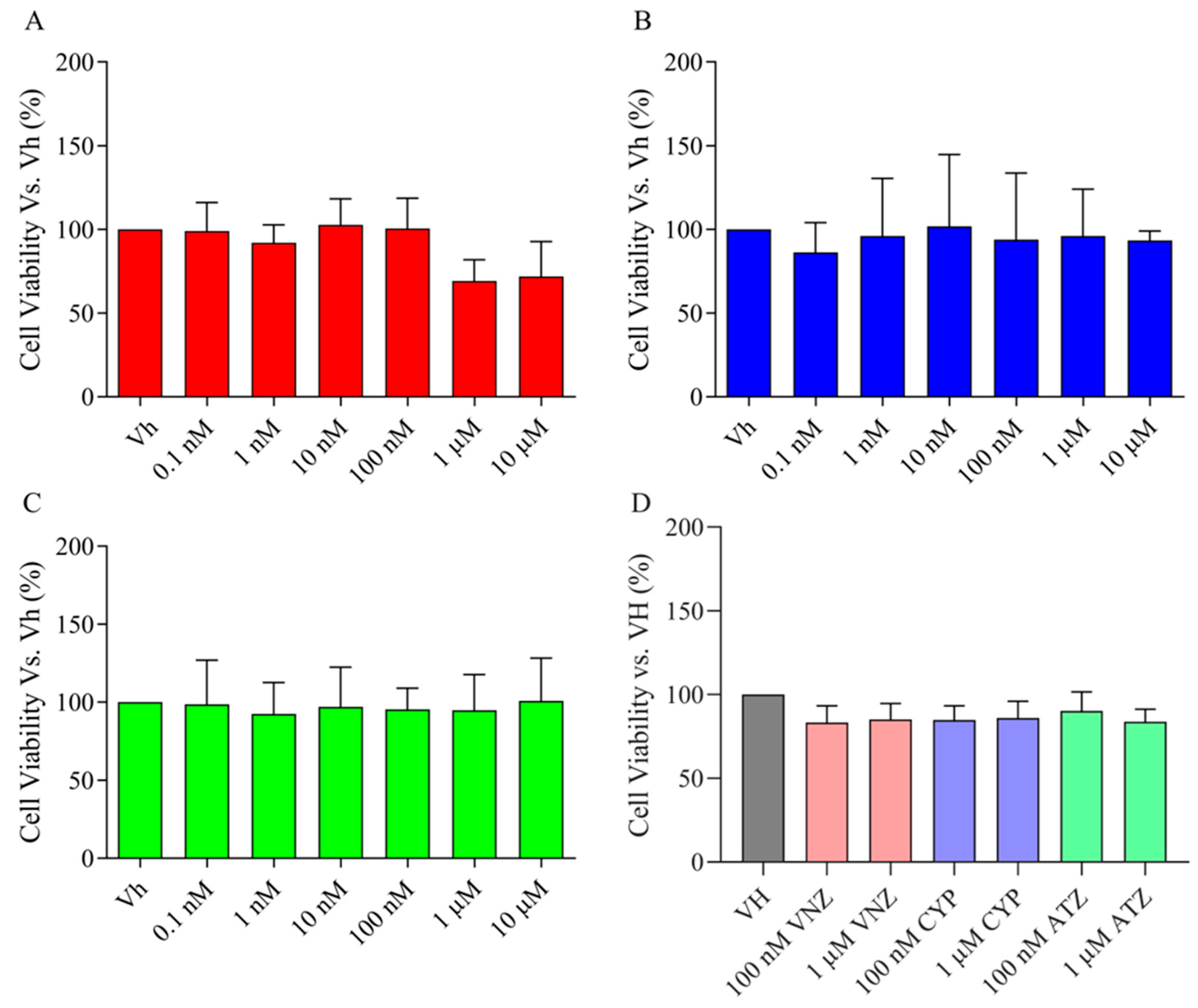
2.1. ATZ, CYP and VNZ at Subtoxic Concentrations Have No Significant Cytotoxicity to SH-SY5Y Cells
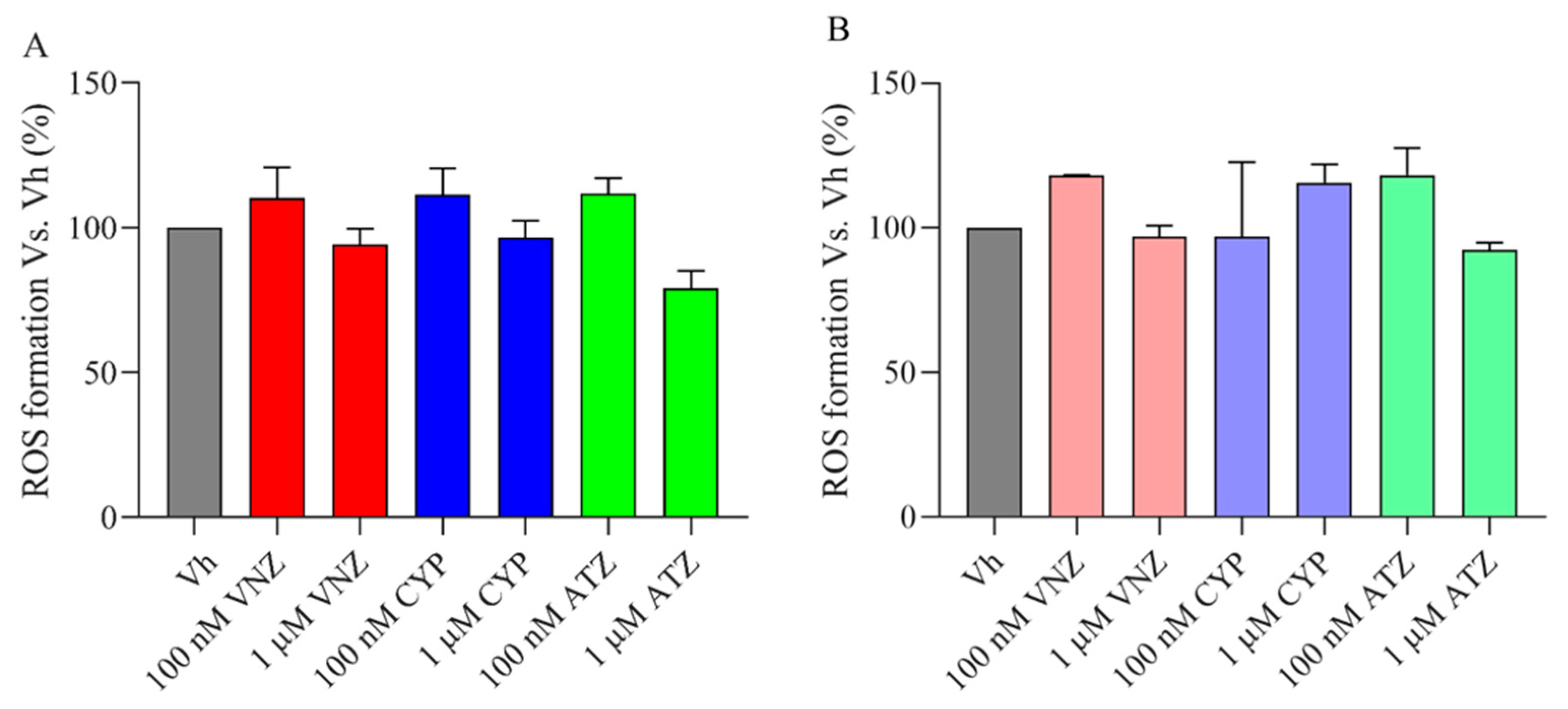
2.2. ATZ, CYP and VNZ at Subtoxic Concentrations Have No Significant Effects on Reactive Oxygen Species Formation
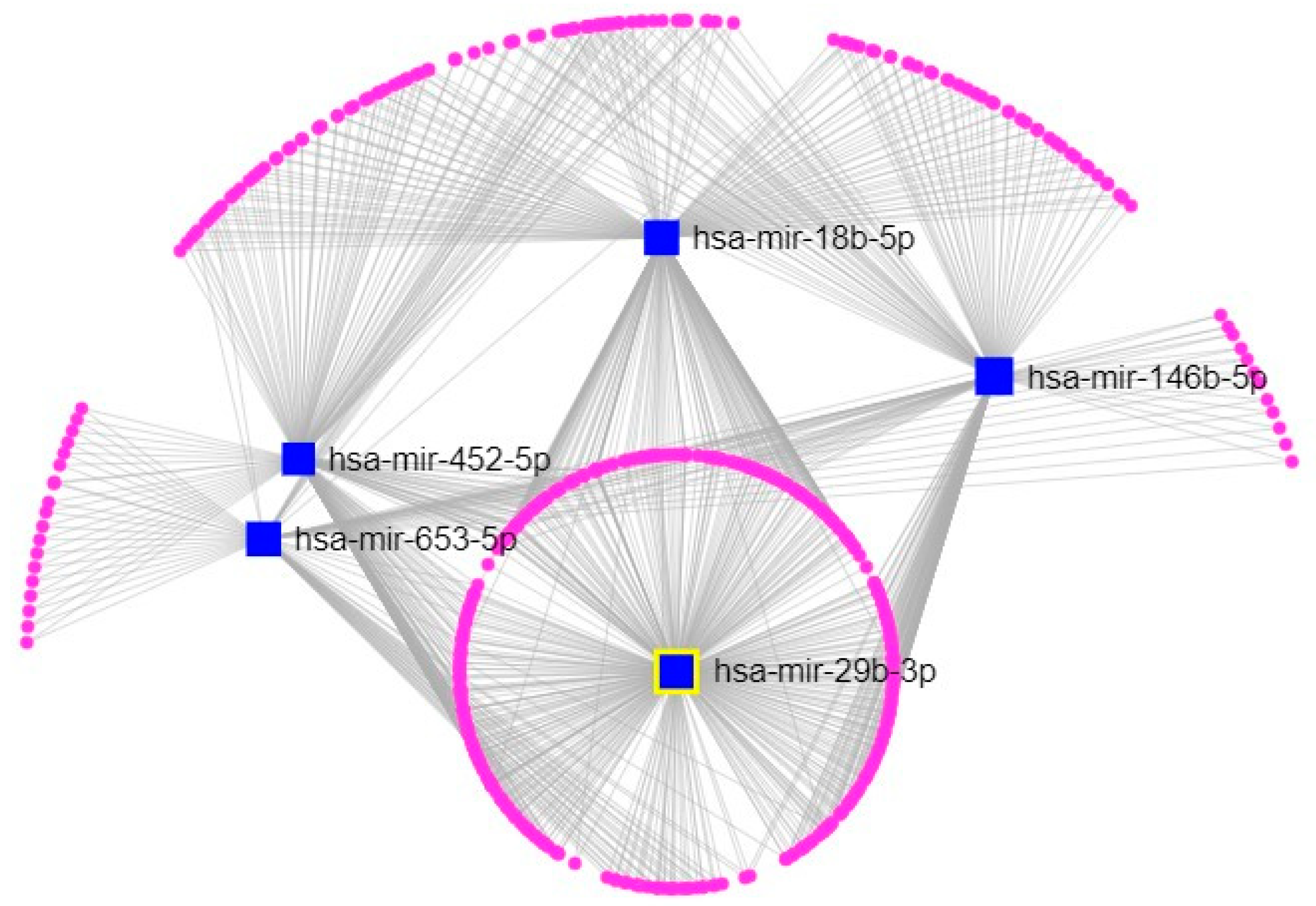
2.3. Exposure to ATZ, CYP and VNZ at Subtoxic Concentrations Induce Alterations of miRNAs Profile
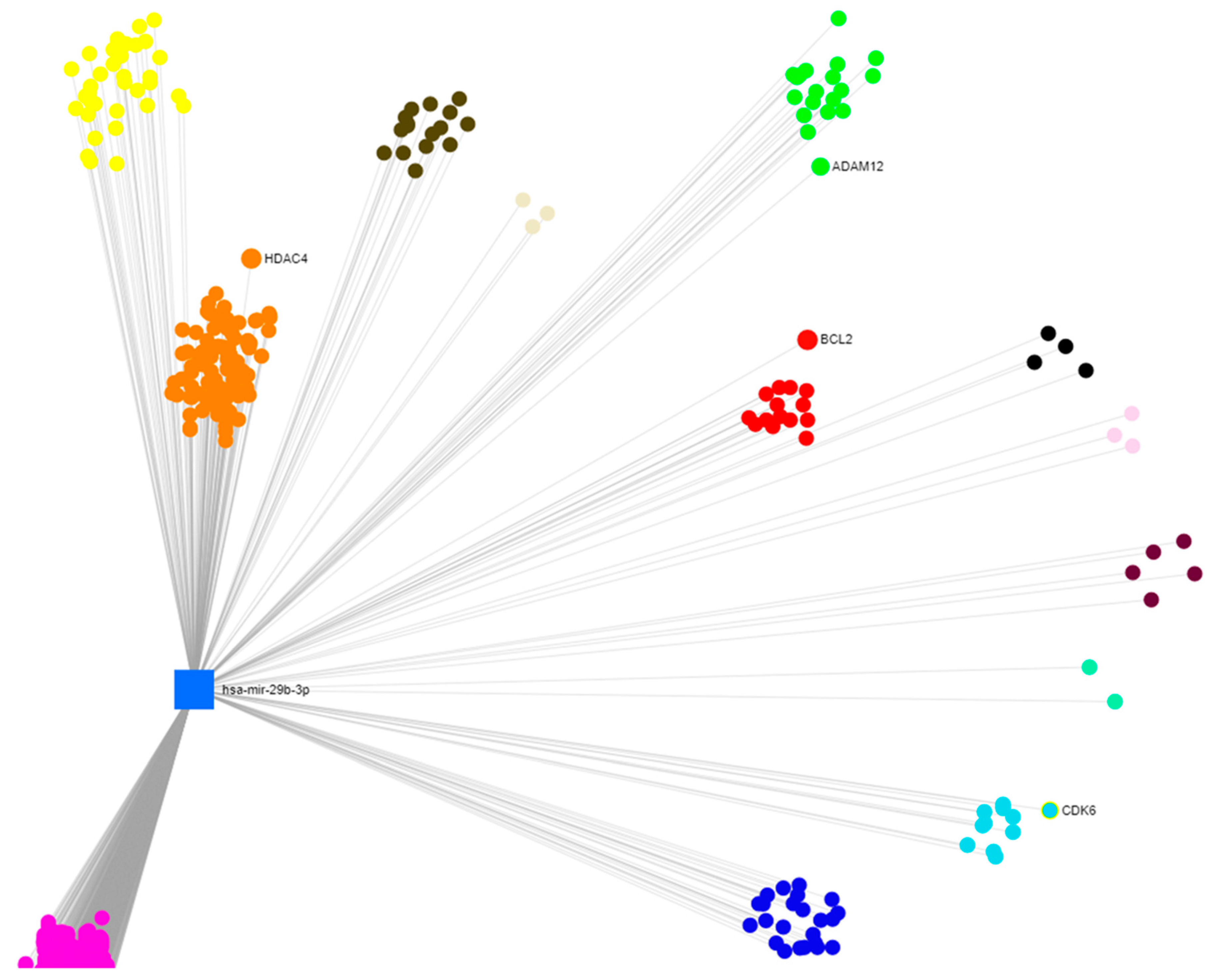
2.4. In Silico Prediction of miR-29b-3p Target Genes
2.5. Subtoxic Concentrations of VNZ Downregulate miR-29b-3p Contributing to the Increased Level of ADAM12 and CDK6 Genes in Differentiated SH-SY5Y Cells
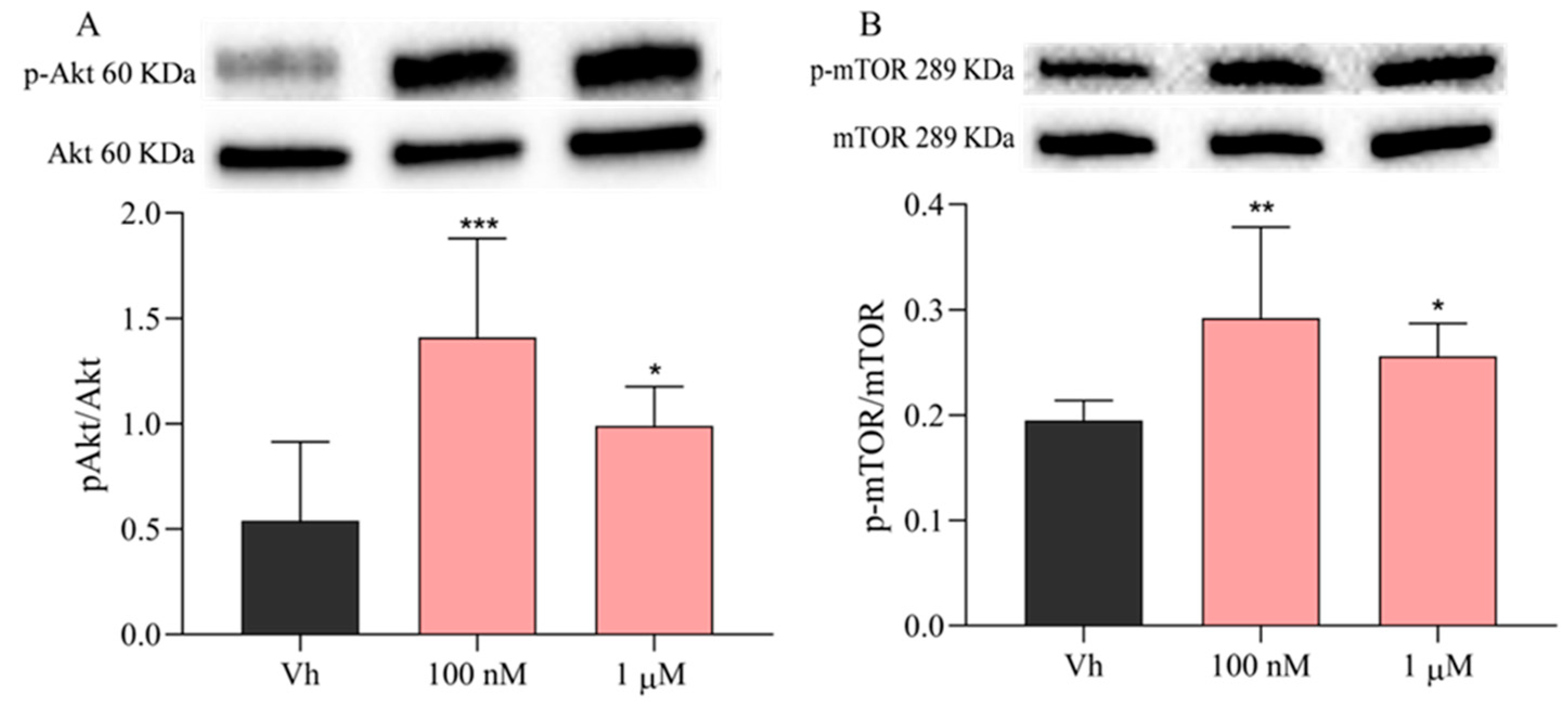
2.6. Subtoxic Concentrations of VNZ Downregulate miR-29b-3p Contributing to the Activation of PI3K/Akt/mTOR Pathway and Suppress p53 Activation in Differentiated SH-SY5Y Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.2.1. SH-SY5Y
4.2.2. SH-SY5Y Differentiation
4.3. EDCs Treatment
4.4. Determination of Neuronal Viability
4.5. Determination of ROS Formation Induced by EDCs
4.6. RNA Extraction
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis of miRNA Expression
4.8. Gene Expression Analysis by RNA Reverse Transcription and qRT-PCR
4.9. Western Blotting
4.10. In Silico Target Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yilmaz, B.; Terekeci, H.; Sandal, S.; Kelestimur, F. Endocrine Disrupting Chemicals: Exposure, Effects on Human Health, Mechanism of Action, Models for Testing and Strategies for Prevention. Rev. Endocr. Metab. Disord. 2020, 21, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Darbre, P.D. Overview of Air Pollution and Endocrine Disorders. Int. J. Gen. Med. 2018, 11, 191–207. [Google Scholar] [CrossRef] [Green Version]
- De Coster, S.; Van Larebeke, N. Endocrine-Disrupting Chemicals: Associated Disorders and Mechanisms of Action. J. Environ. Public Health 2012, 2012, 713696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mnif, W.; Hassine, A.I.H.; Bouaziz, A.; Bartegi, A.; Thomas, O.; Roig, B. Effect of Endocrine Disruptor Pesticides: A Review. Int. J. Environ. Res. Public Health 2011, 8, 2265–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galbiati, V.; Buoso, E.; d’Emmanuele di Villa Bianca, R.; Paola, R.D.; Morroni, F.; Nocentini, G.; Racchi, M.; Viviani, B.; Corsini, E. Immune and Nervous Systems Interaction in Endocrine Disruptors Toxicity: The Case of Atrazine. Front. Toxicol. 2021, 3, 649024. [Google Scholar] [CrossRef]
- De Albuquerque, F.P.; de Oliveira, J.L.; Moschini-Carlos, V.; Fraceto, L.F. An Overview of the Potential Impacts of Atrazine in Aquatic Environments: Perspectives for Tailored Solutions Based on Nanotechnology. Sci. Total Environ. 2020, 700, 134868. [Google Scholar] [CrossRef]
- He, H.; Chen, W.; Wei, Y.; Zhang, T.; Geng, W.; Zhai, J. Effects of Perinatal Exposure to Endocrine-Disrupting Chemicals on the Reproductive System of F3 Generation Male Rodents: A Meta-Analysis. Environ. Sci. Pollut. Res. Int. 2022, 29, 33218–33229. [Google Scholar] [CrossRef]
- Agnihotri, N.P.; Jain, H.K.; Gajbhiye, V.T. Persistence of Some Synthetic Pyrethroid Insecticides in Soil, Water and Sediment. I. J. Entomol. Res. 1986, 10, 147–151. [Google Scholar]
- Liu, C.; Qu, J.; Wu, M.; Huang, X.; Li, L. Cypermethrin Triggers YY1-Mediated Testosterone Biosynthesis Suppression. Ecotoxicol. Environ. Saf. 2021, 225, 112792. [Google Scholar] [CrossRef]
- Environmental Protection Agency (EPA). R.E.D. FACTS Vinclozolin; EPA-738-F-00-021. 2000. Available online: https://www3.epa.gov/pesticides/chem_search/reg_actions/reregistration/fs_PC-113201_1-Oct-00.pdf (accessed on 10 October 2022).
- Kelce, W.R.; Monosson, E.; Gamcsik, M.P.; Laws, S.C.; Gray, L.E. Environmental Hormone Disruptors: Evidence That Vinclozolin Developmental Toxicity Is Mediated by Antiandrogenic Metabolites. Toxicol. Appl. Pharmacol. 1994, 126, 276–285. [Google Scholar] [CrossRef]
- van Ravenzwaay, B.; Kolle, S.N.; Ramirez, T.; Kamp, H.G. Vinclozolin: A Case Study on the Identification of Endocrine Active Substances in the Past and a Future Perspective. Toxicol. Lett. 2013, 223, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Genovese, T.; Siracusa, R.; Fusco, R.; D’amico, R.; Impellizzeri, D.; Peritore, A.F.; Crupi, R.; Gugliandolo, E.; Morabito, R.; Cuzzocrea, S.; et al. Atrazine Inhalation Causes Neuroinflammation, Apoptosis and Accelerating Brain Aging. Int. J. Mol. Sci. 2021, 22, 7938. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B. Endocrine Disruptors as a Threat to Neurological Function. J. Neurol. Sci. 2011, 305, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stradtman, S.C.; Freeman, J.L. Mechanisms of Neurotoxicity Associated with Exposure to the Herbicide Atrazine. Toxics 2021, 9, 207. [Google Scholar] [CrossRef]
- Environmental Protection Agency (EPA). Atrazine—Draft Human Health Risk Assessment for Registration Review 2018; EPA-HQ-OPP-2013-0266-1159. 2018. Available online: https://www.regulations.gov/document/EPA-HQ-OPP-2013-0266-1159 (accessed on 10 October 2022).
- Zhou, L.; Zhou, M.; Tan, H.; Xiao, M. Cypermethrin-Induced Cortical Neurons Apoptosis via the Nrf2/ARE Signaling Pathway. Pestic. Biochem. Physiol. 2020, 165, 104547. [Google Scholar] [CrossRef]
- Fleisch, A.F.; Wright, R.O.; Baccarelli, A.A. Environmental Epigenetics: A Role in Endocrine Disease? J. Mol. Endocrinol. 2012, 49, R61–R67. [Google Scholar] [CrossRef] [Green Version]
- Shivdasani, R.A. MicroRNAs: Regulators of Gene Expression and Cell Differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef]
- Catellani, C.; Ravegnini, G.; Sartori, C.; Angelini, S.; Street, M.E. GH and IGF System: The Regulatory Role of MiRNAs and LncRNAs in Cancer. Front. Endocrinol. 2021, 12, 701246. [Google Scholar] [CrossRef]
- Li, Z.; Ru, X.; Wang, S.; Cao, G. MiR-24-3p Regulation of Retinol Binding Protein 4 in Trophoblast Biofunction and Preeclampsia. Mol. Reprod. Dev. 2022, 89, 423–430. [Google Scholar] [CrossRef]
- Qiu, Z.; He, L.; Yu, F.; Lv, H.; Zhou, Y. LncRNA FAM13A-AS1 Regulates Proliferation and Apoptosis of Cervical Cancer Cells by Targeting MiRNA-205-3p/DDI2 Axis. J. Oncol. 2022, 2022, 8411919. [Google Scholar] [CrossRef]
- Lema, C.; Cunningham, M.J. MicroRNAs and Their Implications in Toxicological Research. Toxicol. Lett. 2010, 198, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Wirbisky, S.E.; Weber, G.J.; Schlotman, K.E.; Sepúlveda, M.S.; Freeman, J.L. Embryonic Atrazine Exposure Alters Zebrafish and Human MiRNAs Associated with Angiogenesis, Cancer, and Neurodevelopment. Food Chem. Toxicol. 2016, 98, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.; Chen, Z.; Chen, H.; Lu, W.; Xie, S.; Meng, Q.H.; Wu, Y.; Xia, D. Cypermethrin Promotes Lung Cancer Metastasis via Modulation of Macrophage Polarization by Targeting MicroRNA-155/Bcl6. Toxicol. Sci. 2018, 163, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yang, J.; Zhang, Y.; Fu, H.; Yan, Z.; Zhu, Y. Vinclozolin-Induced Mouse Penile Malformation and “Small Testis” via MiR132, MiR195a Together with the Hippo Signaling Pathway. Toxicology 2021, 460, 152842. [Google Scholar] [CrossRef]
- Pruccoli, L.; Morroni, F.; Sita, G.; Hrelia, P.; Tarozzi, A. Esculetin as a Bifunctional Antioxidant Prevents and Counteracts the Oxidative Stress and Neuronal Death Induced by Amyloid Protein in Sh-Sy5y Cells. Antioxidants 2020, 9, 551. [Google Scholar] [CrossRef]
- Lavogina, D.; Lust, H.; Tahk, M.-J.; Laasfeld, T.; Vellama, H.; Nasirova, N.; Vardja, M.; Eskla, K.-L.; Salumets, A.; Rinken, A.; et al. Revisiting the Resazurin-Based Sensing of Cellular Viability: Widening the Application Horizon. Biosensors 2022, 12, 196. [Google Scholar] [CrossRef]
- Morroni, F.; Sita, G.; Graziosi, A.; Turrini, E.; Fimognari, C.; Tarozzi, A.; Hrelia, P. Protective Effects of 6-(Methylsulfinyl)Hexyl Isothiocyanate on Aβ1-42-Induced Cognitive Deficit, Oxidative Stress, Inflammation, and Apoptosis in Mice. Int. J. Mol. Sci. 2018, 19, 2083. [Google Scholar] [CrossRef] [Green Version]
- Bokobza, C.; Joshi, P.; Schang, A.L.; Csaba, Z.; Faivre, V.; Montané, A.; Galland, A.; Benmamar-Badel, A.; Bosher, E.; Lebon, S.; et al. MiR-146b Protects the Perinatal Brain against Microglia-Induced Hypomyelination. Ann. Neurol. 2022, 91, 48–65. [Google Scholar] [CrossRef]
- Kim, K.Y.; Kim, Y.R.; Choi, K.W.; Lee, M.; Lee, S.; Im, W.; Shin, J.Y.; Kim, J.Y.; Hong, Y.H.; Kim, M.; et al. Downregulated MiR-18b-5p Triggers Apoptosis by Inhibition of Calcium Signaling and Neuronal Cell Differentiation in Transgenic SOD1 (G93A) Mice and SOD1 (G17S and G86S) ALS Patients. Transl. Neurodegener. 2020, 9, 23. [Google Scholar] [CrossRef]
- Peng, Z.; Yang, X.; Zhang, H.; Yin, M.; Luo, Y.; Xie, C. MiR-29b-3p Aggravates NG108-15 Cell Apoptosis Triggered by Fluorine Combined with Aluminum. Ecotoxicol. Environ. Saf. 2021, 224, 112658. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Z.; Wang, X.; Ma, Q.; Ji, H.; Wang, Y.; Yu, R. PBX2-Mediated CircTLK1 Activates JAK/STAT Signaling to Promote Gliomagenesis via MiR-452-5p/SSR1 Axis. Front. Genet. 2021, 12, 698831. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Shu, T.; Peng, H.; Liu, J.; Li, C.; Wang, M.; Wu, P.; Liu, Y. LncRNA H19 Inhibits Oxidative Stress Injury of Cochlear Hair Cells by Regulating MiR-653-5p/SIRT1 Axis. Acta Biochim. Biophys. Sin. 2022, 54, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. MiRNet 2.0: Network-Based Visual Analytics for MiRNA Functional Analysis and Systems Biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Marttila, S.; Rovio, S.; Mishra, P.P.; Seppälä, I.; Lyytikäinen, L.P.; Juonala, M.; Waldenberger, M.; Oksala, N.; Ala-Korpela, M.; Harville, E.; et al. Adulthood Blood Levels of Hsa-MiR-29b-3p Associate with Preterm Birth and Adult Metabolic and Cognitive Health. Sci. Rep. 2021, 11, 9203. [Google Scholar] [CrossRef]
- Pereira, P.A.; Tomás, J.F.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. Recombinant Pre-MiR-29b for Alzheimer’s Disease Therapeutics. Sci. Rep. 2016, 6, 19946. [Google Scholar] [CrossRef] [Green Version]
- Roshan, R.; Shridhar, S.; Sarangdhar, M.A.; Banik, A.; Chawla, M.; Garg, M.; Singh, V.P.; Pillai, B. Brain-Specific Knockdown of MiR-29 Results in Neuronal Cell Death and Ataxia in Mice. RNA 2014, 20, 1287–1297. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.; Lu, J.; Zhang, C.; You, R.; Shi, L.; Jiang, N.; Nie, D.; Zhu, J.; Li, M.; Guo, J. MiR-29b Inhibits the Growth of Glioma via MYCN Dependent Way. Oncotarget 2017, 8, 45224–45233. [Google Scholar] [CrossRef] [Green Version]
- Rodgers, K.M.; Udesky, J.O.; Rudel, R.A.; Brody, J.G. Environmental Chemicals and Breast Cancer: An Updated Review of Epidemiological Literature Informed by Biological Mechanisms. Env. Res. 2018, 160, 152–182. [Google Scholar] [CrossRef]
- Hardesty, J.E.; Al-Eryani, L.; Wahlang, B.; Falkner, K.C.; Shi, H.; Jin, J.; Vivace, B.J.; Ceresa, B.P.; Prough, R.A.; Cave, M.C. Epidermal Growth Factor Receptor Signaling Disruption by Endocrine and Metabolic Disrupting Chemicals. Toxicol. Sci. 2018, 162, 622–634. [Google Scholar] [CrossRef] [Green Version]
- Luzio, A.; Matos, M.; Santos, D.; Fontaínhas-Fernandes, A.A.; Monteiro, S.M.; Coimbra, A.M. Disruption of Apoptosis Pathways Involved in Zebrafish Gonad Differentiation by 17α-Ethinylestradiol and Fadrozole Exposures. Aquat. Toxicol. 2016, 177, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, L.; Del Pozzo, G.; Barba, P.; Caputo, L.; Mita, L.; Viggiano, E.; Russo, G.L.; Nicolucci, C.; Rossi, S.; Bencivenga, U.; et al. Effects of Some Endocrine Disruptors on Cell Cycle Progression and Murine Dendritic Cell Differentiation. Gen. Comp. Endocrinol. 2012, 178, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qian, L.; Wang, C.; Teng, M.; Duan, M.; Zhou, Y.; Chen, X.; Bo, R.; Wang, C.; Li, X. Dysregulation of Endocrine Disruption, Apoptosis and the Transgenerational Toxicity Induced by Spirotetramat. Chemosphere 2020, 240, 124900. [Google Scholar] [CrossRef] [PubMed]
- Tehranian, C.; Fankhauser, L.; Harter, P.N.; Ratcliffe, C.D.H.; Zeiner, P.S.; Messmer, J.M.; Hoffmann, D.C.; Frey, K.; Westphal, D.; Ronellenfitsch, M.W.; et al. The PI3K/Akt/MTOR Pathway as a Preventive Target in Melanoma Brain Metastasis. Neuro Oncol. 2022, 24, 213–225. [Google Scholar] [CrossRef]
- Wang, L.; Tan, Z.; Zhang, Y.; Kady Keita, N.; Liu, H.; Zhang, Y. ADAM12 Silencing Promotes Cellular Apoptosis by Activating Autophagy in Choriocarcinoma Cells. Int. J. Oncol. 2020, 56, 1162–1174. [Google Scholar] [CrossRef] [Green Version]
- Morroni, F.; Sita, G.; Graziosi, A.; Turrini, E.; Fimognari, C.; Tarozzi, A.; Hrelia, P. Neuroprotective Effect of Caffeic Acid Phenethyl Ester in a Mouse Model of Alzheimer’s Disease Involves Nrf2/HO-1 Pathway. Aging Dis. 2018, 9, 605. [Google Scholar] [CrossRef] [Green Version]
- Bellutti, F.; Tigan, A.-S.; Nebenfuehr, S.; Dolezal, M.; Zojer, M.; Grausenburger, R.; Hartenberger, S.; Kollmann, S.; Doma, E.; Prchal-Murphy, M.; et al. CDK6 Antagonizes P53-Induced Responses during Tumorigenesis. Cancer Discov. 2018, 8, 884–897. [Google Scholar] [CrossRef] [Green Version]
- Sanz, G.; Singh, M.; Peuget, S.; Selivanova, G. Inhibition of P53 Inhibitors: Progress, Challenges and Perspectives. J. Mol. Cell Biol. 2019, 11, 586–599. [Google Scholar] [CrossRef] [Green Version]
- Bergman, Å.; Heindel, J.J.; Jobling, S.; Kidd, K.; Zoeller, T.R.; World Health Organization. State of the Science of Endocrine Disrupting Chemicals 2012; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Jauhari, A.; Singh, T.; Yadav, S. Neurodevelopmental Disorders and Neurotoxicity: MicroRNA in Focus. J. Chem. Neuroanat. 2022, 120, 102072. [Google Scholar] [CrossRef]
- Sabry, R.; Yamate, J.; Favetta, L.; LaMarre, J. MicroRNAs: Potential Targets and Agents of Endocrine Disruption in Female Reproduction. J. Toxicol. Pathol. 2019, 32, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Santanam, U.; Cimmino, A.; Palamarchuk, A.; Efanov, A.; Maximov, V.; Volinia, S.; Alder, H.; Liu, C.-G.; Rassenti, L.; et al. Tcl1 Expression in Chronic Lymphocytic Leukemia Is Regulated by MiR-29 and MiR-181. Cancer Res. 2006, 66, 11590–11593. [Google Scholar] [CrossRef] [Green Version]
- Porkka, K.P.; Pfeiffer, M.J.; Waltering, K.K.; Vessella, R.L.; Tammela, T.L.J.; Visakorpi, T. MicroRNA Expression Profiling in Prostate Cancer. Cancer Res. 2007, 67, 6130–6135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique MicroRNA Molecular Profiles in Lung Cancer Diagnosis and Prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shipley, M.M.; Mangold, C.A.; Szpara, M.L. Differentiation of the SH-SY5Y Human Neuroblastoma Cell Line. J. Vis. Exp. 2016, 108, e53193. [Google Scholar] [CrossRef]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of MicroRNA Cluster MiR-29a/b-1 in Sporadic Alzheimer’s Disease Correlates with Increased BACE1/Beta-Secretase Expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [Green Version]
- Klatt, C.L.; Theis, V.; Hahn, S.; Theiss, C.; Matschke, V. Deregulated MiR-29b-3p Correlates with Tissue-Specific Activation of Intrinsic Apoptosis in An Animal Model of Amyotrophic Lateral Sclerosis. Cells 2019, 8, 1077. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Shim, H.G.; Hwang, T.; Kim, H.; Kang, S.-H.; Dho, Y.-S.; Park, S.-H.; Kim, S.J.; Park, C.-K. Restoration of MiR-29b Exerts Anti-Cancer Effects on Glioblastoma. Cancer Cell Int. 2017, 17, 104. [Google Scholar] [CrossRef]
- Kveiborg, M.; Albrechtsen, R.; Couchman, J.R.; Wewer, U.M. Cellular Roles of ADAM12 in Health and Disease. Int. J. Biochem. Cell Biol. 2008, 40, 1685–1702. [Google Scholar] [CrossRef]
- Zahnow, C.A. ErbB Receptors and Their Ligands in the Breast. Expert Rev. Mol. Med. 2006, 8, 1–21. [Google Scholar] [CrossRef]
- Cui, D.; Arima, M.; Takubo, K.; Kimura, T.; Horiuchi, K.; Minagawa, T.; Matsuda, S.; Ikeda, E. ADAM12 and ADAM17 Are Essential Molecules for Hypoxia-Induced Impairment of Neural Vascular Barrier Function. Sci. Rep. 2015, 5, 12796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harold, D.; Jehu, L.; Turic, D.; Hollingworth, P.; Moore, P.; Summerhayes, P.; Moskvina, V.; Foy, C.; Archer, N.; Hamilton, B.A.; et al. Interaction between the ADAM12 and SH3MD1 Genes May Confer Susceptibility to Late-Onset Alzheimer’s Disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144B, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, D.; Lendeckel, U.; Theodosiou, P.; Dobrowolny, H.; Mawrin, C.; Keilhoff, G.; Bukowska, A.; Dietzmann, K.; Bogerts, B.; Bernstein, H.-G. ADAM 12: A Putative Marker of Oligodendrogliomas? Dis. Mrk. 2013, 34, 81–91. [Google Scholar] [CrossRef]
- Toft-Hansen, H.; Nuttall, R.K.; Edwards, D.R.; Owens, T. Key Metalloproteinases Are Expressed by Specific Cell Types in Experimental Autoimmune Encephalomyelitis. J. Immunol. 2004, 173, 5209–5218. [Google Scholar] [CrossRef] [Green Version]
- Luna, C.; Li, G.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Role of MiR-29b on the Regulation of the Extracellular Matrix in Human Trabecular Meshwork Cells under Chronic Oxidative Stress. Mol. Vis. 2009, 15, 2488–2497. [Google Scholar]
- Ramdas, V.; McBride, M.; Denby, L.; Baker, A.H. Canonical Transforming Growth Factor-β Signaling Regulates Disintegrin Metalloprotease Expression in Experimental Renal Fibrosis via MiR-29. Am. J. Pathol. 2013, 183, 1885–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duhachek-Muggy, S.; Zolkiewska, A. ADAM12-L Is a Direct Target of the MiR-29 and MiR-200 Families in Breast Cancer. BMC Cancer 2015, 15, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nebenfuehr, S.; Kollmann, K.; Sexl, V. The Role of CDK6 in Cancer. Int. J. Cancer 2020, 147, 2988–2995. [Google Scholar] [CrossRef]
- Ji, W.; Zhang, W.; Wang, X.; Shi, Y.; Yang, F.; Xie, H.; Zhou, W.; Wang, S.; Guan, X. C-Myc Regulates the Sensitivity of Breast Cancer Cells to Palbociclib via c-Myc/MiR-29b-3p/CDK6 Axis. Cell Death Dis. 2020, 11, 760. [Google Scholar] [CrossRef]
- Zhao, J.-J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. MicroRNA Expression Profile and Identification of MiR-29 as a Prognostic Marker and Pathogenetic Factor by Targeting CDK6 in Mantle Cell Lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef]
- Li, Y.; Chitnis, N.; Nakagawa, H.; Kita, Y.; Natsugoe, S.; Yang, Y.; Li, Z.; Wasik, M.; Klein-Szanto, A.J.P.; Rustgi, A.K.; et al. PRMT5 Is Required for Lymphomagenesis Triggered by Multiple Oncogenic Drivers. Cancer Discov. 2015, 5, 288–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, B.P.; Liao, Y.; Xia, W.; Zou, Y.; Spohn, B.; Hung, M.C. HER-2/Neu Induces P53 Ubiquitination via Akt-Mediated MDM2 Phosphorylation. Nat. Cell Biol. 2001, 3, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, C.; Wu, R.; Hu, W. Tumor Suppressor P53 Meets MicroRNAs. J. Mol. Cell Biol. 2011, 3, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hong, Y.; Scherer, S.J.; Schartl, M. Lack of Ultraviolet-Light Inducibility of the Medakafish (Oryzias Latipes) Tumor Suppressor Gene P53. Gene 2001, 264, 197–203. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Rhee, J.-S.; Hwang, D.-S.; Kim, I.-C.; Raisuddin, S.; Lee, J.-S. P53 Gene Expression Is Modulated by Endocrine Disrupting Chemicals in the Hermaphroditic Fish, Kryptolebias Marmoratus. Comp. Biochem. Physiol. C Toxicol. Pharm. 2008, 147, 150–157. [Google Scholar] [CrossRef]
- Morroni, F.; Sita, G.; Graziosi, A.; Ravegnini, G.; Molteni, R.; Paladini, M.S.; Dias, K.S.T.; Dos Santos, A.F.; Viegas, C.; Camps, I.; et al. PQM130, a Novel Feruloyl-Donepezil Hybrid Compound, Effectively Ameliorates the Cognitive Impairments and Pathology in a Mouse Model of Alzheimer’s Disease. Front. Pharmacol. 2019, 10, 658. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER Version 14: More Genomes, a New PANTHER GO-Slim and Improvements in Enrichment Analysis Tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]

| Undifferentiated Cells | ||||||
|---|---|---|---|---|---|---|
| Target Name | ATZ 100 nM | ATZ 1 µM | CYP 100 nM | CYP 1 µM | VNZ 100 nM | VNZ 1 µM |
| miR-18b-5p | 1.88 | 1.26 | 1.28 | 1.13 | 1.70 | 1.74 |
| miR-29b-3p | 1.21 | 0.63 | 1.53 | 0.95 | 1.49 | 0.78 |
| miR-146b-5p | 1.28 | 1.22 | 1.47 | 1.18 | 1.99 | 0.93 |
| miR-452-5p | 1.94 | 1.24 | 1.41 | 1.45 | 1.46 | 1.90 |
| miR-653-5p | 1.12 | 0.80 | 1.17 | 1.16 | 0.95 | 0.91 |
| Differentiated cells | ||||||
| Target name | ATZ 100 nM | ATZ 1 µM | CYP 100 nM | CYP 1 µM | VNZ 100 nM | VNZ 1 µM |
| miR-18b-5p | 0.30 * | 0.65 | 0.95 | 0.67 | 0.26 * | 0.32 * |
| miR-29b-3p | 0.42 * | 0.92 | 0.65 | 0.87 | 0.38 * | 0.70 |
| miR-146b-5p | 0.49 * | 0.56 | 1.06 | 1.32 | 1.51 | 0.97 |
| miR-452-5p | 0.55 | 1.09 | 0.81 | 0.86 | 0.57 | 0.25 * |
| miR-653-5p | 0.41 * | 0.59 | 1.15 | 1.07 | 0.26 * | 0.32 * |
| ADAM12 | Bcl2 | CDK6 | HDAC4 | |
|---|---|---|---|---|
| ATZ 100 nM | 1.20 | 1.31 | 1.28 | 1.12 |
| CYP 100 nM | 1.43 | 1.56 | 1.98 | 1.30 |
| VNZ 100 nM | 2.11 * | 1.99 | 2.13 * | 1.46 |
| ATZ 1 µM | 1.38 | 1.13 | 1.58 | 1.32 |
| CYP 1 µM | 1.34 | 1.05 | 1.74 | 1.46 |
| VNZ 1 µM | 2.58 * | 1.60 | 2.97 * | 1.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graziosi, A.; Sita, G.; Corrieri, C.; Angelini, S.; d’Emmanuele di Villa Bianca, R.; Mitidieri, E.; Sorrentino, R.; Hrelia, P.; Morroni, F. Effects of Subtoxic Concentrations of Atrazine, Cypermethrin, and Vinclozolin on microRNA-Mediated PI3K/Akt/mTOR Signaling in SH-SY5Y Cells. Int. J. Mol. Sci. 2022, 23, 14538. https://doi.org/10.3390/ijms232314538
Graziosi A, Sita G, Corrieri C, Angelini S, d’Emmanuele di Villa Bianca R, Mitidieri E, Sorrentino R, Hrelia P, Morroni F. Effects of Subtoxic Concentrations of Atrazine, Cypermethrin, and Vinclozolin on microRNA-Mediated PI3K/Akt/mTOR Signaling in SH-SY5Y Cells. International Journal of Molecular Sciences. 2022; 23(23):14538. https://doi.org/10.3390/ijms232314538
Chicago/Turabian StyleGraziosi, Agnese, Giulia Sita, Camilla Corrieri, Sabrina Angelini, Roberta d’Emmanuele di Villa Bianca, Emma Mitidieri, Raffaella Sorrentino, Patrizia Hrelia, and Fabiana Morroni. 2022. "Effects of Subtoxic Concentrations of Atrazine, Cypermethrin, and Vinclozolin on microRNA-Mediated PI3K/Akt/mTOR Signaling in SH-SY5Y Cells" International Journal of Molecular Sciences 23, no. 23: 14538. https://doi.org/10.3390/ijms232314538
APA StyleGraziosi, A., Sita, G., Corrieri, C., Angelini, S., d’Emmanuele di Villa Bianca, R., Mitidieri, E., Sorrentino, R., Hrelia, P., & Morroni, F. (2022). Effects of Subtoxic Concentrations of Atrazine, Cypermethrin, and Vinclozolin on microRNA-Mediated PI3K/Akt/mTOR Signaling in SH-SY5Y Cells. International Journal of Molecular Sciences, 23(23), 14538. https://doi.org/10.3390/ijms232314538









