Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas
Abstract
1. Introduction
2. Results
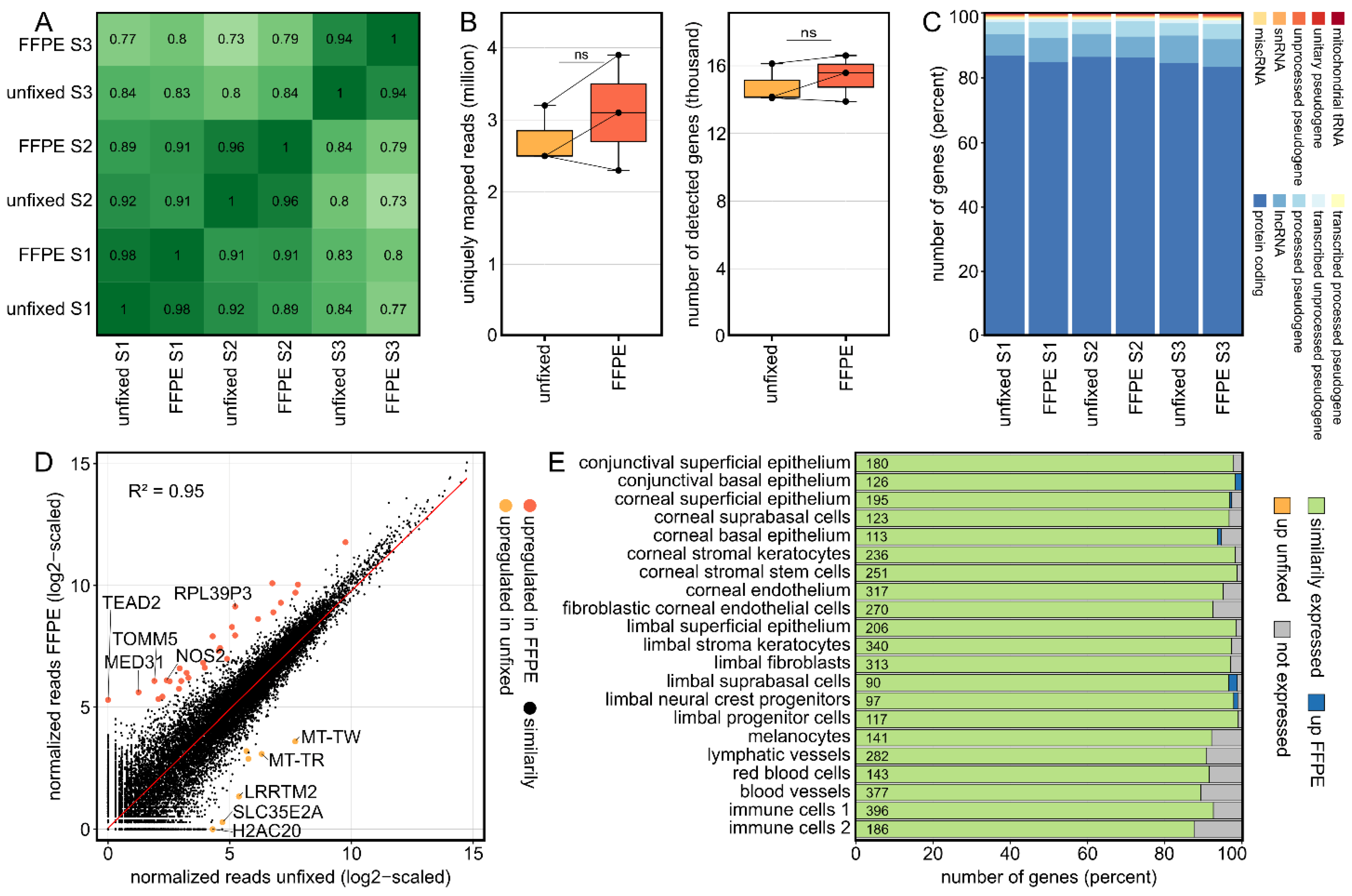
2.1. Formalin Fixation Does Not Have a Significant Impact on the Transcriptional Profile of Human Corneal Tissue
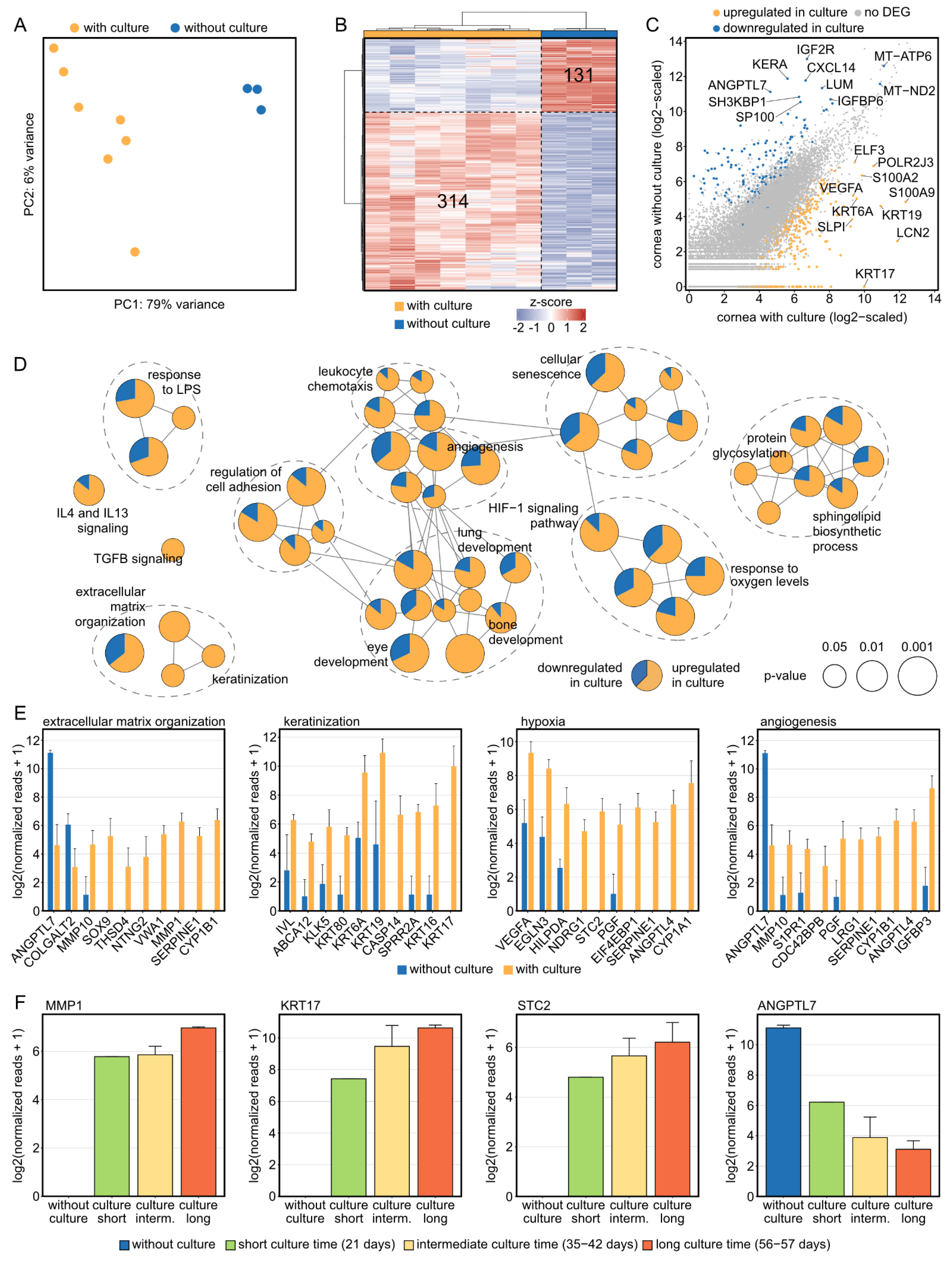
2.2. Corneal Tissue Culture Significantly Modulates the Transcriptional Profile of Human Corneas
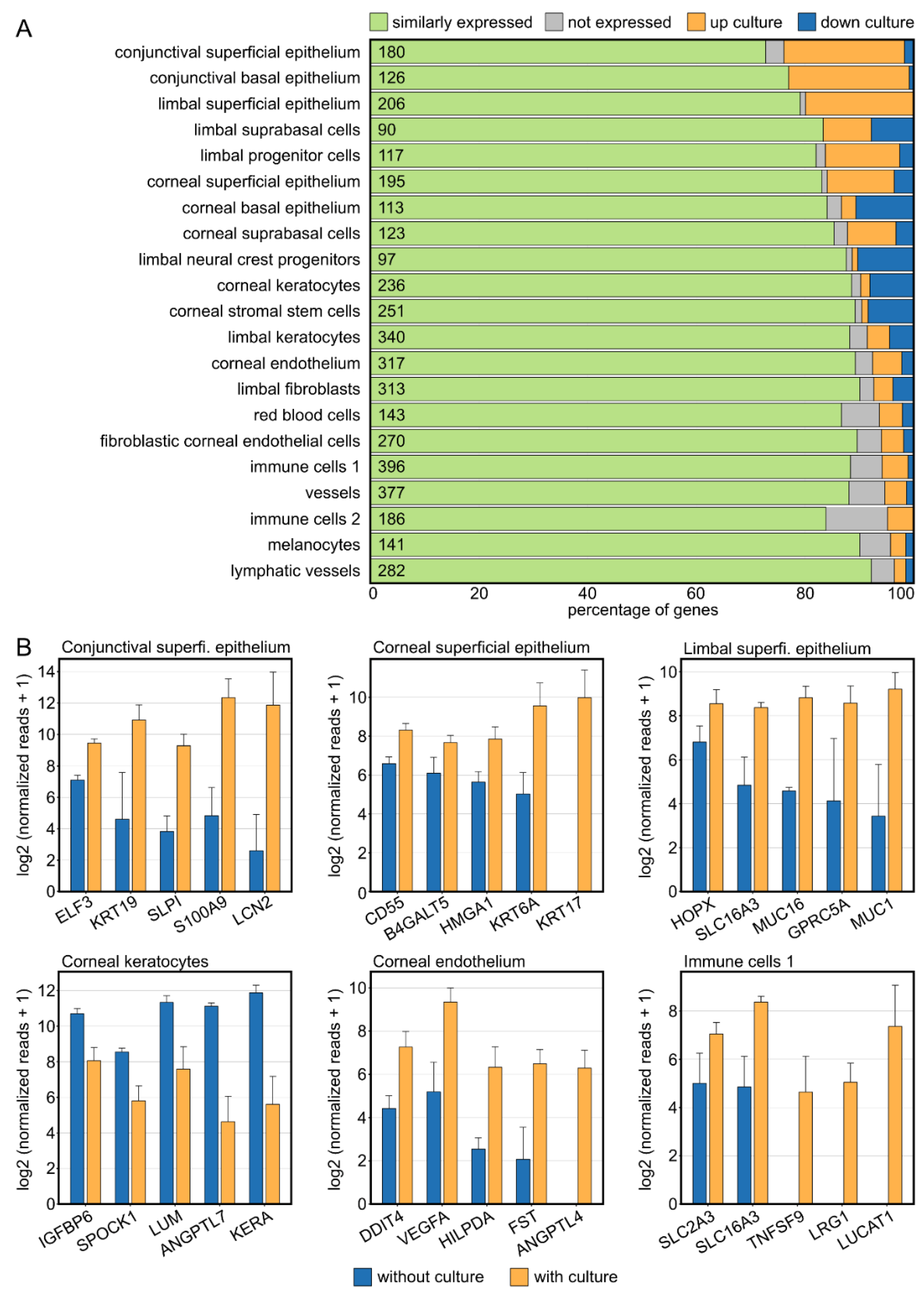
2.3. Corneal Tissue Culture Significantly Modulates the Corneal Cellular Composition
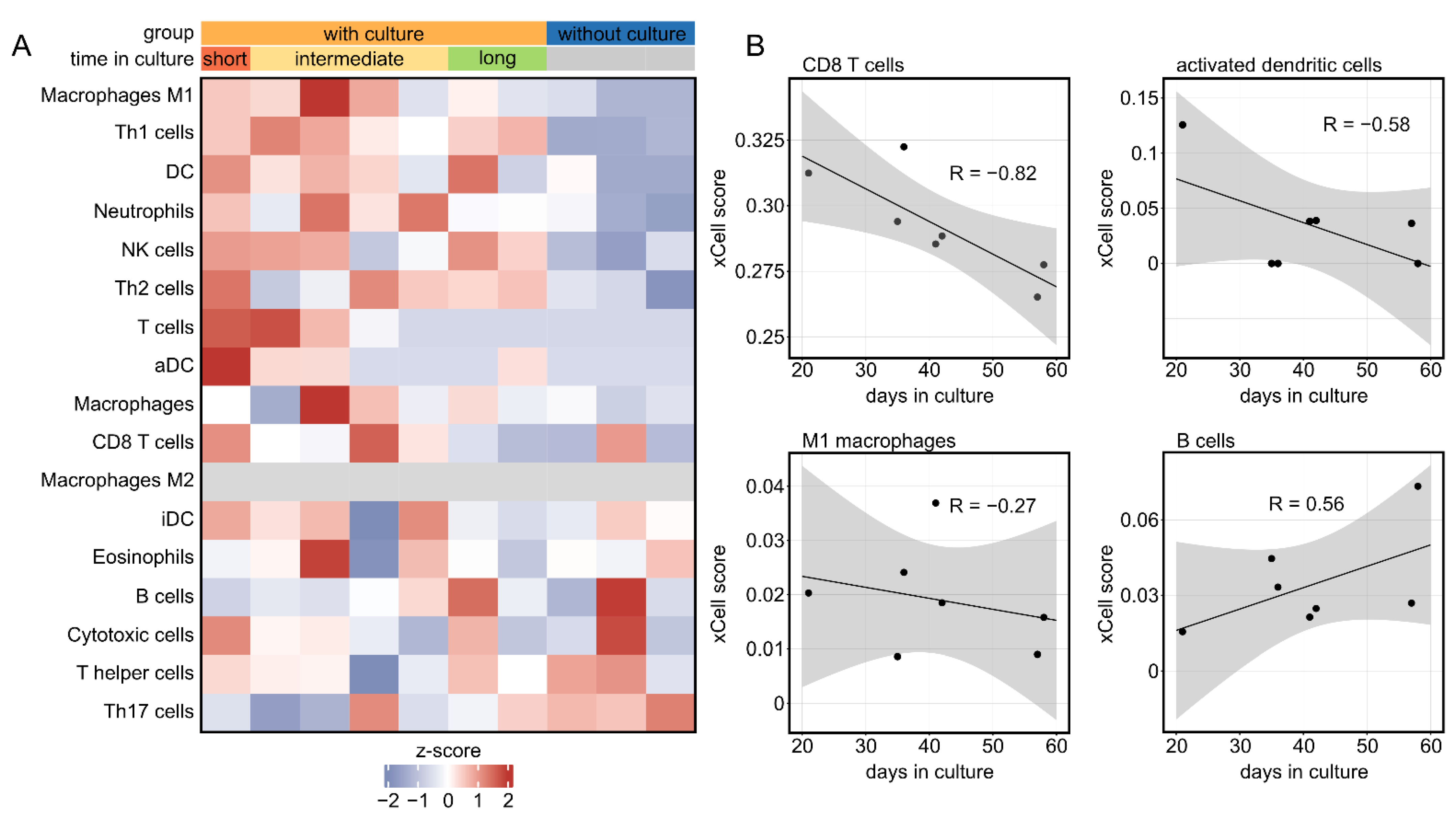
2.4. The Corneal Immune Cell Profile Significantly Changes in Tissue Culture in a Time-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Tissue Specimens
4.3. Tissue Processing
4.4. RNA Extraction
4.5. RNA Sequencing
4.6. Bioinformatics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Swamy, V.; McGaughey, D. Eye in a Disk: EyeIntegration Human Pan-Eye and Body Transcriptome Database Version 1.0. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3236–3246. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Boneva, S.; Schlecht, A.; Lapp, T.; Auw-Haedrich, C.; Lagrèze, W.; Agostini, H.; Reinhard, T.; Schlunck, G.; Lange, C. The Human Eye Transcriptome Atlas: A searchable comparative transcriptome database for healthy and diseased human eye tissue. Genomics 2022, 114, 110286. [Google Scholar] [CrossRef]
- Collin, J.; Queen, R.; Zerti, D.; Bojic, S.; Dorgau, B.; Moyse, N.; Molina, M.M.; Yang, C.; Dey, S.; Reynolds, G.; et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 2021, 21, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Boneva, S.; Schlecht, A.; Böhringer, D.; Mittelviefhaus, H.; Reinhard, T.; Agostini, H.; Auw-Haedrich, C.; Schlunck, G.; Wolf, J.; Lange, C. 3′ MACE RNA-sequencing allows for transcriptome profiling in human tissue samples after long-term storage. Lab. Investig. 2020, 100, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- EEBA. Minimum Medical Standards (MMS). Published Online 1 February 2020. Available online: https://www.eeba.eu/files/pdf/EEBA%20Minimum%20Medical%20Standards%20Revision%205%20Final.pdf (accessed on 31 May 2022).
- Chu, W. The past twenty-five years in eye banking. Cornea 2000, 19, 754–765. [Google Scholar] [CrossRef]
- Moshirfar, M.; Odayar, V.S.; McCabe, S.E.; Ronquillo, Y.C. Corneal Donation: Current Guidelines and Future Direction. Clin. Ophthalmol. 2021, 15, 2963–2973. [Google Scholar] [CrossRef]
- Maier, P.; Heinzelmann, S.; Böhringer, D.; Reinhard, T. Prolonged organ culture reduces the incidence of endothelial immune reactions. Eye 2016, 30, 127–132. [Google Scholar] [CrossRef]
- Zhuang, X.; Schlunck, G.; Wolf, J.; Rosmus, D.-D.; Bleul, T.; Luo, R.; Böhringer, D.; Wieghofer, P.; Lange, C.A.K.; Reinhard, T.; et al. Time- and Stimulus-Dependent Characteristics of Innate Immune Cells in Organ-Cultured Human Corneal Tissue. J. Innate Immun. 2021, 14, 1–14. [Google Scholar] [CrossRef]
- Tahvildari, M.; Emami-Naeini, P.; Omoto, M.; Mashaghi, A.; Chauhan, S.K.; Dana, R. Treatment of donor corneal tissue with immunomodulatory cytokines: A novel strategy to promote graft survival in high-risk corneal transplantation. Sci. Rep. 2017, 7, 971. [Google Scholar] [CrossRef]
- Yang, B.; Zhang, W.; Zhang, M.; Wang, X.; Peng, S.; Zhang, R. KRT6A Promotes EMT and Cancer Stem Cell Transformation in Lung Adenocarcinoma. Technol. Cancer Res. Treat. 2020, 19, 1533033820921248. [Google Scholar] [CrossRef]
- Kawakita, T.; Espana, E.M.; Higa, K.; Kato, N.; Li, W.; Tseng, S.C.G. Activation of Smad-mediated TGF-β signaling triggers epithelial-mesenchymal transitions in murine cloned corneal progenitor cells. J. Cell. Physiol. 2013, 228, 225–234. [Google Scholar] [CrossRef]
- di Martino, E.; Ali, M.; Inglehearn, C.F. Matrix metalloproteinases in keratoconus—Too much of a good thing? Exp. Eye Res. 2019, 182, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Onochie, O.E.; Onyejose, A.J.; Rich, C.B.; Trinkaus-Randall, V. The Role of Hypoxia in Corneal Extracellular Matrix Deposition and Cell Motility. Anat. Rec. 2020, 303, 1703–1716. [Google Scholar] [CrossRef] [PubMed]
- Ail, D.; Samardzija, M.; Chang, A.C.M.; Keck, J.; Reddel, R.R.; Grimm, C. Stanniocalcin2, but Not Stanniocalcin1, Responds to Hypoxia in a HIF1-Dependent Manner in the Retina. Front. Neurosci. 2022, 16, 882559. [Google Scholar] [CrossRef]
- Law, A.Y.S.; Wong, C.K.C. Stanniocalcin-2 promotes epithelial-mesenchymal transition and invasiveness in hypoxic human ovarian cancer cells. Exp. Cell Res. 2010, 316, 3425–3434. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Bao, L.; Chen, Y.; Wang, C.; Luo, M.; Zhang, B.; Zhou, M.; Wang, J.E.; Fang, Y.V.; Kumar, A.; et al. HIF2-Induced Long Noncoding RNA RAB11B-AS1 Promotes Hypoxia-Mediated Angiogenesis and Breast Cancer Metastasis. Cancer Res. 2020, 80, 964–975. [Google Scholar] [CrossRef]
- Qiu, Z.; Yang, J.; Deng, G.; Li, D.; Zhang, S. Angiopoietin-like 4 promotes angiogenesis and neurogenesis in a mouse model of acute ischemic stroke. Brain Res. Bull. 2021, 168, 156–164. [Google Scholar] [CrossRef]
- Toyono, T.; Usui, T.; Yokoo, S.; Taketani, Y.; Nakagawa, S.; Kuroda, M.; Yamagami, S.; Amano, S. Angiopoietin-like 7 is an anti-angiogenic protein required to prevent vascularization of the cornea. PLoS ONE 2015, 10, e0116838. [Google Scholar] [CrossRef]
- Wang, X.; Abraham, S.; McKenzie, J.A.G.; Jeffs, N.; Swire, M.; Tripathi, V.B.; Luhmann, U.F.O.; Lange, C.A.K.; Zhai, Z.; Arthur, H.M.; et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature 2013, 499, 306–311. [Google Scholar] [CrossRef]
- Uyar, B.; Palmer, D.; Kowald, A.; Escobar, H.M.; Barrantes, I.; Möller, S.; Akalin, A.; Fuellen, G. Single-cell analyses of aging, inflammation and senescence. Ageing Res. Rev. 2020, 64, 101156. [Google Scholar] [CrossRef]
- Polisetti, N.; Islam, M.M.; Griffith, M. The artificial cornea. Methods Mol. Biol. 2013, 1014, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xue, Q.; Li, J.; Ma, L.; Yao, Y.; Ye, H.; Cui, Z.; Yang, H. 3D bioprinting for artificial cornea: Challenges and perspectives. Med. Eng. Phys. 2019, 71, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Auw-Haedrich, C.; Schlecht, A.; Boneva, S.; Mittelviefhaus, H.; Lapp, T.; Agostini, H.; Reinhard, T.; Schlunck, G.; Lange, C.A.K. Transcriptional characterization of conjunctival melanoma identifies the cellular tumor microenvironment and prognostic gene signatures. Sci. Rep. 2020, 10, 17022. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Schlecht, A.; Rosmus, D.D.; Boneva, S.; Agostini, H.; Schlunck, G.; Wieghofer, P.; Lange, C. Comparative transcriptome analysis of human and murine choroidal neovascularization identifies fibroblast growth factor inducible-14 as phylogenetically conserved mediator of neovascular age-related macular degeneration. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166340. [Google Scholar] [CrossRef]
- Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Boneva, S.; Rosmus, D.D.; Agostini, H.; Schlunck, G.; Wieghofer, P.; Schlecht, A.; Lange, C. Deciphering the Molecular Signature of Human Hyalocytes in Relation to Other Innate Immune Cell Populations. Investig. Ophthalmol. Vis. Sci. 2022, 63, 9. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
| Donor | Age at Death [years] | Sex | Endothelial Cell Count [cells/mm²] | Necrosis [%] | Time between Death and Corneal Extraction [days] | Time in Culture [days] | Specifics | Tissue Culture (TC) | Corneal Graft Diameter |
|---|---|---|---|---|---|---|---|---|---|
| Corneal tissue in culture unfixed samples | |||||||||
| 1 | 60 | M | 1898 | 20 | 1 | 42 | / | M2 | 9.0 mm |
| 2 | 59 | M | 949 | 10 | 1 | 36 | Pseudophakic | M2 | 9.0 mm |
| 3 | 87 | F | 1241 | 5 | 2 | 35 | Corneal scar | M2 | 9.0 mm |
| 4 | 82 | M | 803 | 50 | 0 | 21 | / | M2 | 9.0 mm |
| 5a | 88 | M | 1752 | 3 | 1 | 57 | Pseudophakic, Arcus lipoides | M2 | ½ 9.0 mm |
| 6a | 78 | M | 1825 | 3 | 1 | 56 | Arcus lipoides | M2 | ½ 9.0 mm |
| 7a | 87 | M | 2117 | 10 | 1 | 40 | Arcus lipoides | M2 | ½ 9.0 mm |
| Corneal tissue in culture FFPE fixed samples | |||||||||
| 5b | As in 5a | ½ 9.0 mm | |||||||
| 6b | As in 6a | ½ 9.0 mm | |||||||
| 7b | As in 7a | ½ 9.0 mm | |||||||
| Healthy corneal samples (FFPE fixed) | |||||||||
| 8 | 55 | M | Not measured | Not measured | Fixed directly after enucleation | NA | Uveal melanoma | No TC contact | 9.0 mm |
| 9 | 81 | M | Not measured | Not measured | Fixed directly after enucleation | NA | Uveal melanoma | No TC contact | 9.0 mm |
| 10 | 73 | F | Not measured | Not measured | Fixed directly after enucleation | NA | Uveal melanoma | No TC contact | 9.0 mm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolf, J.; Kammrath Betancor, P.; Maier, P.; Heinzelmann, S.U.; Jiang, J.; Lange, C.; Reinhard, T.; Schlunck, G.; Lapp, T. Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas. Int. J. Mol. Sci. 2022, 23, 14507. https://doi.org/10.3390/ijms232314507
Wolf J, Kammrath Betancor P, Maier P, Heinzelmann SU, Jiang J, Lange C, Reinhard T, Schlunck G, Lapp T. Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas. International Journal of Molecular Sciences. 2022; 23(23):14507. https://doi.org/10.3390/ijms232314507
Chicago/Turabian StyleWolf, Julian, Paola Kammrath Betancor, Philip Maier, Sonja Ute Heinzelmann, Jana Jiang, Clemens Lange, Thomas Reinhard, Günther Schlunck, and Thabo Lapp. 2022. "Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas" International Journal of Molecular Sciences 23, no. 23: 14507. https://doi.org/10.3390/ijms232314507
APA StyleWolf, J., Kammrath Betancor, P., Maier, P., Heinzelmann, S. U., Jiang, J., Lange, C., Reinhard, T., Schlunck, G., & Lapp, T. (2022). Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas. International Journal of Molecular Sciences, 23(23), 14507. https://doi.org/10.3390/ijms232314507





