BDE-47, -99, -209 and Their Ternary Mixture Disrupt Glucose and Lipid Metabolism of Hepg2 Cells at Dietary Relevant Concentrations: Mechanistic Insight through Integrated Transcriptomics and Proteomics Analysis
Abstract
1. Introduction
2. Results
2.1. Genes Differentially Expressed by PBDE Treatment
2.2. Proteins Differentially Expressed by PBDE Treatment
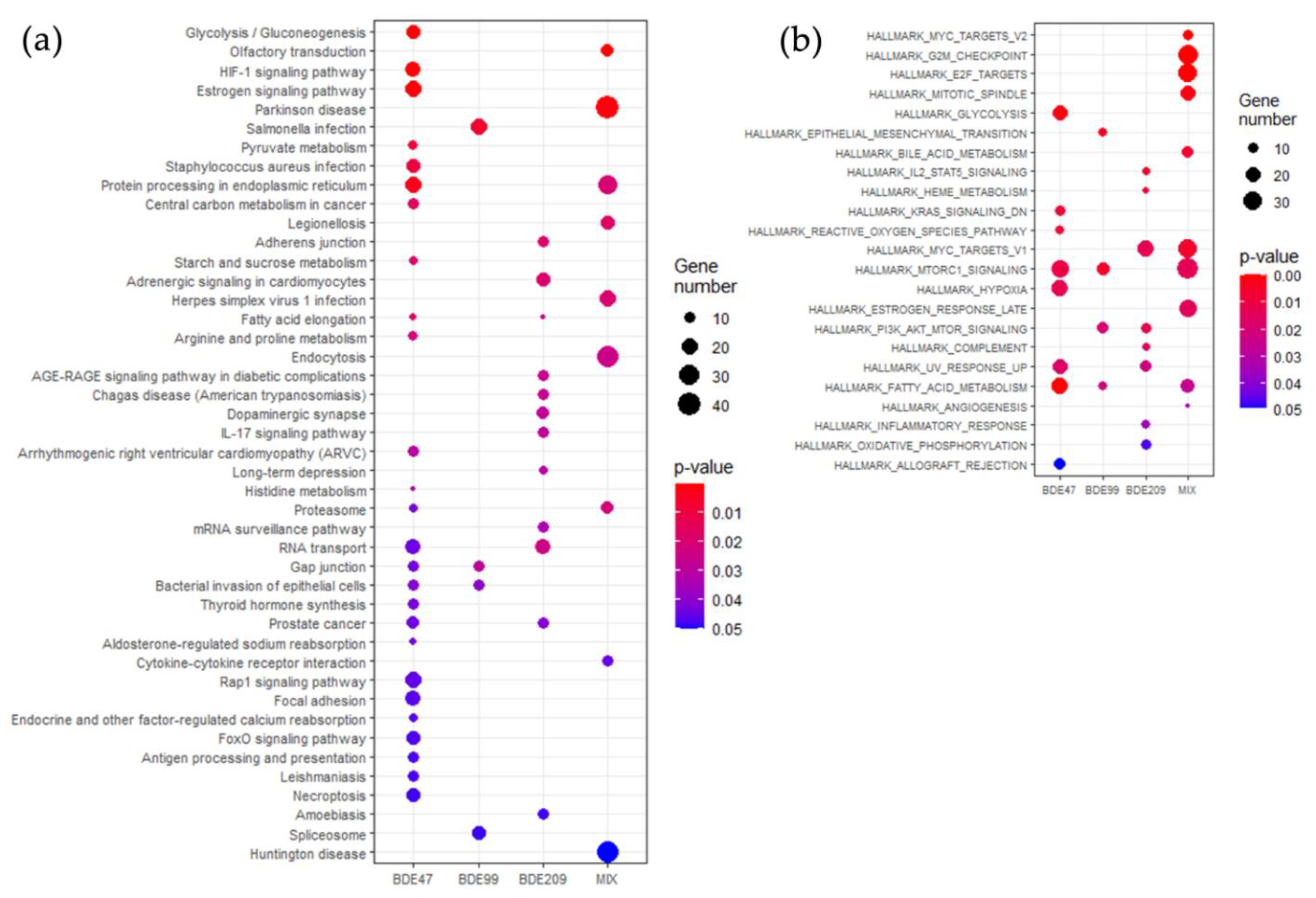
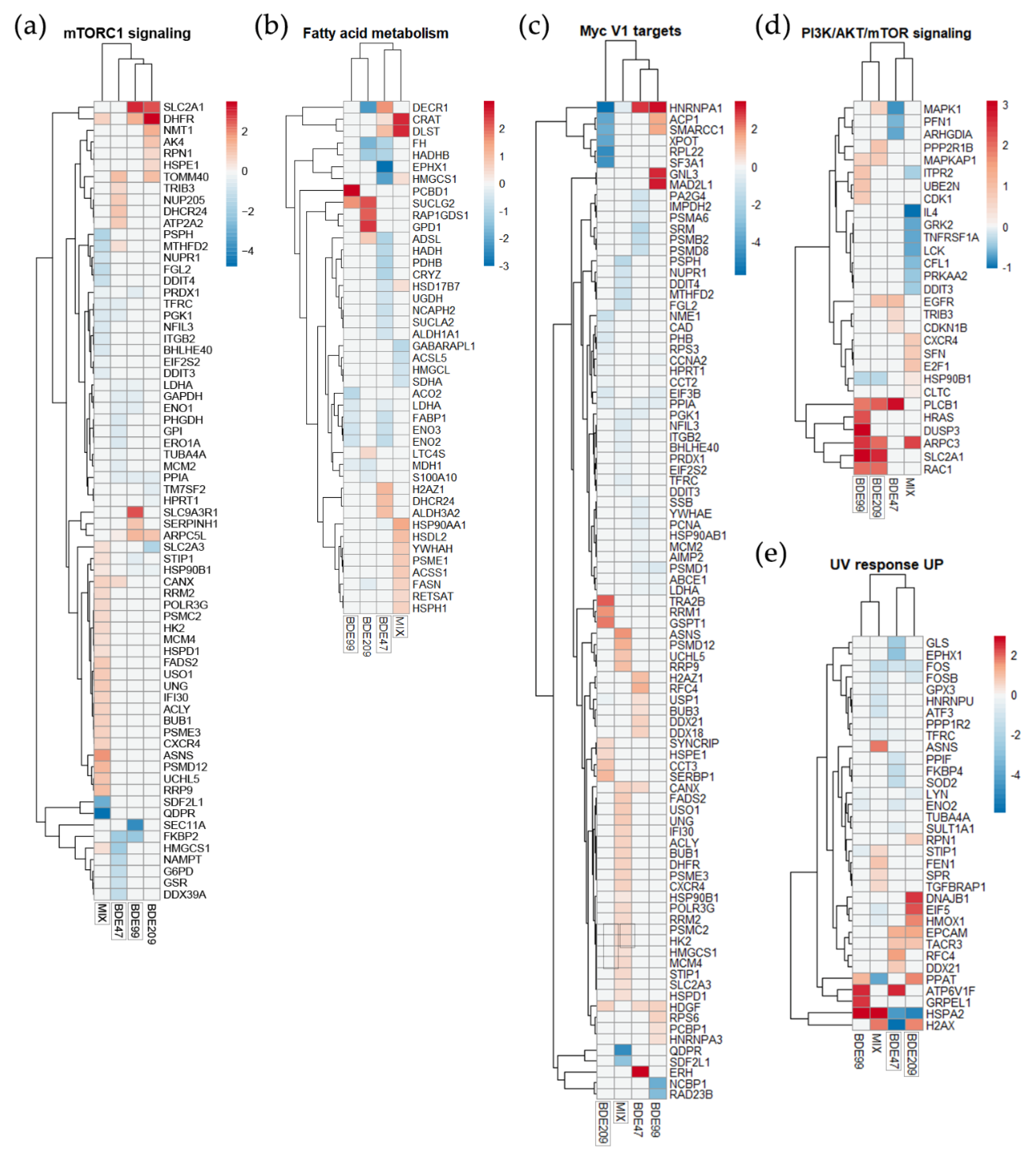
2.3. KEGG Pathways and GSEA Hallmarks Enriched by PBDEs
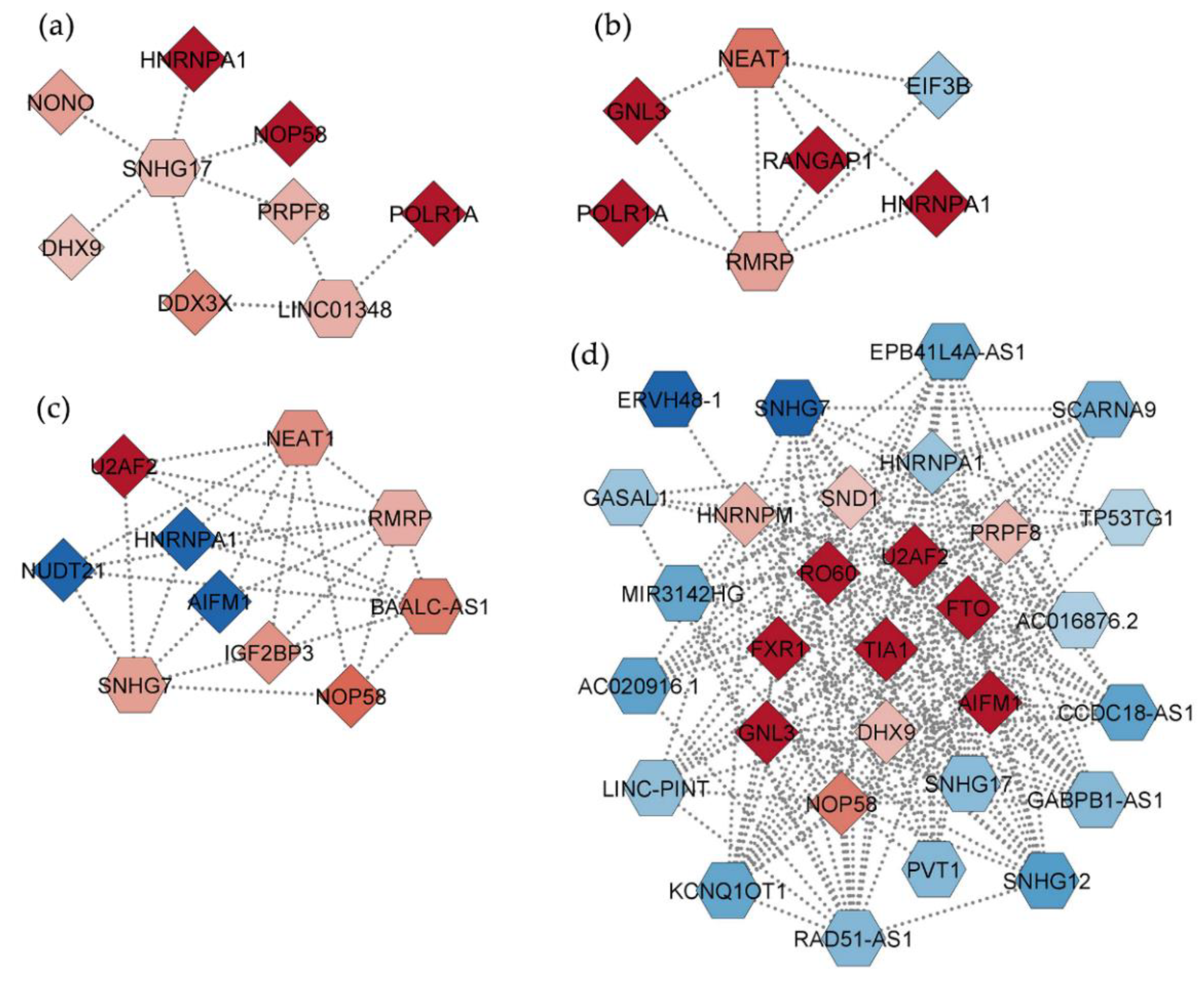
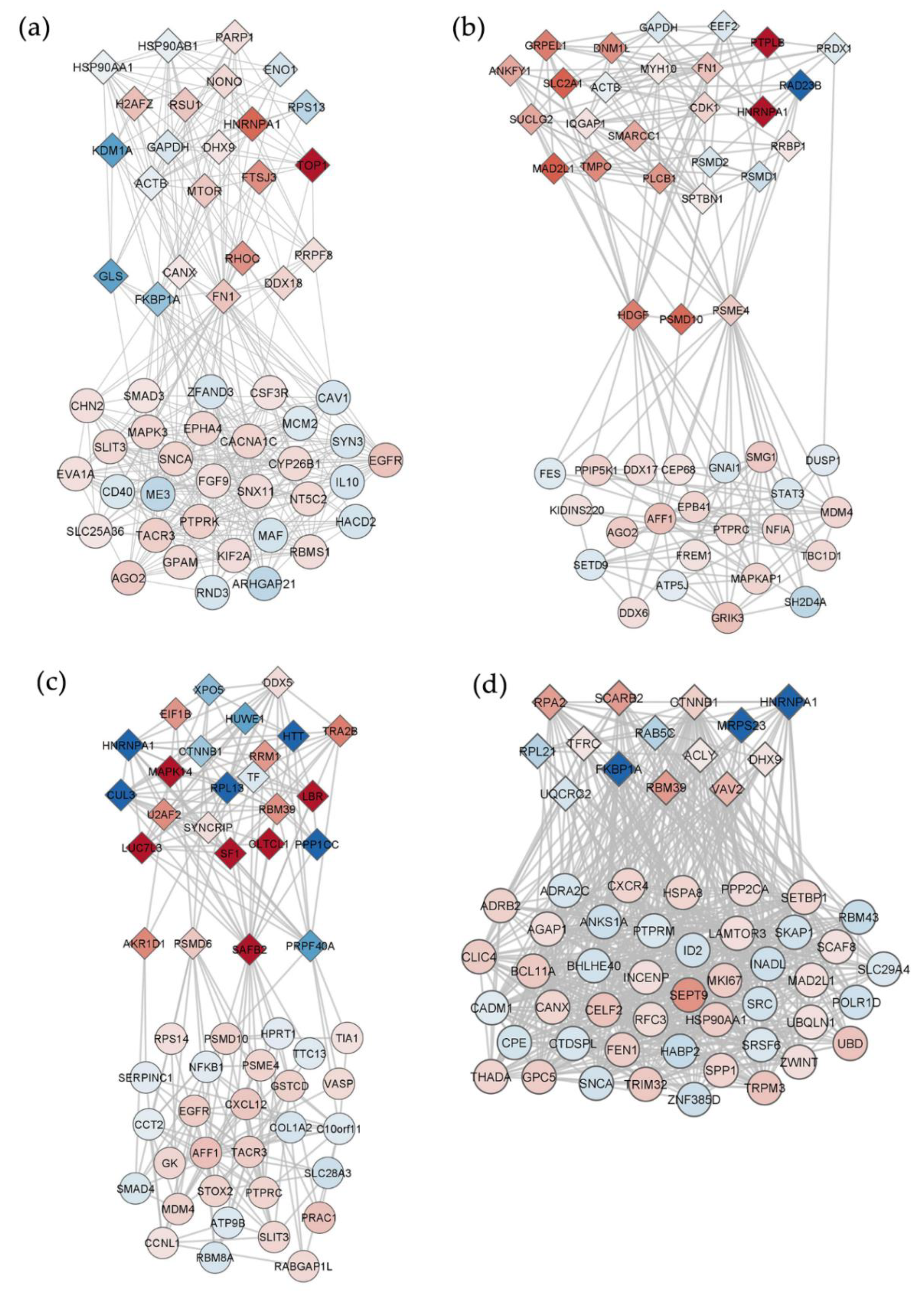
2.4. Interaction Networks of lncRNAs Deregulated by PBDEs
2.5. Interaction Networks among DEGs and DEPs and Relevant Hubs
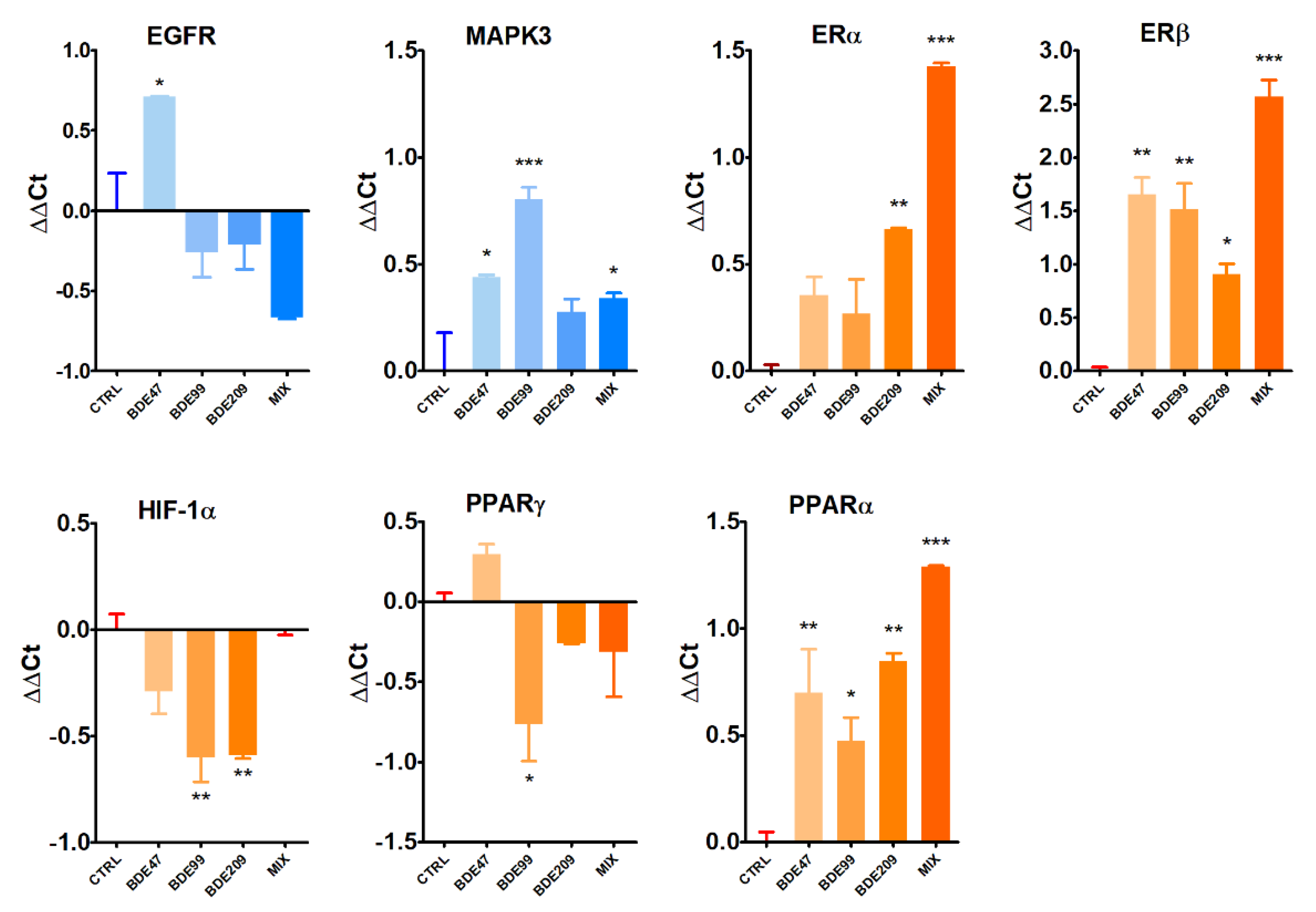
2.6. Real-Time PCR of Selected Markers
2.7. Glucose Secretion
2.8. Oil Red Staining of Intracellular Lipids
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Treatment
4.3. Transcriptomic Analysis
4.4. Proteomic Analysis
4.5. LncRNA Interaction Analysis
4.6. Functional Analysis
4.7. Interaction Network Analysis
4.8. Real-Time PCR
4.9. Glucose Assay
4.10. Oil Red O Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- European Parliament. Directive 2003/11/EC of the European Parliament and of the Council Amending for the 24th time Council Directive 76/769/EEC Relating to Restrictions on the Marketing and use of Certain Dangerous Substances and Preparations (Pentabromodiphenyl Ether and Octabrodiphenyl Ether). 2003; Volume 42, pp. 45–46. Available online: https://op.europa.eu/en/publication-detail/-/publication/0a566c4b-75ab-459b-8546-dbae21493f1b/language-en (accessed on 7 October 2022).
- ECJ. Judgement of the European Court of Justice in Case C-14/06 Concerning ‘DecaBDE’. Off. J. Eur. Union 2008, C116/2. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A62006CJ0014 (accessed on 7 October 2022).
- EFSA. Scientific Opinion on Polybrominated Diphenyl Ethers (PBDEs) in Food. EFSA J. 2011, 9, 2156. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. An Exposure Assessment of Polybrominated Diphenyl Ethers (PBDE). Available online: https://cfpub.epa.gov/ncea/risk/recordisplay.cfm?deid=210404 (accessed on 7 October 2022).
- Malliari, E.; Kalantzi, O. Children’s Exposure to Brominated Flame Retardants in Indoor Environments—A Review. Environ. Int. 2017, 108, 146–169. [Google Scholar] [CrossRef] [PubMed]
- Fromme, H.; Becher, G.; Hilger, B.; Völkel, W. Brominated Flame retardants–Exposure and Risk Assessment for the General Population. Int. J. Hyg. Environ. Health 2016, 219, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, L.E.; Hansen, P.W.; Mizrak, S.; Hansen, H.K.; Mørck, T.A.; Nielsen, F.; Siersma, V.; Mathiesen, L. Biomonitoring of Danish School Children and Mothers Including Biomarkers of PBDE and Glyphosate. Rev. Environ. Health 2017, 32, 279–290. [Google Scholar] [CrossRef]
- Lunder, S.; Hovander, L.; Athanassiadis, I.; Bergman, Å. Significantly Higher Polybrominated Diphenyl Ether Levels in Young US Children than in their Mothers. Environ. Sci. Technol. 2010, 44, 5256–5262. [Google Scholar] [CrossRef]
- Vuong, A.M.; Yolton, K.; Xie, C.; Webster, G.M.; Sjödin, A.; Braun, J.M.; Dietrich, K.N.; Lanphear, B.P.; Chen, A. Childhood Polybrominated Diphenyl Ether (PBDE) Exposure and Neurobehavior in Children at 8 Years. Environ. Res. 2017, 158, 677–684. [Google Scholar] [CrossRef]
- Drobná, B.; Fabišiková, A.; Čonka, K.; Gago, F.; Oravcová, P.; Wimmerová, S.; Feiler, M.O.; Šovčíková, E. PBDE Serum Concentration and Preschool Maturity of Children from Slovakia. Chemosphere 2019, 233, 387–395. [Google Scholar] [CrossRef]
- Cowell, W.J.; Margolis, A.; Rauh, V.A.; Sjödin, A.; Jones, R.; Wang, Y.; Garcia, W.; Perera, F.; Wang, S.; Herbstman, J.B. Associations between Prenatal and Childhood PBDE Exposure and Early Adolescent Visual, Verbal and Working Memory. Environ. Int. 2018, 118, 9–16. [Google Scholar] [CrossRef]
- Hamers, T.; Kamstra, J.H.; Sonneveld, E.; Murk, A.J.; Kester, M.H.; Andersson, P.L.; Legler, J.; Brouwer, A. In Vitro Profiling of the Endocrine-Disrupting Potency of Brominated Flame Retardants. Toxicol. Sci. 2006, 92, 157–173. [Google Scholar] [CrossRef]
- Legler, J. New Insights into the Endocrine Disrupting Effects of Brominated Flame Retardants. Chemosphere 2008, 73, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Meerts, I.A.; Van Zanden, J.J.; Luijks, E.A.; van Leeuwen-Bol, I.; Marsh, G.; Jakobsson, E.; Bergman, Å.; Brouwer, A. Potent Competitive Interactions of some Brominated Flame Retardants and Related Compounds with Human Transthyretin in Vitro. Toxicol. Sci. 2000, 56, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Chi, Q.; Zhang, W.; Wang, L.; Huang, J.; Yuan, M.; Xiao, H.; Wang, X. Evaluation of Structurally Different Brominated Flame Retardants Interacting with the Transthyretin and their Toxicity on HepG2 Cells. Chemosphere 2020, 246, 125749. [Google Scholar] [CrossRef] [PubMed]
- Prezioso, G.; Giannini, C.; Chiarelli, F. Effect of Thyroid Hormones on Neurons and Neurodevelopment. Horm. Res. Paediatr. 2018, 90, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Krassas, G.; Poppe, K.; Glinoer, D. Thyroid Function and Human Reproductive Health. Endocr. Rev. 2010, 31, 702–755. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.A.; Singh, B.K.; Yen, P.M. Direct Effects of Thyroid Hormones on Hepatic Lipid Metabolism. Nat. Rev. Endocrinol. 2018, 14, 259–269. [Google Scholar] [CrossRef]
- Oulhote, Y.; Chevrier, J.; Bouchard, M.F. Exposure to Polybrominated Diphenyl Ethers (PBDEs) and Hypothyroidism in Canadian Women. J. Clin. Endocrinol. Metab. 2016, 101, 590–598. [Google Scholar] [CrossRef]
- Jacobson, M.H.; Barr, D.B.; Marcus, M.; Muir, A.B.; Lyles, R.H.; Howards, P.P.; Pardo, L.; Darrow, L.A. Serum Polybrominated Diphenyl Ether Concentrations and Thyroid Function in Young Children. Environ. Res. 2016, 149, 222–230. [Google Scholar] [CrossRef]
- Abdelouahab, N.; Ainmelk, Y.; Takser, L. Polybrominated Diphenyl Ethers and Sperm Quality. Reprod. Toxicol. 2011, 31, 546–550. [Google Scholar] [CrossRef]
- Peltier, M.R.; Fassett, M.J.; Arita, Y.; Chiu, V.Y.; Shi, J.M.; Takhar, H.S.; Mahfuz, A.; Garcia, G.S.; Menon, R.; Getahun, D. Women with High Plasma Levels of PBDE-47 are at Increased Risk of Preterm Birth. J. Perinat. Med. 2021, 49, 439–447. [Google Scholar] [CrossRef]
- Boutot, M.E.; Whitcomb, B.W.; Abdelouahab, N.; Baccarelli, A.A.; Boivin, A.; Caku, A.; Gillet, V.; Martinez, G.; Pasquier, J.; Zhu, J. In Utero Exposure to Persistent Organic Pollutants and Childhood Lipid Levels. Metabolites 2021, 11, 657. [Google Scholar] [CrossRef] [PubMed]
- Ongono, J.S.; Dow, C.; Gambaretti, J.; Severi, G.; Boutron-Ruault, M.; Bonnet, F.; Fagherazzi, G.; Mancini, F.R. Dietary Exposure to Brominated Flame Retardants and Risk of Type 2 Diabetes in the French E3N Cohort. Environ. Int. 2019, 123, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, S.; Liu, L.; Wang, L.; Xiao, X.; Sun, Z.; Wang, X.; Wang, C.; Wang, M.; Li, L. Environmental Exposure to BDE47 is Associated with Increased Diabetes Prevalence: Evidence from Community-Based Case-Control Studies and an Animal Experiment. Sci. Rep. 2016, 6, 27854. [Google Scholar] [CrossRef] [PubMed]
- González-Casanova, J.E.; Pertuz-Cruz, S.L.; Caicedo-Ortega, N.H.; Rojas-Gomez, D.M. Adipogenesis Regulation and Endocrine Disruptors: Emerging Insights in Obesity. BioMed. Res. Int. 2020, 2020, 7453786. [Google Scholar] [CrossRef] [PubMed]
- Aaseth, J.; Javorac, D.; Djordjevic, A.B.; Bulat, Z.; Skalny, A.V.; Zaitseva, I.P.; Aschner, M.; Tinkov, A.A. The Role of Persistent Organic Pollutants in Obesity: A Review of Laboratory and Epidemiological Studies. Toxics 2022, 10, 65. [Google Scholar] [CrossRef]
- Tait, S.; Perugini, M.; La Rocca, C. Relative Toxicological Ranking of Eight Polybrominated Diphenyl Ether Congeners using Cytotoxicity, Chemical Properties and Exposure Data. Food Chem. Toxicol. 2017, 108, 74–84. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Zhu, Y.; Jing, L.; Li, X.; Zheng, D.; Zhou, G.; Zhang, Y.; Sang, Y.; Shi, Z.; Sun, Z.; Zhou, X. Decabromodiphenyl Ether Disturbs Hepatic Glycolipid Metabolism by Regulating the PI3K/AKT/GLUT4 and mTOR/PPARγ/RXRα Pathway in Mice and L02 Cells. Sci. Total Environ. 2021, 763, 142936. [Google Scholar] [CrossRef]
- Alimu, A.; Abudureman, H.; Wang, Y.Z.; Li, M.Y.; Wang, J.S.; Liu, Z.L. Decabromodiphenyl Ether Causes Insulin Resistance and Glucose and Lipid Metabolism Disorders in Mice. World J. Diabetes 2021, 12, 1267–1281. [Google Scholar] [CrossRef]
- Suvorov, A.; Battista, M.; Takser, L. Perinatal Exposure to Low-Dose 2, 2′, 4, 4′-Tetrabromodiphenyl Ether Affects Growth in Rat Offspring: What is the Role of IGF-1? Toxicology 2009, 260, 126–131. [Google Scholar] [CrossRef]
- Khalil, A.; Cevik, S.E.; Hung, S.; Kolla, S.; Roy, M.A.; Suvorov, A. Developmental 2, 2′, 4, 4′-Tetrabromodiphenyl Exposure to Ether Permanently Alters Blood-Liver Balance of Lipids in Male Mice. Endocr. Disrupters Metab. 2020. [Google Scholar] [CrossRef] [PubMed]
- Suvorov, A.; Takser, L. Global Gene Expression Analysis in the Livers of Rat Offspring Perinatally Exposed to Low Doses of 2,2′,4,4′-Tetrabromodiphenyl Ether. Environ. Health Perspect. 2010, 118, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Liang, S.; Yin, N.; Faiola, F. Establishment of a Human Embryonic Stem Cell-Based Liver Differentiation Model for Hepatotoxicity Evaluations. Ecotoxicol. Environ. Saf. 2019, 174, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Li, C.Y.; Kelly, E.J.; Sheppard, L.; Cui, J.Y. Transcriptomic Profiling of PBDE-Exposed HepaRG Cells Unveils Critical lncRNA-PCG Pairs Involved in Intermediary Metabolism. PLoS ONE 2020, 15, e0224644. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; Parker, M.; Mpanga, R.; Cevik, S.E.; Thorburn, C.; Suvorov, A. Developmental Exposure to 2, 2′, 4, 4′–tetrabromodiphenyl Ether Induces Long-Lasting Changes in Liver Metabolism in Male Mice. J. Endocr. Soc. 2017, 1, 323–344. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its Impact on Gene Expression at a Glance. J. Cell. Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef]
- Lee, M.; Puthumana, J.; Lee, S.; Kang, H.; Park, J.C.; Jeong, C.; Han, J.; Hwang, D.; Seo, J.S.; Park, H.G. BDE-47 Induces Oxidative Stress, Activates MAPK Signaling Pathway, and Elevates De Novo Lipogenesis in the Copepod Paracyclopina Nana. Aquat. Toxicol. 2016, 181, 104–112. [Google Scholar] [CrossRef]
- Mansouri, A.; Gattolliat, C.; Asselah, T. Mitochondrial Dysfunction and Signaling in Chronic Liver Diseases. Gastroenterology 2018, 155, 629–647. [Google Scholar] [CrossRef]
- Dowling, R.J.; Topisirovic, I.; Fonseca, B.D.; Sonenberg, N. Dissecting the Role of mTOR: Lessons from mTOR Inhibitors. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 433–439. [Google Scholar] [CrossRef]
- Ruiz, C.E.; Manuguerra, S.; Cuesta, A.; Esteban, M.A.; Santulli, A.; Messina, C.M. Sub-Lethal Doses of Polybrominated Diphenyl Ethers Affect some Biomarkers Involved in Energy Balance and Cell Cycle, Via Oxidative Stress in the Marine Fish Cell Line SAF-1. Aquat. Toxicol. 2019, 210, 1–10. [Google Scholar] [CrossRef]
- Manuguerra, S.; Espinosa Ruiz, C.; Santulli, A.; Messina, C.M. Sub-Lethal Doses of Polybrominated Diphenyl Ethers, in Vitro, Promote Oxidative Stress and Modulate Molecular Markers Related to Cell Cycle, Antioxidant Balance and Cellular Energy Management. Int. J. Environ. Res. Public Health 2019, 16, 588. [Google Scholar] [CrossRef] [PubMed]
- Karpeta, A.; Maniecka, A.; Gregoraszczuk, E.Ł. Different Mechanisms of Action of 2, 2′, 4, 4′-tetrabromodiphenyl Ether (BDE-47) and its Metabolites (5-OH-BDE-47 and 6-OH-BDE-47) on Cell Proliferation in OVCAR-3 Ovarian Cancer Cells and MCF-7 Breast Cancer Cells. J. Appl. Toxicol. 2016, 36, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, H.; Sun, H.; Shen, O.; Wang, X.; Lam, M.H.; Giesy, J.P.; Zhang, X.; Yu, H. Endocrine Effects of Methoxylated Brominated Diphenyl Ethers in Three in Vitro Models. Mar. Pollut. Bull. 2011, 62, 2356–2361. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging Functions of the EGFR in Cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]
- Pouysségur, J.; Lenormand, P. ERK1 and ERK2 Map Kinases: Specific Roles Or Functional Redundancy? Front. Cell Dev. Biol. 2016, 4, 53. [Google Scholar] [CrossRef]
- Levin, E.R. Bidirectional Signaling between the Estrogen Receptor and the Epidermal Growth Factor Receptor. Mol. Endocrinol. 2003, 17, 309–317. [Google Scholar] [CrossRef]
- Zhang, F.; Peng, L.; Huang, Y.; Lin, X.; Zhou, L.; Chen, J. Chronic BDE-47 Exposure Aggravates Malignant Phenotypes and Chemoresistance by Activating ERK through ERα and GPR30 in Endometrial Carcinoma. Front. Oncol. 2019, 9, 1079. [Google Scholar] [CrossRef]
- Luo, Y.; Lin, J.; Zhang, J.; Song, Z.; Zheng, D.; Chen, F.; Zhuang, X.; Li, A.; Liu, X. LncRNA SNHG17 Contributes to Proliferation, Migration, and Poor Prognosis of Hepatocellular Carcinoma. Can. J. Gastroenterol. Hepatol. 2021, 2021, 9990338. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, M.; Liu, Y.; Lyu, P.; Yeh, C.; Lin, K. LINC01348 Suppresses Hepatocellular Carcinoma Metastasis through Inhibition of SF3B3-Mediated EZH2 Pre-mRNA Splicing. Oncogene 2021, 40, 4675–4685. [Google Scholar] [CrossRef]
- Hongfeng, Z.; Andong, J.; Liwen, S.; Mingping, B.; Xiaowei, Y.; Mingyong, L.; Aimin, Y. lncRNA RMRP Knockdown Suppress Hepatocellular Carcinoma Biological Activities Via Regulation miRNA-206/TACRJ. Cell. Biochem. 2020, 121, 1690–1702. [Google Scholar] [CrossRef]
- Sun, Y.; Song, Y.; Liu, C.; Geng, J. LncRNA NEAT1-MicroRNA-140 Axis Exacerbates Nonalcoholic Fatty Liver through Interrupting AMPK/SREBP-1 Signaling. Biochem. Biophys. Res. Commun. 2019, 516, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Chen, X.; Zhang, F.; Zhao, M. RMRP Inhibition Prevents NAFLD Progression in Rats Via Regulating miR-206/PTPN1 Axis. Mamm. Genome 2022, 33, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Lin, Y.; Yu, Y.; Sun, D. LncRNA NEAT1/microRNA-129-5p/SOCS2 Axis Regulates Liver Fibrosis in Alcoholic Steatohepatitis. J. Transl. Med. 2020, 18, 445. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; He, M.; Chen, J.; Li, C.; Zhang, Q. Long non-coding RNA SNHG7 inhibits NLRP3-dependent pyroptosis by targeting the miR-34a/SIRT1 axis in liver cancer. Oncol. Lett. 2020, 20, 893–901. [Google Scholar] [CrossRef]
- Yang, X.; Sun, L.; Wang, L.; Yao, B.; Mo, H.; Yang, W. LncRNA SNHG7 Accelerates the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cells Via Regulating miR-122-5p and RPL. Biomed. Pharmacother. 2019, 118, 109386. [Google Scholar] [CrossRef]
- Liao, M.; Liao, W.; Xu, N.; Li, B.; Liu, F.; Zhang, S.; Wang, Y.; Wang, S.; Zhu, Y.; Chen, D. LncRNA EPB41L4A-AS1 Regulates Glycolysis and Glutaminolysis by Mediating Nucleolar Translocation of HDACE. BioMedicine 2019, 41, 200–213. [Google Scholar] [CrossRef]
- Li, C.; Wang, P.; Du, J.; Chen, J.; Liu, W.; Ye, K. LncRNA RAD51-AS1/miR-29b/C-3p/NDRG2 Crosstalk Repressed Proliferation, Invasion and Glycolysis of Colorectal Cancer. IUBMB Life 2021, 73, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Khatun, M.; Sur, S.; Steele, R.; Ray, R.; Ray, R.B. Inhibition of Long Noncoding RNA Linc-Pint by Hepatitis C Virus in Infected Hepatocytes Enhances Lipogenesis. Hepatology 2021, 74, 41–54. [Google Scholar] [CrossRef]
- Hanamura, A.; Cáceres, J.F.; Mayeda, A.; Franza, B.R., Jr.; Krainer, A.R. Regulated Tissue-Specific Expression of Antagonistic Pre-mRNA Splicing Factors. RNA 1998, 4, 430–444. [Google Scholar]
- Miyaso, H.; Nakamura, N.; Naito, M.; Hirai, S.; Matsuno, Y.; Itoh, M.; Mori, C. Early Postnatal Exposure to a Low Dose of Decabromodiphenyl Ether Affects Expression of Androgen and Thyroid Hormone Receptor-Alpha and its Splicing Variants in Mouse Sertoli Cells. PLoS ONE 2014, 9, e114487. [Google Scholar] [CrossRef]
- Gui, W.; Zhu, W.F.; Zhu, Y.; Tang, S.; Zheng, F.; Yin, X.; Lin, X.; Li, H. LncRNAH19 Improves Insulin Resistance in Skeletal Muscle by Regulating Heterogeneous Nuclear Ribonucleoprotein A1. Cell Commun. Signal. 2020, 18, 173. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Wang, Y.; Chen, X.; Shi, Y.; Wang, J.; Zhang, J.; Qian, J.; Li, R.; Tao, T.; Wei, W. PKM2 Promotes Glucose Metabolism and Cell Growth in Gliomas through a Mechanism Involving a Let-7a/C-Myc/hnRNPA1 Feedback Loop. Oncotarget 2015, 6, 13006–13018. [Google Scholar] [CrossRef] [PubMed]
- Chettouh, H.; Fartoux, L.; Aoudjehane, L.; Wendum, D.; Clapéron, A.; Chrétien, Y.; Rey, C.; Scatton, O.; Soubrane, O.; Conti, F. Mitogenic Insulin Receptor-A is Overexpressed in Human Hepatocellular Carcinoma due to EGFR-Mediated Dysregulation of RNA Splicing Factors. Cancer Res. 2013, 73, 3974–3986. [Google Scholar] [CrossRef] [PubMed]
- Damiano, F.; Rochira, A.; Tocci, R.; Alemanno, S.; Gnoni, A.; Siculella, L. hnRNP A1 Mediates the Activation of the IRES-Dependent SREBP-1a mRNA Translation in Response to Endoplasmic Reticulum Stress. Biochem. J. 2013, 449, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Larsson, P.; Engqvist, H.; Biermann, J.; Werner Rönnerman, E.; Forssell-Aronsson, E.; Kovács, A.; Karlsson, P.; Helou, K.; Parris, T.Z. Optimization of Cell Viability Assays to Improve Replicability and Reproducibility of Cancer Drug Sensitivity Screens. Sci. Rep. 2020, 10, 5798. [Google Scholar] [CrossRef]
- Tait, S.; La Rocca, C.; Mantovani, A. Exposure of Human Fetal Penile Cells to Different PCB Mixtures: Transcriptome Analysis Points to Diverse Modes of Interference on External Genitalia Programming. Reprod. Toxicol. 2011, 32, 1–14. [Google Scholar] [CrossRef]
- Tait, S.; Tassinari, R.; Maranghi, F.; Mantovani, A. Bisphenol A Affects Placental Layers Morphology and Angiogenesis during Early Pregnancy Phase in Mice. J. Appl. Toxicol. 2015, 35, 1278–1291. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Anonymous; Springer: Berlin/Heidelberg, Germany, 2005; pp. 397–420. [Google Scholar]
- R Core Team. The Comprehensive R Archive Network; R Core Team: Vienna, Austria, 2019; Available online: https://cran.r-project.org/ (accessed on 7 October 2022).
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package biomaRt. Nat. Protoc. 2009, 4, 1184. [Google Scholar] [CrossRef]
- Oliveros, J. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams 2007. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. 2016. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 7 October 2022).
- Snevechenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins Silver-Stained Polyacrilamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-Seq Data using Factor Analysis of Control Genes Or Samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA Interactome Repository with Increased Coverage and Annotation. Nucleic Acids Res. 2020, 48, D189–D197. [Google Scholar] [CrossRef]
- Li, J.; Liu, S.; Zhou, H.; Qu, L.; Yang, J. starBase V2. 0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant graphics for data analysis. Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
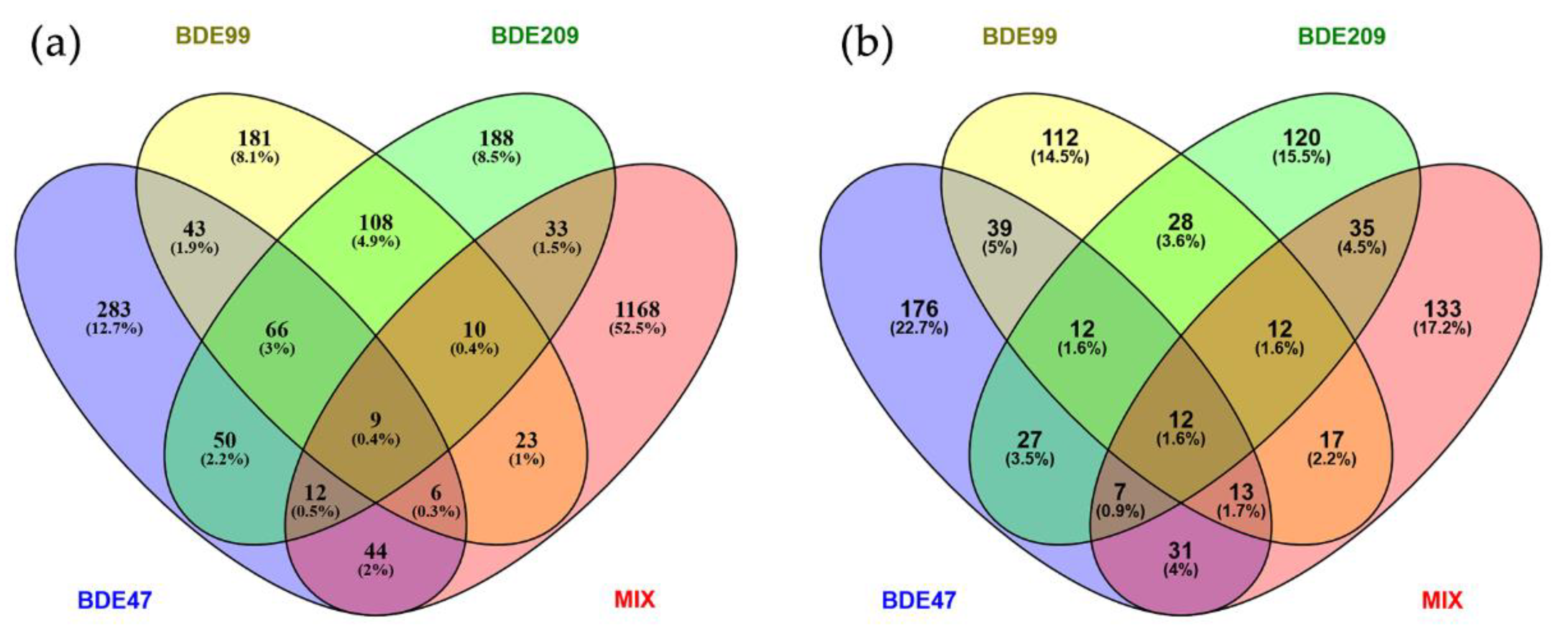

| BDE-47 | BDE-99 | BDE-209 | MIX | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DEGs | up | down | tot | up | down | tot | up | down | tot | up | down | tot |
| 309 | 204 | 513 | 324 | 122 | 446 | 330 | 146 | 476 | 446 | 860 | 1306 | |
| DEPs | up | down | tot | up | down | tot | up | down | tot | up | down | tot |
| 169 | 148 | 317 | 190 | 55 | 245 | 154 | 99 | 253 | 203 | 58 | 261 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casella, M.; Lori, G.; Coppola, L.; La Rocca, C.; Tait, S. BDE-47, -99, -209 and Their Ternary Mixture Disrupt Glucose and Lipid Metabolism of Hepg2 Cells at Dietary Relevant Concentrations: Mechanistic Insight through Integrated Transcriptomics and Proteomics Analysis. Int. J. Mol. Sci. 2022, 23, 14465. https://doi.org/10.3390/ijms232214465
Casella M, Lori G, Coppola L, La Rocca C, Tait S. BDE-47, -99, -209 and Their Ternary Mixture Disrupt Glucose and Lipid Metabolism of Hepg2 Cells at Dietary Relevant Concentrations: Mechanistic Insight through Integrated Transcriptomics and Proteomics Analysis. International Journal of Molecular Sciences. 2022; 23(22):14465. https://doi.org/10.3390/ijms232214465
Chicago/Turabian StyleCasella, Marialuisa, Gabriele Lori, Lucia Coppola, Cinzia La Rocca, and Sabrina Tait. 2022. "BDE-47, -99, -209 and Their Ternary Mixture Disrupt Glucose and Lipid Metabolism of Hepg2 Cells at Dietary Relevant Concentrations: Mechanistic Insight through Integrated Transcriptomics and Proteomics Analysis" International Journal of Molecular Sciences 23, no. 22: 14465. https://doi.org/10.3390/ijms232214465
APA StyleCasella, M., Lori, G., Coppola, L., La Rocca, C., & Tait, S. (2022). BDE-47, -99, -209 and Their Ternary Mixture Disrupt Glucose and Lipid Metabolism of Hepg2 Cells at Dietary Relevant Concentrations: Mechanistic Insight through Integrated Transcriptomics and Proteomics Analysis. International Journal of Molecular Sciences, 23(22), 14465. https://doi.org/10.3390/ijms232214465







