Evaluation of Fusarium Head Blight Resistance Effects by Haplotype-Based Genome-Wide Association Study in Winter Wheat Lines Derived by Marker Backcrossing Approach
Abstract
1. Introduction
2. Results
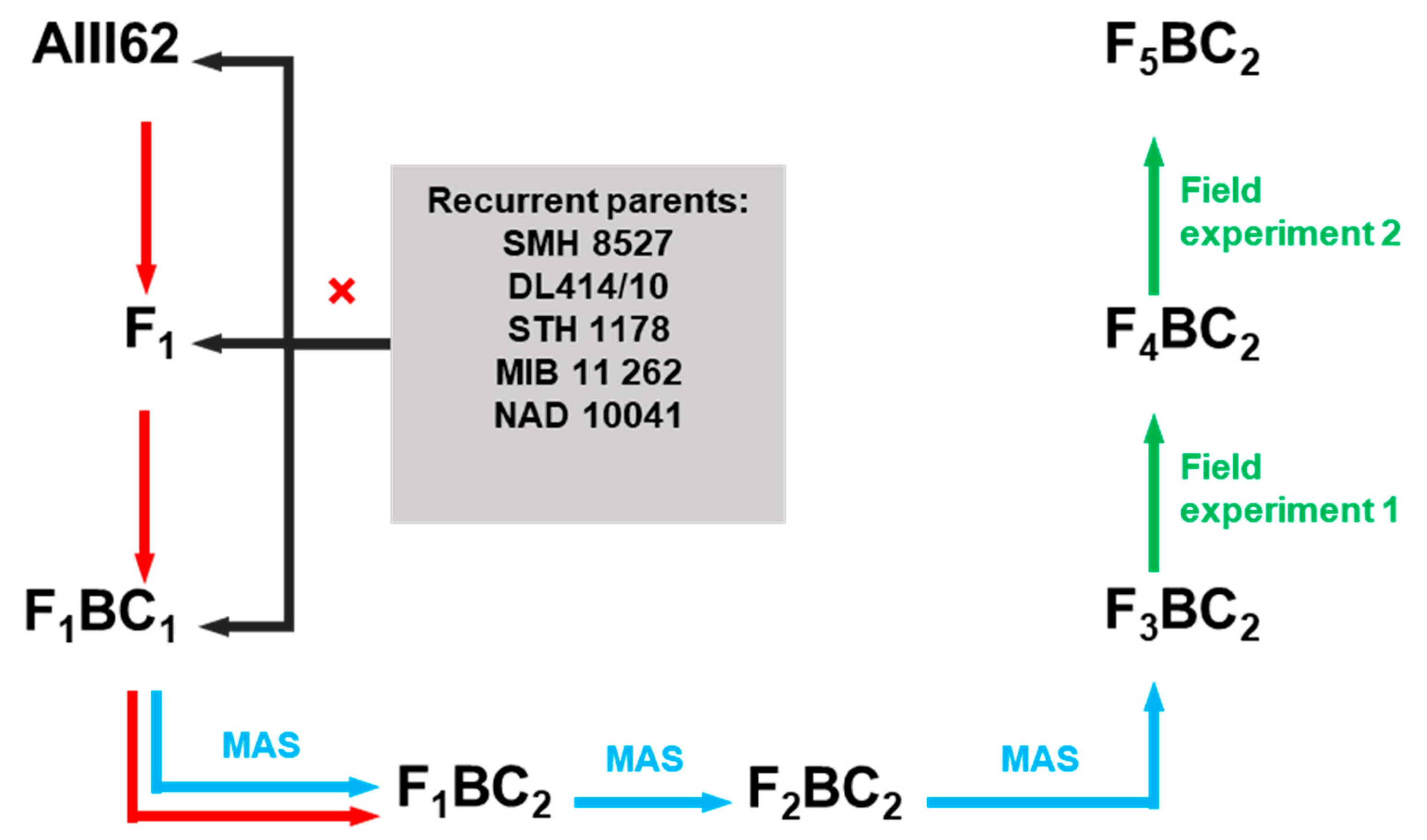
2.1. Fusarium Head Blight Response Lines
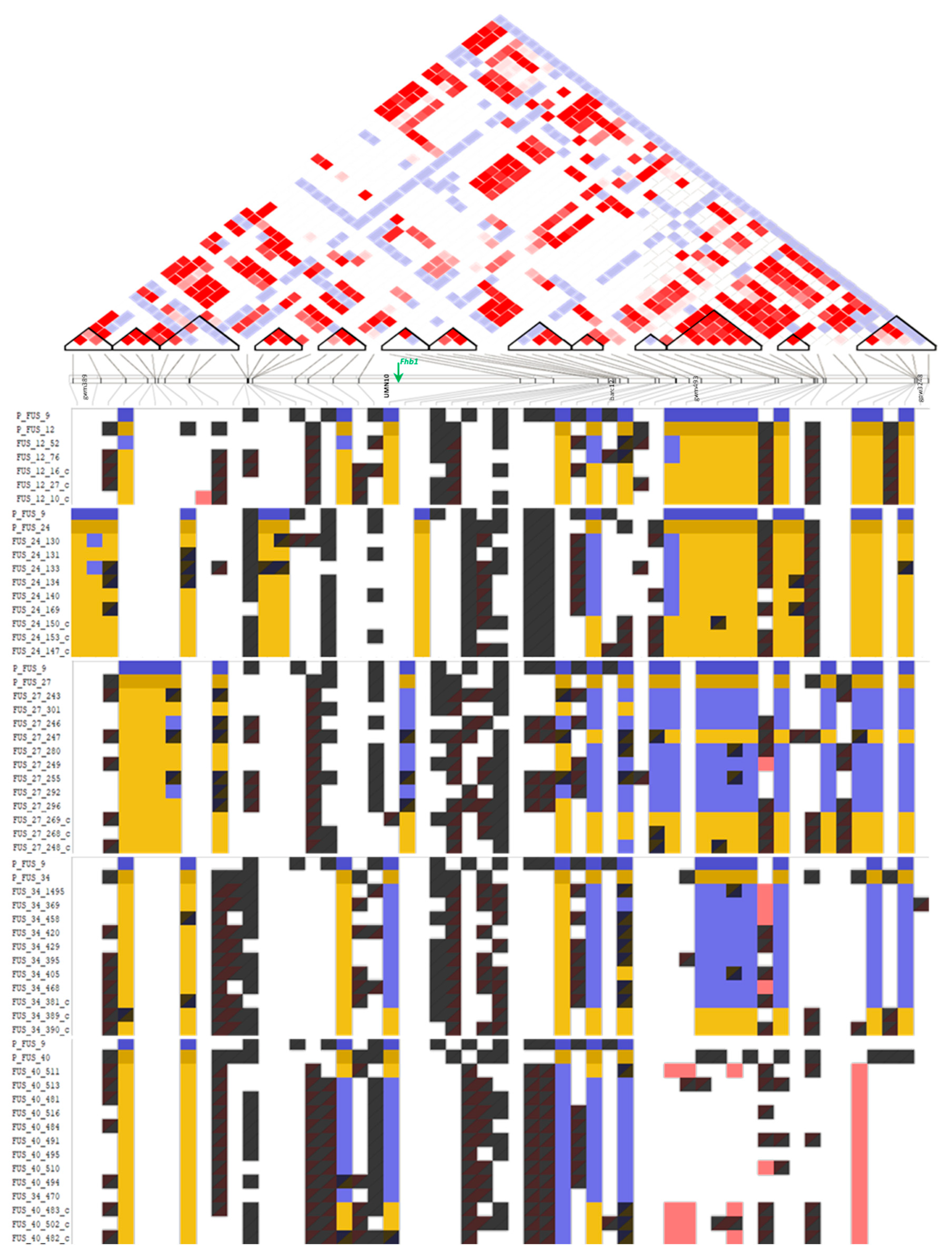
2.2. SSR Marker-Based Selection
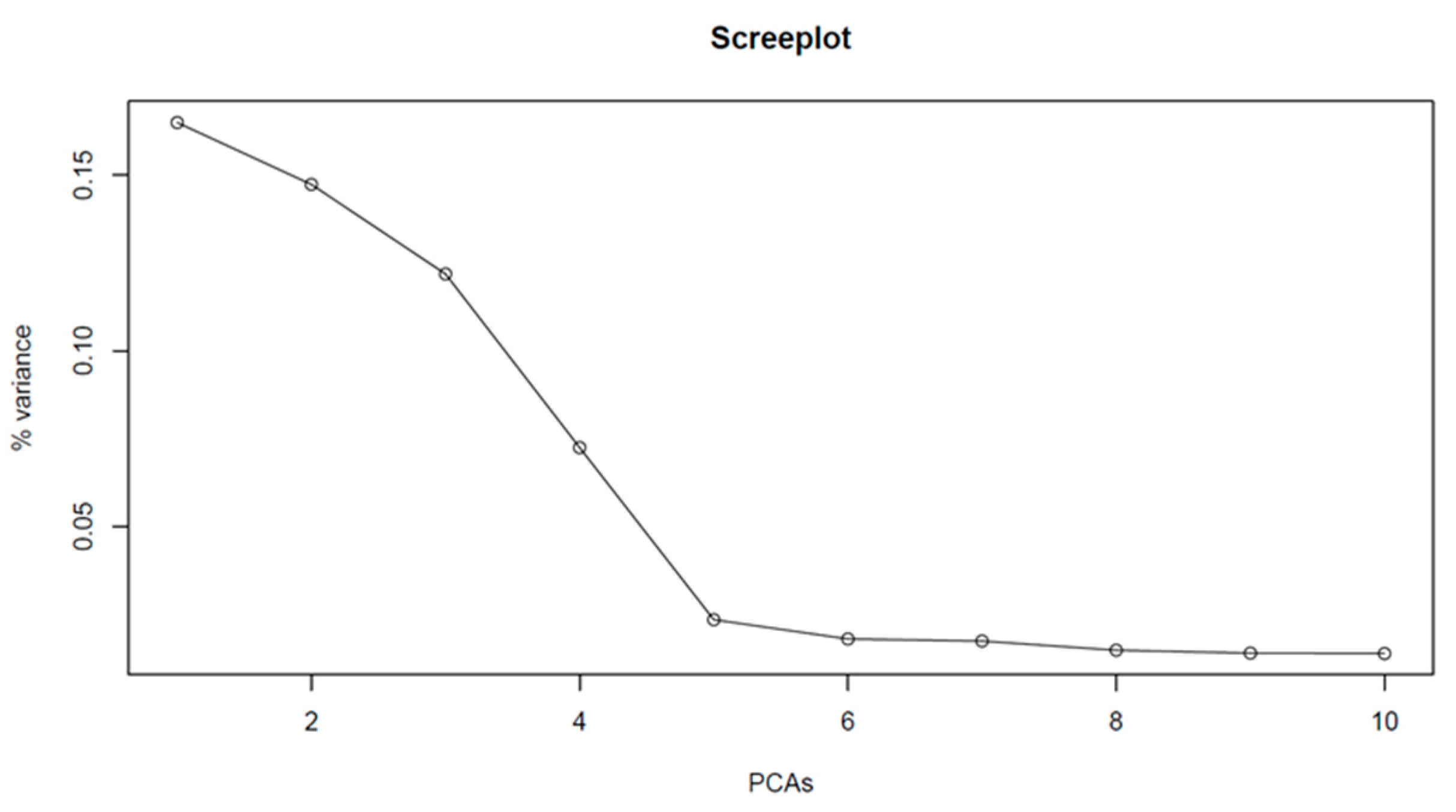
2.3. Population Structure Analysis and Haploblock Calling
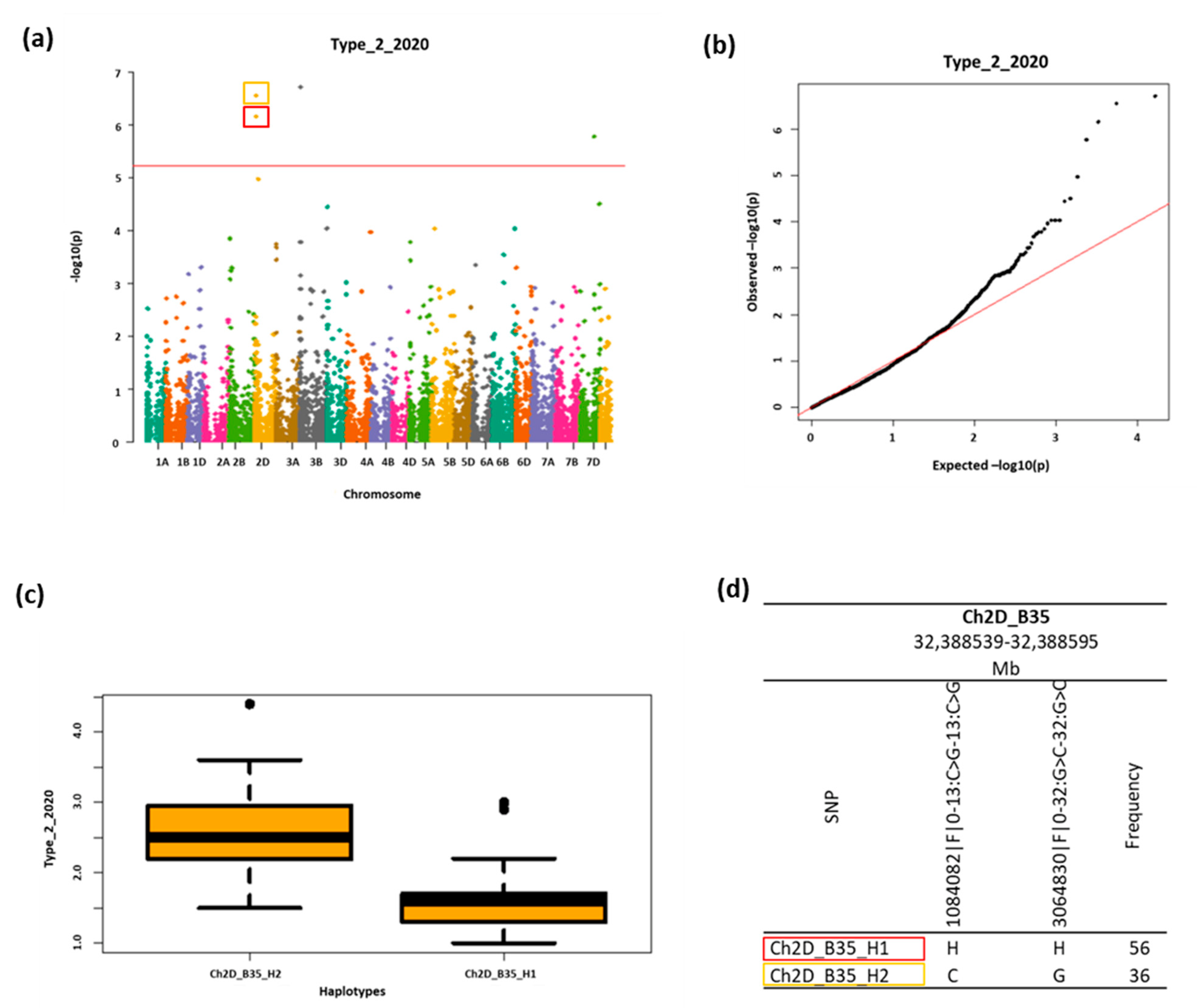
2.4. GWAS Analysis
2.5. Fhb1 Locus
2.6. Association Weight Matrix Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
- Winter wheat cultivars: Belenus (France), Belissa (Poland), Błyskawica (Poland), Ceres—durum (Poland), Euforia (Poland), Opcja (Poland), Plejada (Poland), Reduta (Poland), Sfera (Poland), Tobak (Germany), Wilejka (Poland) [54];
- Wheat resistant checks, wheat lines carrying the Fhb1 gene: UNG 136.6.1.1 [Fhb1+] (Hungary), S 10 [Fhb1+] (Poland), S 30 [Fhb1+] (Poland), S 32 [Fhb1+] (Poland) and wheat breeding lines without the Fhb1 gene: 20,828 [Fhb1-] (Austria) and A40-19-1-2 (Austria) and cultivars Arina (Switzerland) and Fregata (Poland) [54];
- Wheat susceptible checks, Polish breeding lines: SMH 8694 (S) (Poland), SMH 8816 (S) (Poland), DL325/11/3 (S) (Poland), KBP 14 16 (S) (Poland) [55].
4.2. Fungal Material and Field Experiments
4.3. Statistical Analysis
4.4. Genetic Marker Selection
4.5. Haploblock Construction and GWAS Analysis
4.6. Association Weight Matrix Pipeline
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parry, D.W.; Jenkinson, P.; McLeod, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Kang, Z.; Buchenauer, H. Immunocytochemical localization of Fusarium toxins in infected wheat spikes by Fusarium culmorum. Physiol. Mol. Plant Pathol. 1999, 55, 275–288. [Google Scholar] [CrossRef]
- Pestka, J.J. Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin J. 2010, 3, 323–347. [Google Scholar] [CrossRef]
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef]
- Juroszek, P.; von Tiedemann, A. Linking plant disease models to climate change scenarios to project future risks of crop diseases: A review. J. Plant Dis. Prot. 2015, 122, 3–15. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Steiner, B.; Buerstmayr, H. Breeding for Fusarium head blight resistance in wheat—Progress and challenges. Plant Breed. 2020, 139, 429–454. [Google Scholar] [CrossRef]
- Shah, L.; Ali, A.; Zhu, Y.; Wang, S.; Si, H.; Ma, C. Wheat Resistance to Fusarium Head Blight and possibilities of its Improvement using Molecular Marker-Assisted Selection. Czech J. Plant Breed. 2017, 53, 47–54. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Wagner, C.; Nosenko, T.; Omony, J.; Steiner, B.; Nussbaumer, T.; Mayer, K.F.X.; Buerstmayr, H. Fusarium head blight resistance in European winter wheat: Insights from genome-wide transcriptome analysis. BMC Genom. 2021, 22, 470. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Christensen, J.J. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Mesterhazy, A. Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Mesterhazy, A.; Bartok, T.; Mirocha, C.G.; Komoroczy, R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999, 118, 97–110. [Google Scholar] [CrossRef]
- Ma, Z.; Xie, Q.; Li, G.; Jia, H.; Zhou, J.; Kong, Z.; Li, N.; Yuan, Y. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 2020, 133, 1541–1568. [Google Scholar] [CrossRef] [PubMed]
- Steiner, B.; Buerstmayr, M.; Michel, S.; Schweiger, W.; Lemmens, M.; Buerstmayr, H. Breeding strategies and advances in line selection for Fusarium head blight resistance in wheat. Trop Plant Pathol. 2017, 42, 165–174. [Google Scholar] [CrossRef]
- Anderson, J.A.; Stack, R.W.; Liu, S.; Waldron, B.L.; Fjeld, A.D.; Coyne, C.; Moreno-Sevilla, B.; Mitchel Fetch, J.; Song, Q.J.; Cregan, P.B.; et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor. Appl. Genet. 2001, 102, 1164–1168. [Google Scholar] [CrossRef]
- Anderson, J.A. Marker-assisted selection for fusarium head blight resistance in wheat. Int. J. Food Microbiol. 2007, 119, 51–53. [Google Scholar] [CrossRef]
- Pumphrey, M.O.; Bernardo, R.; Anderson, J.A. Validating the Fhb1 QTL for Fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop Sci. 2007, 47, 200–206. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, X.; Pumphrey, M.O.; Stack, R.W.; Gill, B.S.; Anderson, J.A. Complex microcolinearity among wheat, rice and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct. Integr. Genomics 2006, 6, 83–89. [Google Scholar] [CrossRef]
- Liu, S.; Pumphrey, M.; Gill, B.; Trick, H.; Zhang, J.; Dolezel, J.; Chalhoub, B.; Anderson, J. Toward positional cloning of FHB1, a major QTL for fusarium head blight resistance in wheat. Cereal Res. Commun. 2008, 36, 195–201. [Google Scholar] [CrossRef]
- Hao, C.Y.; Perretant, M.R.; Choulet, F.; Wang, L.F.; Paux, E.; Sourdille, P.; Zhang, X.Y.; Feuillet, C.; Balfourier, F. Genetic diversity and linkage disequilibrium studies on a 3.1-Mb genomic region of chromosome 3B in European and Asian bread wheat (Triticum aestivum L.) populations. Theor. Appl. Genet. 2010, 121, 1209–1225. [Google Scholar] [CrossRef]
- Hao, C.; Wang, Y.; Hou, J.; Feuillet, C.; Balfourier, F.; Zhang, X. Association mapping and haplotype analysis of a 3.1-Mb genomic region involved in fusarium head blight resistance on wheat chromosome 3BS. PLoS ONE 2012, 7, e46444. [Google Scholar] [CrossRef] [PubMed]
- Lagudah, E.S.; Krattinger, S.G. A new player contributing to durable Fusarium resistance. Nat. Genet. 2019, 51, 1070–1071. [Google Scholar] [CrossRef] [PubMed]
- Von der Ohe, C.; Ebmeyer, E.; Korzun, V.; Miedaner, T. Agronomic and quality performance of winter wheat backcross populations carrying non-adapted Fusarium head blight resistance QTL. Crop Sci. 2010, 50, 2283–2290. [Google Scholar] [CrossRef]
- Salameh, A.; Buerstmayr, M.; Steiner, B.; Neumayer, A.; Lemmens, M.; Buerstmayr, H. Effects of introgression of two QTL for Fusarium head blight resistance from Asian spring wheat by marker-assisted backcrossing into European winter wheat on Fusarium head blight resistance, yield and quality traits. Mol. Breed. 2011, 28, 485–494. [Google Scholar] [CrossRef]
- Suzuki, T.; Sato, M.; Takeuchi, T. Evaluation of the effects of five QTL regions on Fusarium head blight resistance and agronomic traits in spring wheat (Triticum aestivum L.). Breed. Sci. 2012, 62, 11–17. [Google Scholar] [CrossRef]
- Chrpova, J.; Sip, V.; Sedlacek, T.; Stockova, L.; Veskrna, O.; Horcicka, P. Effectiveness of Marker-based selection for Fusarium head blight resistance in spring wheat. Czech J. Genet. Plant Breed. 2011, 47, S123–S129. [Google Scholar] [CrossRef]
- Zwart, R.S.; Muylle, H.; Van Bockstaele, E.; Roldán-Ruiz, I. Evaluation of genetic diversity of Fusarium head blight resistance in European winter wheat. Theor. Appl. Genet. 2008, 117, 813–828. [Google Scholar] [CrossRef]
- Miedaner, T.; Wuerschum, T.; Maurer, H.P.; Korzun, V.; Ebmeyer, E.; Reif, J.C. Association mapping for Fusarium head blight resistancein European soft winter wheat. Mol. Breed. 2011, 28, 647–655. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Pozniak, C.J.; Clarke, J.M.; Clarke, F.R. Potential for detection of marker–trait associations in durum wheat using unbalanced, historical phenotypic datasets. Mol. Breed. 2012, 30, 1537–1550. [Google Scholar] [CrossRef]
- Wang, H.; Smith, K.P.; Combs, E.; Blake, T.; Horsley, R.D.; Muehlbauer, G.J. Effect of population size and unbalanced data sets on QTL detection using genome-wide association mapping in barley breeding germplasm. Theor. Appl. Genet. 2012, 124, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Kollers, S.; Rodemann, B.; Ling, J.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Plieske, J.; Kulosa, D.; Ganal, M.W.; et al. Whole genome association mapping of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). PLoS ONE 2013, 8, e57500. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, V.; Liu, G.; Zhao, Y.; Miedaner, T.; Longin, C.F.H.; Gowda, M.; Mette, M.F.; Reif, J.C. Genetic architecture is more complex for resistance to Septoria tritici blotch than to Fusarium head blight in Central European winter wheat. BMC Genom. 2015, 16, 430. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.P.; Brown, P.; Brown-Guedira, G.; Krill, A.M.; Thurber, C.; Merrill, K.R.; Foresman, B.J.; Kolbet, F.L. Genome-wide association mapping of Fusarium head blight resistance in wheat using genotyping-by-sequencing. Plant Genome 2016, 9, 1–14. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Anderson, J.A.; Zhang, J.; Zhao, W.; Wheeler, J.; Klassen, N.; See, D.R.; Dong, Y. Genome-wide association mapping of Fusarium head blight resistance in spring wheat lines developed in the Pacific Northwest and CIMMYT. Phytopathology 2017, 107, 1486–1495. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, L.; Zhang, W.; Yang, L.; Zhu, W.; Li, J.; Liu, Y.; Tong, H.; Fu, L.; Liu, J.; et al. Genome-Wide Association Analysis of Fusarium Head Blight Resistance in Chinese Elite Wheat Lines. Front. Plant Sci. 2020, 11, 206. [Google Scholar] [CrossRef]
- Waldron, B.L.; Moreno-Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP mapping of QTL for Fusarium head blight resistance in wheat. Crop Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Bai, G.; Kolb, F.L.; Shaner, G.; Domier, L.L. Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathology 1999, 89, 343–348. [Google Scholar] [CrossRef]
- Lemmens, M.; Koutnik, A.; Steiner, B.; Buerstmayr, H.; Berthiller, F.; Schuhmacher, R.; Maier, F.; Schäfer, W. Investigations on the ability of Fhb1 to protect wheat against nivalenol and deoxynivalenol. Cereal Res. Commun. 2008, 36, 429–435. [Google Scholar] [CrossRef]
- Mesterházy, A.; Lehoczki-Krsjak, S.; Varga, M.; Szabó-Hevér, Á.; Tóth, B.; Lemmens, M. Breeding for FHB Resistance via Fusarium Damaged Kernels and Deoxynivalenol Accumulation as Well as Inoculation Methods in Winter Wheat. Agric. Sci. 2015, 6, 970–1002. [Google Scholar] [CrossRef]
- Li, T.; Bai, G.; Wu, S.; Gu, S. Quantitative trait loci for resistance to fusarium head blight in a Chinese wheat landrace Haiyanzhong. Theor. Appl. Genet. 2011, 122, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Nordborg, M.; Weigel, D. Next-generation genetics in plants. Nature 2008, 456, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Lorenz, A.J.; Hamblin, M.T.; Jannink, J.L. Performance of Single Nucleotide Polymorphisms versus Haplotypes for Genome-Wide Association Analysis in Barley. PLoS ONE 2010, 5, e14079. [Google Scholar] [CrossRef]
- Chia, J.M.; Song, C.; Bradbury, P.J.; Costich, D.; de Leon, N.; Doebley, J.; Elshire, R.J.; Gaut, B.; Geller, L.; Glaubitz, J.C.; et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 2012, 44, 803–807. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; Liu, J.; Zhang, Y.; Cao, S.; He, Z.; Rasheed, A.; Jin, H.; Zhang, C.; Yan, J.; et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019, 19, 168. [Google Scholar] [CrossRef]
- EnsemblPlants. Release 54. Available online: https://plants.ensembl.org (accessed on 14 September 2022).
- Li, T.; Zhang, D.D.; Zhou, X.L.; Bai, G.H.; Li, L.; Gu, S.L. Fusarium head blight resistance loci in a stratified population of wheat landraces and varieties. Euphytica 2016, 207, 551–561. [Google Scholar] [CrossRef]
- Jiang, G.L.; Shi, J.; Ward, R. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. I. Resistance to fungal spread. Theor. Appl. Genet. 2007, 116, 3–13. [Google Scholar] [CrossRef]
- Somers, D.J.; Fedak, G.; Savard, M. Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome 2003, 46, 555–564. [Google Scholar] [CrossRef]
- Agostinelli, A.M.; Clark, A.J.; Brown-Guedira, G.; Van Sanford, D.A. Optimizing phenotypic and genotypic selection for Fusarium head blight resistance in wheat. Euphytica 2012, 186, 115–126. [Google Scholar] [CrossRef]
- Balut, A.L.; Clark, A.J.; Brown-Guedira, G.; Souza, E.; Van Sanford, D.A. Validation of Fhb1 and QFhs.nau-2DL in Several Soft Red Winter Wheat Populations. Crop Sci. 2013, 53, 934–945. [Google Scholar] [CrossRef]
- Eckard, J.T.; Gonzalez-Hernandez, J.L.; Caffe, M.; Berzonsky, W.; Bockus, W.W.; Marais, G.F.; Baenziger, P.S. Native Fusarium head blight resistance from winter wheat cultivars ‘Lyman’, ‘Overland’, ‘Ernie’, and ‘Freedom’ mapped and pyramided onto ‘Wesley’-Fhb1backgrounds. Mol. Breed. 2015, 35, 6. [Google Scholar] [CrossRef]
- Góral, T.; Wiśniewska, H.; Walentyn-Góral, D.; Radecka-Janusik, M.; Czembor, P. Resistance to Fusarium head blight [Fusarium culmorum (W.G. Sm.) Sacc.] of winter wheat lines generated from crosses between winter type cultivars and resistant spring wheat Sumai3. Prog. Plant Prot. 2016, 56, 285–295. [Google Scholar] [CrossRef][Green Version]
- Góral, T.; Wisniewska, H.; Ochodzki, P.; Twardowska, A.; Walentyn-Góral, D. Resistance to Fusarium Head Blight, Kernel Damage, and Concentration of Fusarium Mycotoxins in Grain of Winter Triticale (x Triticosecale Wittmack) Lines. Agronomy 2021, 11, 16. [Google Scholar] [CrossRef]
- Wiśniewska, H.; Kowalczyk, K. Resistance of cultivars and breeding lines of spring wheat to Fusarium culmorum and powdery mildew. J. Appl. Genet. 2005, 46, 35–40. [Google Scholar]
- Góral, T.; Wiśniewska, H.; Ochodzki, P.; Walentyn-Góral, D.; Kwiatek, M. Reaction of winter triticale breeding lines to Fusarium head blight and accumulation of Fusarium metabolites in grain in two environments under drought conditions. Cereal Res. Commun. 2013, 41, 106–115. [Google Scholar] [CrossRef]
- Góral, T.; Wiśniewska, H.; Ochodzki, P.; Nielsen, L.K.; Walentyn-Góral, D.; Stępień, Ł. Relationship between Fusarium Head Blight, Kernel Damage, Concentration of Fusarium Biomass, and Fusarium Toxins in Grain of Winter Wheat Inoculated with Fusarium culmorum. Toxins 2019, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Ochodzki, P.; Twardawska, A.; Wiśniewska, H.; Góral, T. Resistance to Fusarium Head Blight, Kernel Damage, and Concentrations of Fusarium Mycotoxins in the Grain of Winter Wheat Lines. Agronomy 2021, 11, 1690. [Google Scholar] [CrossRef]
- Kubo, K.; Kawada, N.; Fujita, M. Evaluation of Fusarium head blight resistance in wheat and the development of a new variety by integrating type I and II resistance. Jpn. Agric. Res. Q. 2013, 47, 9–19. [Google Scholar] [CrossRef][Green Version]
- Rudd, J.C.; Horsley, R.D.; McKendry, A.L.; Elias, E.M. Host plant resistance genes for Fusarium head blight: Sources, mechanisms, and utility in conventional breeding systems. Crop Sci. 2001, 41, 620–627. [Google Scholar] [CrossRef]
- Czembor, P.C.; Sejbuk, K.; Kleszcz, R. Evaluation of a partially-automated magnetic bead-based method for DNA extraction for wheat and barley MAS. Cereal Res. Commun. 2014, 42, 27–37. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Rampling, L.R.; Harker, N.; Shariflou, M.R.; Morell, M.K. Detection and analysis systems for microsatellite markers in wheat. Aust. J. Agric. Res. 2001, 52, 1131–1141. [Google Scholar] [CrossRef]
- Von Cruz, M.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the U.S. Collection of the new oilseed crop lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef]
- Isidro-Sánchez, J.; Akdemir, D.; Montilla-Bascón, G. Genome-Wide Association Analysis Using R. Methods Mol. Biol. 2017, 1536, 189–207. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Luján Basile, S.M.; Ramírez, I.A.; Crescente, J.M.; Conde, M.B.; Demichelis, M.; Abbate, P.; Rogers, W.J.; Pontaroli, A.C.; Helguera, M.; Vanzetti, L.S. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019, 19, 553. [Google Scholar] [CrossRef]
- Milne, I.; Shaw, P.; Stephen, G.; Bayer, M.; Cardle, L.; Thomas, W.T.B.; Flavell, A.J.; Marshall, D. Flapjack—Graphical genotype visualization. Bioinformatics 2010, 26, 3133–3134. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Konigorski, S.; Yilmaz, Y.E.; Janke, J.; Bergmann, M.M.; Boeing, H.; Pischon, T. Powerful rare variant association testing in a copula-based joint analysis of multiple phenotypes. Gen. Epidemiol. 2020, 44, 26–40. [Google Scholar] [CrossRef]
- Reverter, A.; Fortes, M.R.S. Association Weight Matrix: A Network-Based Approach Towards Functional Genome-Wide Association Studies. In Genome-Wide Association Studies and Genomic Prediction, Methods in Molecular Biology; Gondro, C., van der Werf, J., Hayes, B., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 1019, pp. 437–447. [Google Scholar] [CrossRef]
- Reverter, A.; Chan, E.K.F. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics 2008, 24, 2491–2497. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Martin, S.; Brown, W.M.; Klavans, R.; Boyack, K. OpenOrd: An Open-Source Toolbox for Large Graph Layout. In Visualization and Data Analysis; SPIE: Bellingham, WA, USA, 2011; Volume 7868, p. 786806. [Google Scholar] [CrossRef]



| Chromosome | SNP 1 | HB 2 | SNPs per HB avg. | SNPs per HB max | HT 3 | HB Size avg. (Mb) | HB Size max. (Mb) | Chromosome Coverage (%) |
|---|---|---|---|---|---|---|---|---|
| Chr1A | 340 | 82 | 3.5 | 12 | 288 | 3.1 | 36.2 | 42.7 |
| Chr1B | 424 | 96 | 3.8 | 16 | 363 | 4.1 | 54.9 | 57.7 |
| Chr1D | 331 | 85 | 3.3 | 12 | 330 | 3.8 | 62.7 | 64.8 |
| Chr2A | 593 | 115 | 4.6 | 26 | 318 | 3.1 | 28.6 | 45.4 |
| Chr2B | 717 | 126 | 5.2 | 58 | 444 | 3.6 | 30.6 | 57.1 |
| Chr2D | 581 | 139 | 3.5 | 19 | 513 | 1.8 | 26.1 | 38.9 |
| Chr3A | 451 | 89 | 4.3 | 26 | 346 | 3.8 | 26.2 | 45.5 |
| Chr3B | 633 | 138 | 3.8 | 13 | 477 | 3.5 | 49.2 | 57.3 |
| Chr3D | 475 | 104 | 3.4 | 25 | 403 | 2.3 | 20.3 | 38.6 |
| Chr4A | 340 | 82 | 3.4 | 19 | 306 | 4.9 | 32.6 | 54.1 |
| Chr4B | 338 | 72 | 3.7 | 22 | 261 | 5.0 | 102.9 | 53.9 |
| Chr4D | 223 | 46 | 3.1 | 9 | 199 | 3.3 | 40.8 | 29.6 |
| Chr5A | 529 | 123 | 3.7 | 30 | 444 | 3.2 | 115.1 | 55.7 |
| Chr5B | 583 | 117 | 4.3 | 50 | 412 | 3.4 | 33.2 | 56.6 |
| Chr5D | 409 | 88 | 3.8 | 13 | 354 | 3.3 | 27.1 | 51.6 |
| Chr6A | 373 | 74 | 4.2 | 25 | 284 | 3.5 | 25.4 | 42.0 |
| Chr6B | 580 | 131 | 3.5 | 16 | 480 | 3.2 | 26.2 | 57.7 |
| Chr6D | 404 | 101 | 3.2 | 9 | 395 | 2.2 | 27.9 | 46.8 |
| Chr7A | 667 | 141 | 4.0 | 19 | 543 | 2.6 | 45.9 | 50.2 |
| Chr7B | 513 | 123 | 3.4 | 20 | 477 | 3.0 | 31.7 | 48.7 |
| Chr7D | 516 | 120 | 3.3 | 13 | 476 | 2.1 | 18.3 | 38.7 |
| ChrUn 4 | 231 | 64 | 2.8 | 8 | 260 | 2.0 | 10.9 | 26.5 |
| No | Chromosome | HT * | Locus (bp) | p-Value | Trait | R2 |
|---|---|---|---|---|---|---|
| 1 | 2D | Ch2D_B35_H1 | Chr2D:32388539-32388595 | 6.98948 × 10−7 | Type_2_2020 | 41% |
| 2 | 5.56168 × 10−7 | Type_2_2019&2020 | 43% | |||
| 3 | 3.8519 × 10−6 | FHBi | 6% | |||
| 4 | 2D | Ch2D_B35_H2 | Chr2D:32388539-32388595 | 2.8397 × 10−7 | Type_2_2020 | 41% |
| 5 | 2D | 3.8328 × 10−6 | Type_1&2_2020 | 39% | ||
| 6 | 2D | 1.7512 × 10−7 | Type_2_2019&2020 | 46% | ||
| 7 | 3B | Ch3B_B9_H2 | Chr3B:13276831-13633016 | 1.9527 × 10−7 | Type_2_2020 | 46% |
| 8 | 3B | 2.22 × 10−7 | Type_2_2019&2020 | 49% | ||
| 9 | 7D | Ch7D_B63_H4 | Chr7D:439405257-444190466 | 1.6867 × 10−6 | Type_2_2020 | 34% |
| Trait | Haplotype | Chromosome | Freq. Effect 1 |
|---|---|---|---|
| Type_2_2020 | Ch2D_B35_H1 | 2D | 33 + |
| Ch2D_B35_H2 | 2D | 21 − | |
| Ch3B_B9_H2 | 3B | 21 − | |
| Ch7D_B63_H4 | 7D | 5 − | |
| Type_2_2019&2020 | Ch2D_B35_H1 | 2D | 33 + |
| Ch2D_B35_H2 | 2D | 21 − | |
| Ch3B_B9_H2 | 3B | 21 − | |
| Type_1&2_2020 | Ch2D_B35_H2 | 2D | 21 − |
| FHBi | Ch2D_B35_H1 | 2D | 33 + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radecka-Janusik, M.; Piechota, U.; Piaskowska, D.; Góral, T.; Czembor, P. Evaluation of Fusarium Head Blight Resistance Effects by Haplotype-Based Genome-Wide Association Study in Winter Wheat Lines Derived by Marker Backcrossing Approach. Int. J. Mol. Sci. 2022, 23, 14233. https://doi.org/10.3390/ijms232214233
Radecka-Janusik M, Piechota U, Piaskowska D, Góral T, Czembor P. Evaluation of Fusarium Head Blight Resistance Effects by Haplotype-Based Genome-Wide Association Study in Winter Wheat Lines Derived by Marker Backcrossing Approach. International Journal of Molecular Sciences. 2022; 23(22):14233. https://doi.org/10.3390/ijms232214233
Chicago/Turabian StyleRadecka-Janusik, Magdalena, Urszula Piechota, Dominika Piaskowska, Tomasz Góral, and Paweł Czembor. 2022. "Evaluation of Fusarium Head Blight Resistance Effects by Haplotype-Based Genome-Wide Association Study in Winter Wheat Lines Derived by Marker Backcrossing Approach" International Journal of Molecular Sciences 23, no. 22: 14233. https://doi.org/10.3390/ijms232214233
APA StyleRadecka-Janusik, M., Piechota, U., Piaskowska, D., Góral, T., & Czembor, P. (2022). Evaluation of Fusarium Head Blight Resistance Effects by Haplotype-Based Genome-Wide Association Study in Winter Wheat Lines Derived by Marker Backcrossing Approach. International Journal of Molecular Sciences, 23(22), 14233. https://doi.org/10.3390/ijms232214233







