Computational Biology Dynamics of Mps1 Kinase Molecular Interactions with Isoflavones Reveals a Chemical Scaffold with Potential to Develop New Therapeutics for the Treatment of Cancer
Abstract
1. Introduction
2. Results
2.1. Virtual Screening of Mps1 Using Reversine as Lead Compound
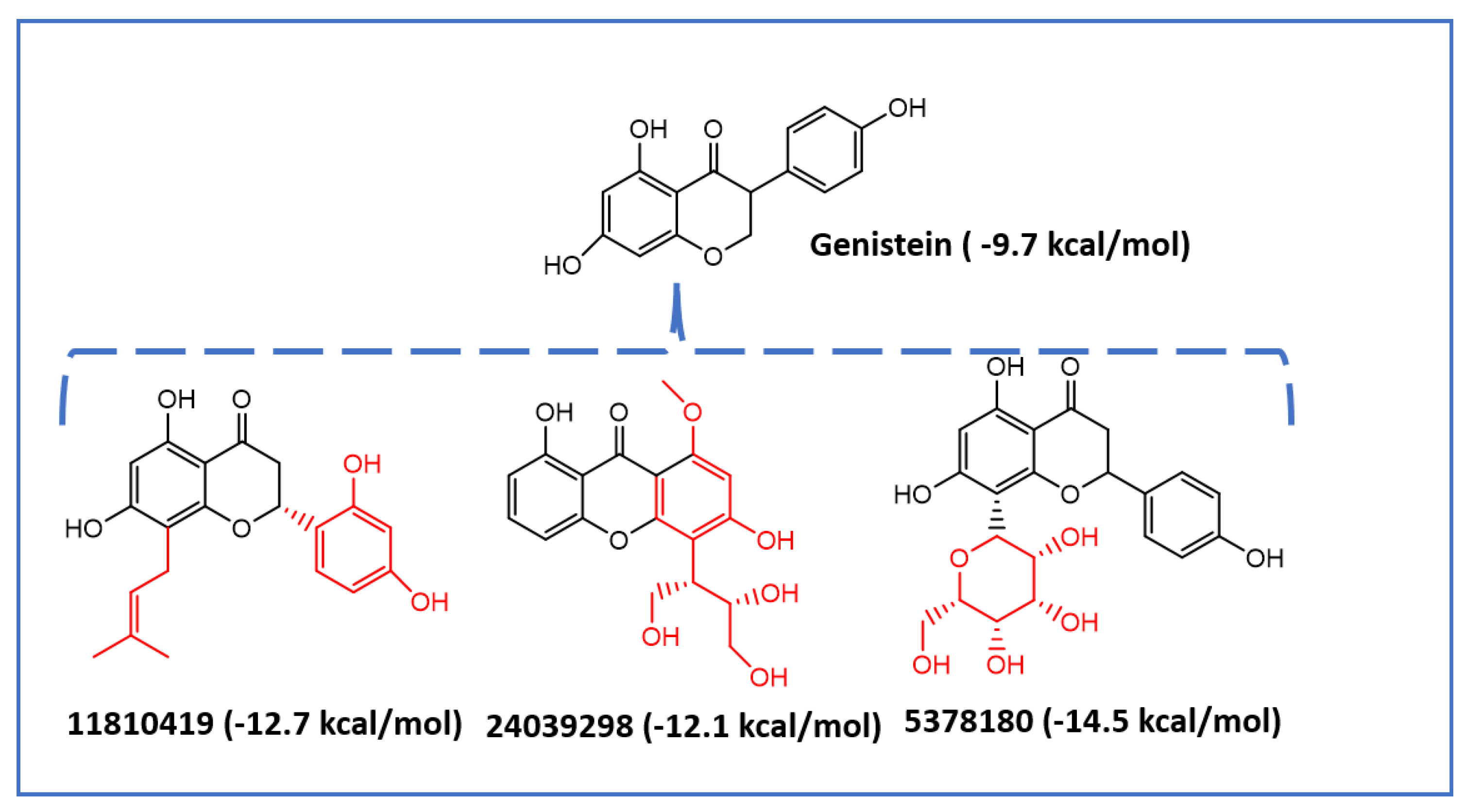
2.2. Virtual Screening of Mps1 Using Genistein as the Lead Compound
3. Discussion
4. Materials and Methods
4.1. Ligand Receptor Structure
4.2. Ligand Preparation
4.3. Molecular Docking Studies
4.3.1. Grid Generation
4.3.2. HTVS and XP Docking
4.4. In Silico Pharmacokinetics Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Koch, A.; Maia, A.; Janssen, A.; Medema, R.H. Molecular basis underlying resistance to Mps1/TTK inhibitors. Oncogene 2016, 35, 2518–2528. [Google Scholar] [CrossRef] [PubMed]
- Broad, A.J.; DeLuca, J.G. The right place at the right time: Aurora B kinase localization to centromeres and kinetochores. Essays Biochem. 2020, 64, 299–311. [Google Scholar] [PubMed]
- Levasseur, M.D.; Thomas, C.; Davies, O.R.; Higgins, J.M.G.; Madgwick, S. Aneuploidy in oocytes is prevented by sustained CDK1 activity through degron masking in cyclin B1. Dev. Cell 2019, 48, 672–684.e5. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Moreno, P.; Quezada-Meza, C.; Chavez-Almarza, C.; Niechi, I.; Silva-Pavez, E.; Trigo-Hidalgo, C.; Aguayo, F.; Jara, L.; Cáceres-Verschae, A.; Varas-Godoy, M.; et al. Phosphorylation of Endothelin-Converting Enzyme-1c at Serines 18 and 20 by CK2 Promotes Aggressiveness Traits in Colorectal Cancer Cells. Front. Oncol. 2020, 10, 1004. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Kim, D.E.; Kim, D.H.; Min, G.H.; Park, J.W.; Kim, Y.B.; Sung, C.K.; Yim, H. Mitotic protein kinase-driven crosstalk of machineries for mitosis and metastasis. Exp. Mol. Med. 2022, 54, 414–425. [Google Scholar] [CrossRef]
- Pachis, S.T.; Kops, G.J. Leader of the SAC: Molecular mechanisms of Mps1/TTK regulation in mitosis. Open Biol. 2018, 8, 180109. [Google Scholar] [CrossRef]
- Combes, G.; Barysz, H.; Garand, C.; Gama Braga, L.; Alharbi, I.; Thebault, P.; Murakami, L.; Bryne, D.P.; Stankovic, S.; Eyers, P.A.; et al. Mps1 Phosphorylates Its N-Terminal Extension to Relieve Autoinhibition and Activate the Spindle Assembly Checkpoint. Curr. Biol. 2018, 28, 872–883.e5. [Google Scholar] [CrossRef]
- Choi, M.; Min, Y.H.; Pyo, J.; Lee, C.W.; Jang, C.Y.; Kim, J.E. Mps1 12, a novel Mps1 inhibitor, suppresses the growth of epatocellular carcinoma cells via the accumulation of chromosomal instability. Br. J. Pharmacol. 2017, 174, 1810–1825. [Google Scholar] [CrossRef]
- Sarangapani, K.K.; Koch, L.B.; Nelson, C.R.; Asbury, C.L.; Biggins, S. Kinetochore-bound Mps1 regulates kinetochore-microtubule attachments via Ndc80 phosphorylation. J. Cell Biol. 2021, 220, e202106130. [Google Scholar] [CrossRef]
- Yamagishi, Y.; Yang, C.H.; Tanno, Y.; Watanabe, Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 2012, 14, 746–752. [Google Scholar] [CrossRef]
- Huang, H.; Yang, Y.; Zhang, W.; Liu, X.; Yang, G. TTK regulates proliferation and apoptosis of gastric cancer cells through the Akt-mTOR pathway. FEBS Open Bio. 2020, 10, 1542–1549. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, B.; Zhu, N.; Tao, M.; Jun, Y.; Chen, X.; Wang, Q.; Luo, C. Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCα/ERK1/2 and PI3K/Akt pathway. Med. Oncol. 2019, 37, 5. [Google Scholar] [CrossRef]
- Thebault, P.; Chirgadze, D.Y.; Dou, Z.; Blundell, T.L.; Elowe, S.; Bolanos-Garcia, V.M. Structural and functional insights into the role of the N-terminal Mps1 TPR domain in the SAC. Biochem. J. 2012, 448, 321–328. [Google Scholar] [CrossRef]
- Nijenhuis, W.; von Castelmur, E.; Littler, D.; De Marco, V.; Tromer, E.; Vleugel, M.; van Osch, M.H.; Snel, B.; Perrakis, A.; Kops, G.J. A Tpr Domain-Containing N-Terminal Module of Mps1 is Required for its Kinetochore Localization by Aurora B. J. Cell Biol. 2013, 201, 217–231. [Google Scholar] [CrossRef]
- Chu, M.L.; Chavas, L.M.; Douglas, K.T.; Eyers, P.; Tabernero, L. Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125. J. Biol. Chem. 2008, 283, 21495–21500. [Google Scholar] [CrossRef]
- Wengner, A.M.; Siemeister, G.; Koppitz, M.; Schulze, V.; Kosemund, D.; Klar, U.; Stoeckigt, D.; Neuhaus, R.; Lienau, P.; Bader, B.; et al. Novel Mps1 Kinase Inhibitors with Potent Antitumor Activity. Mol. Cancer Ther. 2016, 15, 583–592. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, M.; Liang, D.; Sun, W.; Zhang, C.; Jiang, M.; Liu, J.; Li, J.; Li, C.; Yang, X.; et al. Molecular design and anticancer activities of small-molecule monopolar spindle 1 inhibitors: A Medicinal chemistry perspective. Eur. J. Med. Chem. 2019, 175, 247–268. [Google Scholar] [CrossRef]
- Naud, S.; Westwood, I.M.; Faisal, A.; Sheldrake, P.; Bavetsias, V.; Atrash, B.; Cheung, K.M.; Liu, M.; Hayes, A.; Schmitt, J.; et al. Structure-based design of orally bioavailable 1H-pyrrolo[3,2-c]pyridine inhibitors of mitotic kinase monopolar spindle 1 (MPS1). J. Med. Chem. 2013, 56, 10045–10065. [Google Scholar] [CrossRef]
- Schöffski, P.; Awada, A.; de la Bigne, A.M.; Felloussi, Z.; Burbridge, M.; Cantero, F.; Colombo, R.; Maruzzelli, S.; Ammattatelli, K.; de Jonge, M.; et al. First-in-man, first-in-class phase I study with the monopolar spindle 1 kinase inhibitor S81694 administered intravenously in adult patients with advanced, metastatic solid tumours. Eur. J. Cancer 2022, 169, 135–145. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, H.; Kim, H.; Cho, H.Y.; Jee, J.G.; Seo, K.A.; Son, J.B.; Ko, E.; Choi, H.G.; Kim, N.D.; et al. X-ray Crystal Structure-Guided Design and Optimization of 7H-Pyrrolo[2,3-d]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor. J. Med. Chem. 2021, 64, 6985–6995. [Google Scholar] [CrossRef]
- Chen, K.; Duan, W.; Han, Q.; Sun, X.; Li, W.; Hu, S.; Wan, J.; Wu, J.; Ge, Y.; Liu, D. Identification of the hot spot residues for pyridine derivative inhibitor CCT251455 and ATP substrate binding on monopolar spindle 1 (MPS1) kinase by molecular dynamic simulation. J. Biomol. Struct. Dyn. 2019, 37, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, A.; Lin, J.; Wu, L.; Zhang, H.; Yang, X.; Wan, X.; Miao, R.; Sang, X.; Zhao, H. Mps1/TTK: A novel target and biomarker for cancer. J. Drug Target. 2017, 25, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Weinman, S. Mechanisms of doxorubicin resistance in hepatocellular carcinoma. Hepatic Oncol. 2016, 3, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Schulze, V.K.; Klar, U.; Kosemund, D.; Wengner, A.M.; Siemeister, G.; Stöckigt, D.; Neuhaus, R.; Lienau, P.; Bader, B.; Prechtl, S.; et al. Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase. J. Med. Chem. 2020, 63, 8025–8042. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, P.; Chawla, S.P.; Bendell, J.; Shields, A.F.; Shapiro, G.; Rajagopalan, P.; Cyrus, C.; Bruns, I.; Mei, J.; Souza, F.; et al. First-in-human study of the monopolar spindle 1 (Mps1) kinase inhibitor BAY 1161909 in combination with paclitaxel in subjects with advanced malignancies. Ann. Oncol. 2018, 29, VIII138. [Google Scholar] [CrossRef]
- Woodward, H.L.; Innocenti, P.; Cheung, K.J.; Hayes, A.; Roberts, J.; Henley, A.T.; Faisal, A.; Mak, G.W.; Box, G.; Westwood, I.M.; et al. Introduction of a Methyl Group Curbs Metabolism of Pyrido[3,4-d]pyrimidine Monopolar Spindle 1 (MPS1) Inhibitors and Enables the Discovery of the Phase 1 Clinical Candidate N2-(2-Ethoxy-4-(4-methyl-4 H-1,2,4-triazol-3-yl)phenyl)-6-methyl-N8-neopentylpyrido[3,4-d]pyrimidine-2,8-diamine (BOS172722). J. Med. Chem. 2018, 61, 8226–8240. [Google Scholar]
- Chan, C.Y.; Chiu, D.K.; Yuen, V.W.; Law, C.T.; Wong, B.P.; Thu, K.L.; Cescon, D.W.; Soria-Bretones, I.; Cheu, J.W.; Lee, D.; et al. CFI-402257, a TTK inhibitor, effectively suppresses hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2022, 119, e2119514119. [Google Scholar] [CrossRef]
- Mullard, A. Biotech R&D spend jumps by more than 15. Nat. Rev. Drug Discov. 2016, 15, 447. [Google Scholar]
- Thomas, D.W.; Burns, J.; Audette, J.; Carroll, A.; Dow-Hygelund, C.; Hay, M. Clinical Development Success Rates 2006–2015. BIO Ind. Anal. 2016, 1, 25. [Google Scholar]
- El-Demerdash, A.; Metwaly, A.M.; Hassan, A.; Abd El-Aziz, T.M.; Elkaeed, E.B.; Eissa, I.H.; Arafa, R.K.; Stockand, J.D. Comprehensive Virtual Screening of the Antiviral Potentialities of Marine Polycyclic Guanidine Alkaloids against SARS-CoV-2 (COVID-19). Biomolecules 2021, 11, 460. [Google Scholar] [CrossRef]
- Mizera, M.; Szymanowska, D.; Stasiłowicz, A.; Siąkowska, D.; Lewandowska, K.; Miklaszewski, A.; Plech, T.; Tykarska, E.; Cielecka-Piontek, J. Computer-Aided Design of Cefuroxime Axetil/Cyclodextrin System with Enhanced Solubility and Antimicrobial Activity. Biomolecules 2020, 10, 24. [Google Scholar] [CrossRef]
- Rao, A.; Haque, S.; El-Enshasy, H.A.; Singh, V.; Mishra, B.N. RSM–GA Based Optimization of Bacterial PHA Production and In Silico Modulation of Citrate Synthase for Enhancing PHA Production. Biomolecules 2019, 9, 872. [Google Scholar] [CrossRef]
- Rus, A.; Bolanos-Garcia, V.M.; Bastida, A.; Morales, P. Identification of Novel Potential Heparanase Inhibitors Using Virtual Screening. Catalysts 2022, 12, 503. [Google Scholar] [CrossRef]
- Zárate, S.G.; Morales, P.; Świderek, K.; Bolanos-Garcia, V.M.; Bastida, A. A molecular modeling approach to identify novel inhibitors of the major facilitator superfamily of efflux pump transporters. Antibiotics 2019, 8, 25. [Google Scholar] [CrossRef]
- Ogu, C.C.; Maxa, J.L. Drug interactions due to cytochrome p450. Bayl. Univ. Med. Cent. Proc. 2000, 13, 421–423. [Google Scholar] [CrossRef]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic Colorimetric Proliferation Assays: MTT, WST, and Resazurin. Methods Mol. Biol. 2017, 1601, 1–17. [Google Scholar]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Sánchez-Quesada, C.J.; Gaforio, J. Dietary Flavonoids as Cancer Chemopreventive Agents: An Updated Review of Human Studies. Antioxidants 2019, 8, 137. [Google Scholar] [CrossRef]
- Sarkar, F.H.; Adsule, S.; Padhye, S.; Kulkarni, S.; Li, Y. The role of genistein and synthetic derivatives of isoflavone in cancer prevention and therapy. Mini-Rev. Med. Chem. 2006, 6, 401–407. [Google Scholar] [CrossRef]
- Spagnuolo, C.; Russo, G.L.; Orhan, I.E.; Habtemariam, S.; Daglia, M.; Sureda, A.; Nabavi, S.F.; Devi, K.P.; Loizzo, M.R.; Tundis, R.; et al. Genistein and cancer: Current status, challenges, and future directions. Adv. Nutr. 2015, 6, 408–419. [Google Scholar] [CrossRef]
- Takagi, A.; Kano, M.; Kaga, C. Possibility of breast cancer prevention: Use of soy isoflavones and fermented soy beverage produced using probiotics. Int. J. Mol. Sci. 2015, 16, 10907–10920. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Ramachandran, S.; Gupta, N.; Kaushik, I.; Wright, S.; Srivastava, S.; Das, H.; Prasad, S.; Srivastava, S.K. Role of Phytochemicals in Cancer Prevention. Int. J. Mol. Sci. 2019, 20, 4981. [Google Scholar] [CrossRef] [PubMed]
- Křížová, L.; Dadáková, K.; Kašparovská, J.; Kašparovský, T. Isoflavones. Molecules 2019, 24, 1076. [Google Scholar] [CrossRef]
- Ullah, A.; Munir, S.; Badshah, S.L.; Khan, N.; Ghani, L.; Poulson, B.G.; Emwas, A.H.; Jaremko, M. Important Flavonoids and Their Role as a Therapeutic Agent. Molecules 2020, 25, 5243. [Google Scholar] [CrossRef] [PubMed]
- Ziaei, S.; Halaby, R. Dietary isoflavones and breast cancer risk. Medicines 2017, 4, 18. [Google Scholar] [CrossRef]
- Lu, H.; Chang, D.J.; Baratte, B.; Meijer, L.; Schulze-Gahmen, U. Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin. J. Med. Chem. 2005, 48, 737–743. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, C.Z.; Du, G.J.; Qi, L.W.; Calway, T.; He, T.C.; Du, W.; Yuan, C.S. Genistein induces G2/M cell cycle arrest and apoptosis via ATM/p53-dependent pathway in human colon cancer cells. Int. J. Oncol. 2013, 43, 289–296. [Google Scholar] [CrossRef]
- Sarkar, F.H.; Li, Y. Mechanisms of cancer chemoprevention by soy isoflavone genistein. Cancer Metastasis Rev. 2002, 21, 265–280. [Google Scholar] [CrossRef]
- Yokoyama, T.; Kosaka, Y.; Mizuguchi, M. Structural Insight into the Interactions between Death-Associated Protein Kinase 1 and Natural Flavonoids. J. Med. Chem. 2015, 58, 7400–7408. [Google Scholar] [CrossRef]
- Guha, N.; Kwan, M.L.; Quesenberry, C.P.; Weltzien, E.K.; Castillo, A.L.; Caan, B.J. Soy isoflavones and risk of cancer recurrence in a cohort of breast cancer survivors: The Life After Cancer Epidemiology study. Breast Cancer Res. Treat. 2009, 118, 395–405. [Google Scholar] [CrossRef]
- Hiruma, Y.; Koch, A.; Dharadhar, S.; Joosten, R.P.; Perrakis, A. Structural basis of reversine selectivity in inhibiting Mps1 more potently than aurora B kinase. Proteins 2016, 84, 1761–1766. [Google Scholar] [CrossRef]
- Anderhub, S.J.; Mak, G.W.; Gurden, M.D.; Faisal, A.; Drosopoulos, K.; Walsh, K.; Woodward, H.L.; Innocenti, P.; Westwood, I.M.; Naud, S.; et al. High Proliferation Rate and a Compromised Spindle Assembly Checkpoint Confers Sensitivity to the MPS1 Inhibitor BOS172722 in Triple-Negative Breast Cancers. Mol. Cancer Ther. 2019, 18, 1696–1707. [Google Scholar] [CrossRef]
- Liu, Y.; Laufer, R.; Patel, N.K.; Ng, G.; Sampson, P.B.; Li, S.W.; Lang, Y.; Feher, M.; Brokx, R.; Beletskaya, I.; et al. Discovery of Pyrazolo[1,5-a]pyrimidine TTK Inhibitors: CFI-402257 is a Potent, Selective, Bioavailable Anticancer Agent. ACS Med. Chem. Lett. 2016, 7, 671–675. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Capuzzi, S.J.; Muratov, E.N.; Tropsha, A. Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay in terference Compounds. J. Chem. Inf. Model. 2017, 57, 417–427. [Google Scholar] [CrossRef]
- Salmela, A.L.; Pouwels, J.; Varis, A.; Kukkonen, A.M.; Toivonen, P.; Halonen, P.K.; Perälä, M.; Kallioniemi, O.; Gorbsky, G.J.; Kallio, M.J. Dietary flavonoid fisetin induces a forced exit from mitosis by targeting the mitotic spindle checkpoint. Carcinogenesis 2009, 30, 1032–1040. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daniel, J.; Coulter, J.; Woo, J.H.; Wilsbach, K.; Gabrielson, E. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 5384–5389. [Google Scholar] [CrossRef]
- Simon Serrano, S.; Sime, W.; Abassi, Y.; Daams, R.; Massoumi, R.; Jemaà, M. Inhibition of mitotic kinase Mps1 promotes cell death in neuroblastoma. Sci. Rep. 2020, 10, 11997. [Google Scholar] [CrossRef]
- Maachani, U.B.; Kramp, T.; Hanson, R.; Zhao, S.; Celiku, O.; Shankavaram, U.; Colombo, R.; Caplen, N.J.; Camphausen, K.; Tandle, A. Targeting MPS1 Enhances Radiosensitization of Human Glioblastoma by Modulating DNA Repair Proteins. Mol. Cancer Res. 2015, 13, 852–862. [Google Scholar] [CrossRef]
- D’Alise, A.M.; Amabile, G.; Iovino, M.; Di Giorgio, F.P.; Bartiromo, M.; Sessa, F.; Villa, F.; Musacchio, A.; Cortese, R. Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Mol. Cancer Ther. 2008, 7, 1140–1149. [Google Scholar] [CrossRef]
- Mason, J.M.; Wei, X.; Fletcher, G.C.; Kiarash, R.; Brokx, R.; Hodgson, R.; Beletskaya, I.; Bray, M.R.; Mak, T.W. Functional characterization of CFI-402257, a potent and selective Mps1/TTK kinase inhibitor, for the treatment of cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3127–3132. [Google Scholar] [CrossRef] [PubMed]
- Serafim, R.A.; da Silva Santiago, A.; Schwalm, M.P.; Hu, Z.; Dos Reis, C.V.; Takarada, J.E.; Mezzomo, P.; Massirer, K.B.; Kudolo, M.; Gerstenecker, S.; et al. Development of the First Covalent Monopolar Spindle Kinase 1 (MPS1/TTK) Inhibitor. J. Med. Chem. 2022, 65, 3173–3192. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Hoskins, C. Drug development: Lessons from nature. Biomed. Rep. 2017, 6, 612–614. [Google Scholar] [CrossRef] [PubMed]
- George, V.C.; Dellaire, G.; Rupasinghe, H.P.V. Plant flavonoids in cancer chemoprevention: Role in genome stability. J. Nutr. Biochem. 2017, 45, 1–14. [Google Scholar] [CrossRef]
- Blachly, J.S.; Byrd, J.C. Emerging drug profile: Cyclin-dependent kinase inhibitors. Leuk. Lymphoma 2013, 54, 2133–2143. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, X.; Yuan, X.; Shi, J.; Zhang, C.; Yan, N.; Jing, C. Comparison of Phenolic and Flavonoid Compound Profiles and Antioxidant and α-Glucosidase Inhibition Properties of Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja). Plants 2021, 10, 813. [Google Scholar] [CrossRef]
- Zhang, B.; Su, J.P.; Bai, Y.; Li, J.; Liu, Y.H. Inhibitory effects of O-methylated isoflavone glycitein on human breast cancer SKBR-3 cells. Int. J. Clin. Exp. Pathol. 2015, 8, 7809–7817. [Google Scholar]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
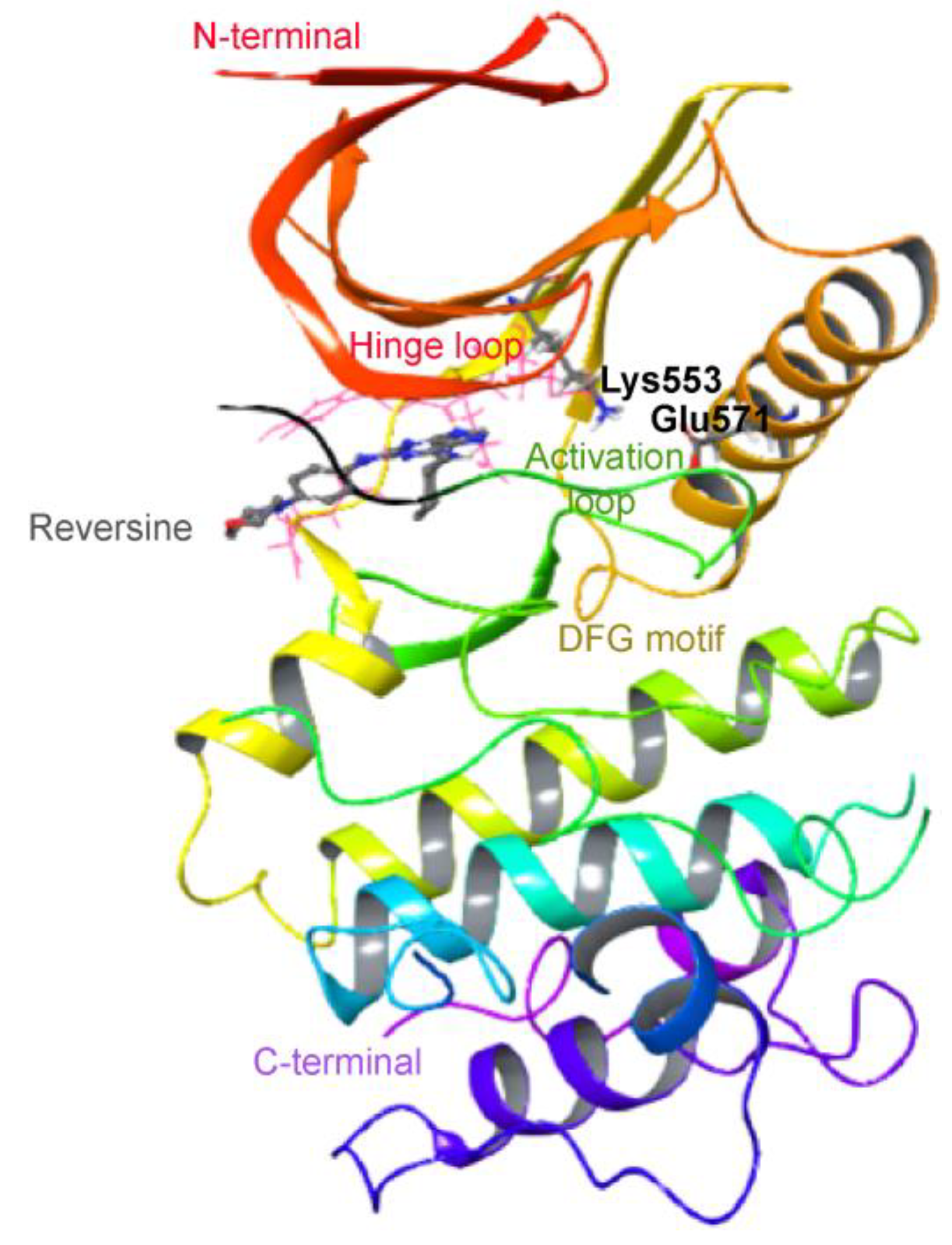
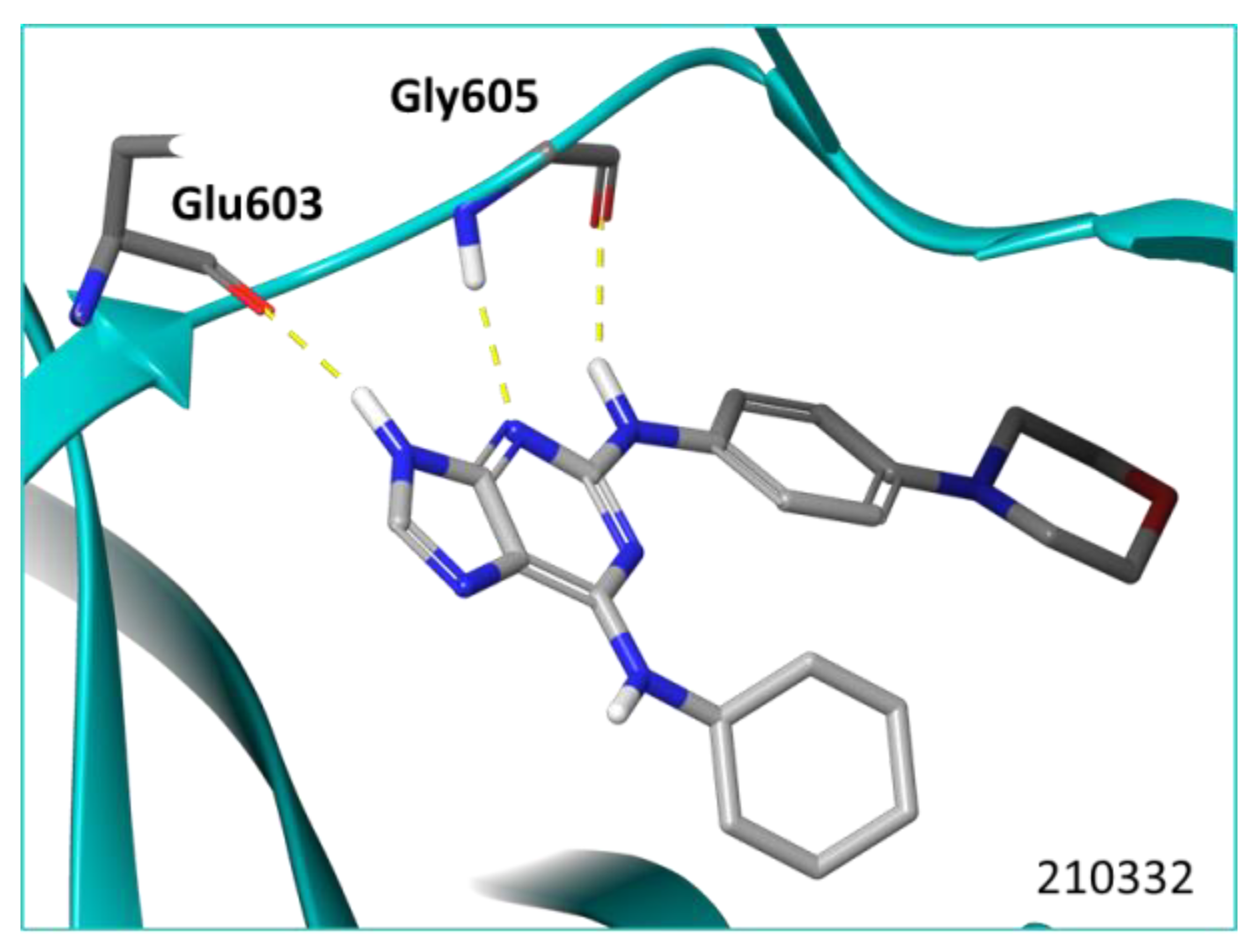

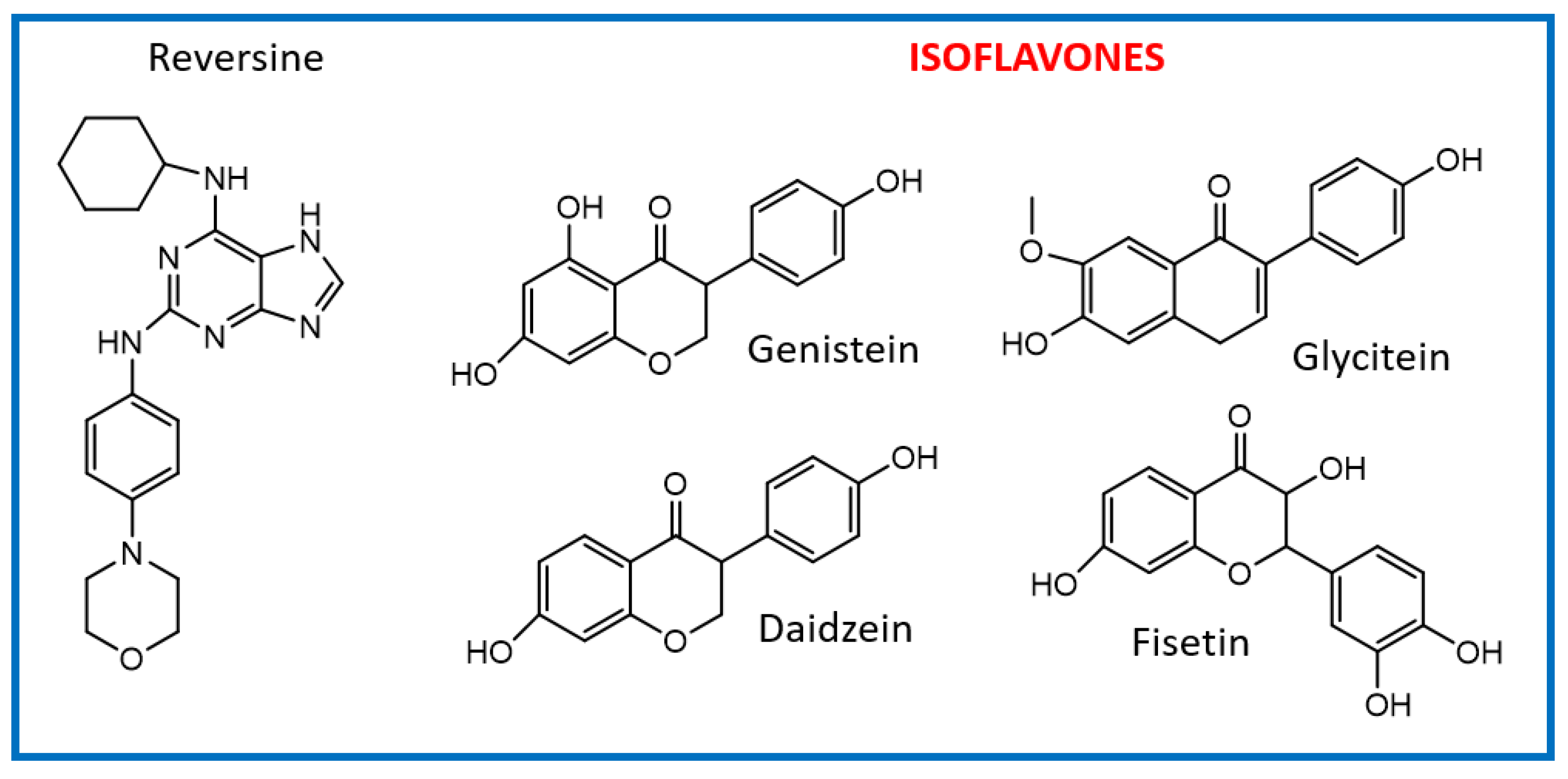

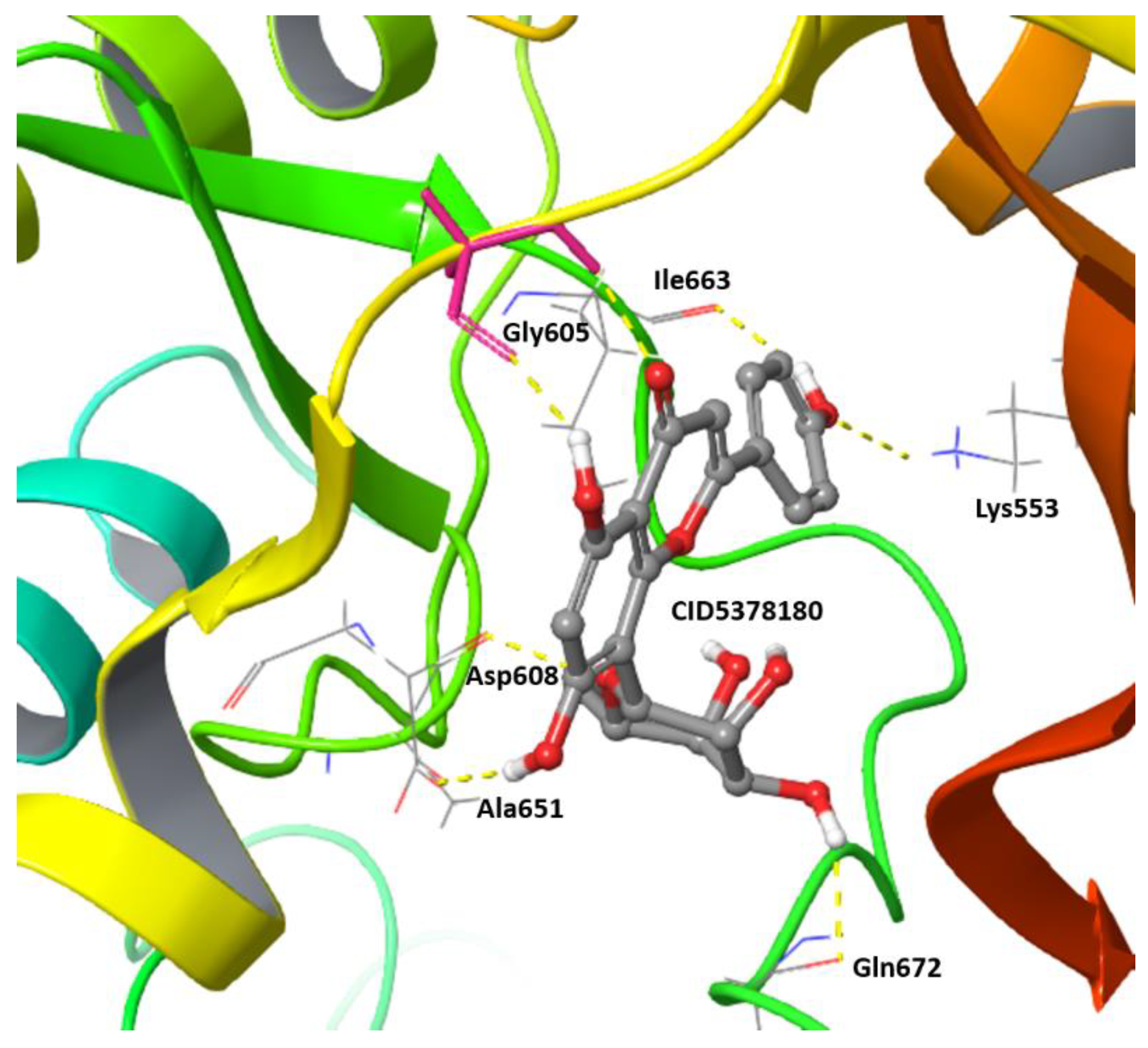
| Compound (PubChem ID) | XP Glide Score (kcal/mol) | IC50 (nM) | Reference |
|---|---|---|---|
Reversine (210332) | −10.97 | 6 | [52] |
Bay1217389 (78320750)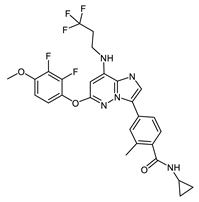 | −10.18 | 50 | [25] |
Bos-172722 (73386890)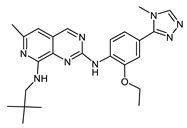 | −9.05 | 3 | [53] |
Bay-1161909 (71599640)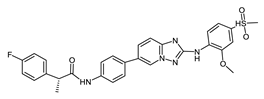 | −8.17 | 0.34 | [25] |
CIF-402257 (118086034)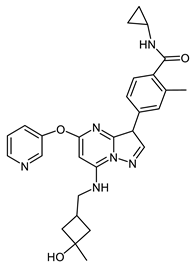 | −8.0 | 10 | [54] |
| Compound ID | 2D Structure | Glide Score (kcal/mol) | Type of Interaction and Mps1 Residues Involved |
|---|---|---|---|
| 210332 Reversine | 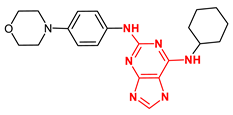 | −10.97 | 2 H-bonds Gly605, and 1H bond Glu603 |
| 123679245 | 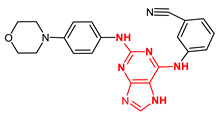 | −11.10 | 2 H-bonds Gly605, and 1H bond Glu603 |
| 68916574 |  | −9.34 | 2 H-bonds Gly605 |
| 117091061 |  | −9.23 | 2 H-bonds Gly605 |
| 57639384 | 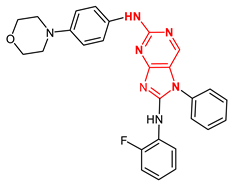 | −9.2 | 2 H-bonds Gly605 |
| 21189870 | 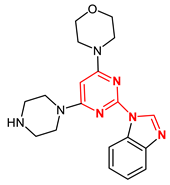 | −9.0 | 1 H-bond Gly605 |
| Compound | QPlogS | QPlogHERG | QPPCaco | QPlogBB | % Human Oral Absorption | PAINS |
|---|---|---|---|---|---|---|
| Reversine | −5.22 | −5.01 | 922 | −0.71 | 100 | 1 |
| 123679245 | −6.37 | −6.29 | 333 | −1.26 | 87.8 | 1 |
| 68916574 | −4.08 | −4.72 | 1296 | −0.23 | 93.8 | 0 |
| 117091061 | −4.11 | −6.08 | 669 | −0.11 | 95.8 | 0 |
| 57639384 | −7.78 | −7.45 | 2334 | −0.35 | 100 | 1 |
| 21189870 | −3.55 | −5.57 | 501 | +0.27 | 89.0 | 0 |
| COMPOUND ID | XP GScore (kcal/mol) | QPlogS | QPlogHERG | QPPCaco | QPlogBB | %(HOA) | PAINS |
|---|---|---|---|---|---|---|---|
| Fisetin | −10.27 | −2.7 | −4.9 | 52 | −1.8 | 60 | 1 |
| Genistein | −9.7 | −2.9 | −4.9 | 171 | −1.2 | 76 | 0 |
| Daidzein | −9.2 | −2.9 | −5.0 | 397 | −0.9 | 83 | 0 |
| Glycitein | −7.9 | −3.2 | −5.0 | 404 | −0.9 | 84 | 0 |
| Compound CID | QPlogS | QPlogHERG | QPPCaco | QPlogBB | %(HOA) | PAINS |
|---|---|---|---|---|---|---|
| 5378180 | −2.66 | −4.83 | 14.83 | −2.62 | 31 | 0 |
| 11810419 | −4.09 | −4.64 | 134.52 | −1.57 | 79 | 0 |
| 24039298 | −2.39 | −4.74 | 61.39 | −2.17 | 61.8 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pugh, L.; Pancholi, A.; Purat, P.C.; Agudo-Alvarez, S.; Benito-Arenas, R.; Bastida, A.; Bolanos-Garcia, V.M. Computational Biology Dynamics of Mps1 Kinase Molecular Interactions with Isoflavones Reveals a Chemical Scaffold with Potential to Develop New Therapeutics for the Treatment of Cancer. Int. J. Mol. Sci. 2022, 23, 14228. https://doi.org/10.3390/ijms232214228
Pugh L, Pancholi A, Purat PC, Agudo-Alvarez S, Benito-Arenas R, Bastida A, Bolanos-Garcia VM. Computational Biology Dynamics of Mps1 Kinase Molecular Interactions with Isoflavones Reveals a Chemical Scaffold with Potential to Develop New Therapeutics for the Treatment of Cancer. International Journal of Molecular Sciences. 2022; 23(22):14228. https://doi.org/10.3390/ijms232214228
Chicago/Turabian StylePugh, Lauren, Alisha Pancholi, Priscila Celeste Purat, Sandra Agudo-Alvarez, Raúl Benito-Arenas, Agatha Bastida, and Victor M. Bolanos-Garcia. 2022. "Computational Biology Dynamics of Mps1 Kinase Molecular Interactions with Isoflavones Reveals a Chemical Scaffold with Potential to Develop New Therapeutics for the Treatment of Cancer" International Journal of Molecular Sciences 23, no. 22: 14228. https://doi.org/10.3390/ijms232214228
APA StylePugh, L., Pancholi, A., Purat, P. C., Agudo-Alvarez, S., Benito-Arenas, R., Bastida, A., & Bolanos-Garcia, V. M. (2022). Computational Biology Dynamics of Mps1 Kinase Molecular Interactions with Isoflavones Reveals a Chemical Scaffold with Potential to Develop New Therapeutics for the Treatment of Cancer. International Journal of Molecular Sciences, 23(22), 14228. https://doi.org/10.3390/ijms232214228








