Rab32/38-Dependent and -Independent Transport of Tyrosinase to Melanosomes in B16-F1 Melanoma Cells
Abstract
1. Introduction
2. Results
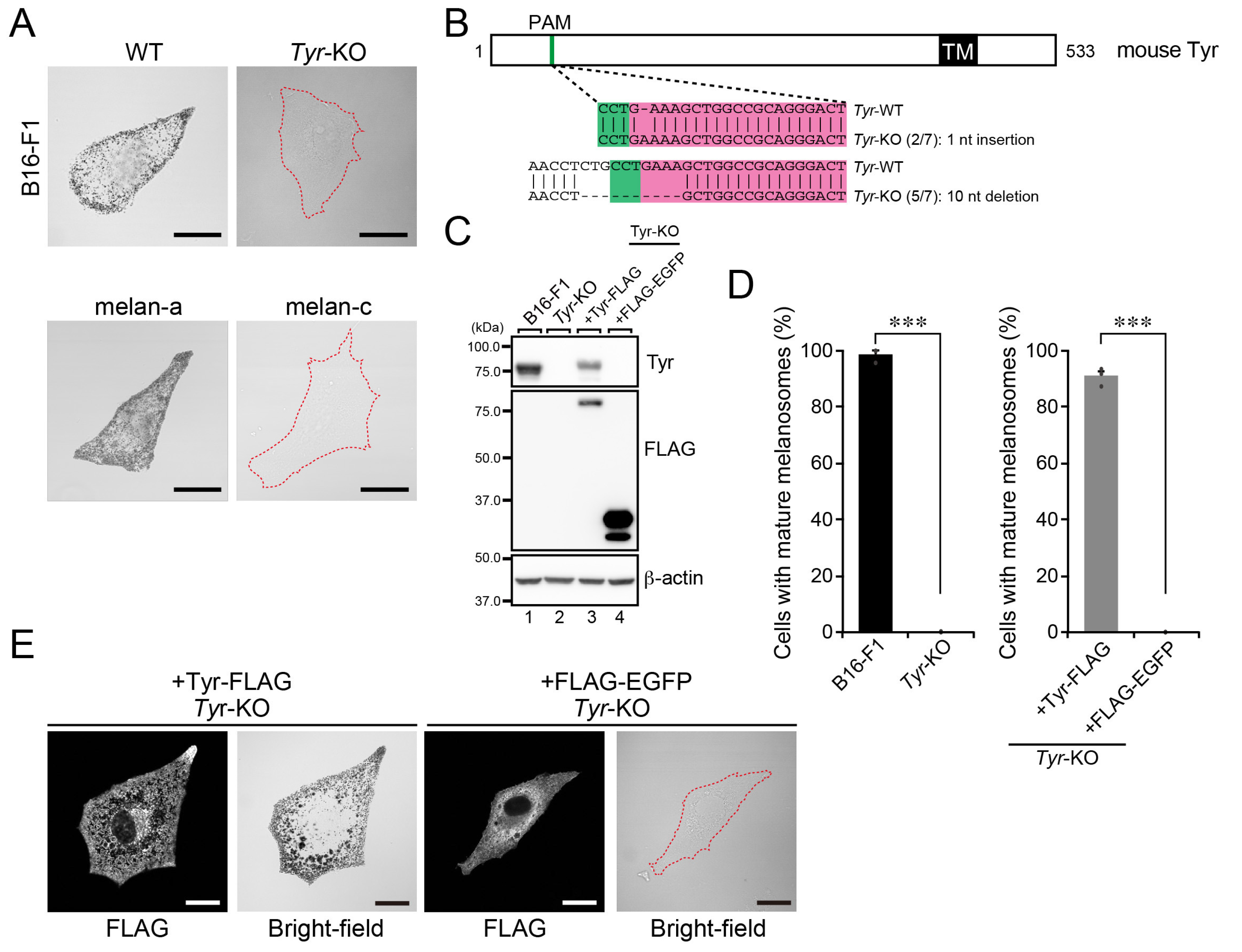
2.1. Establishment and Analysis of Tyr-KO B16-F1 Cells
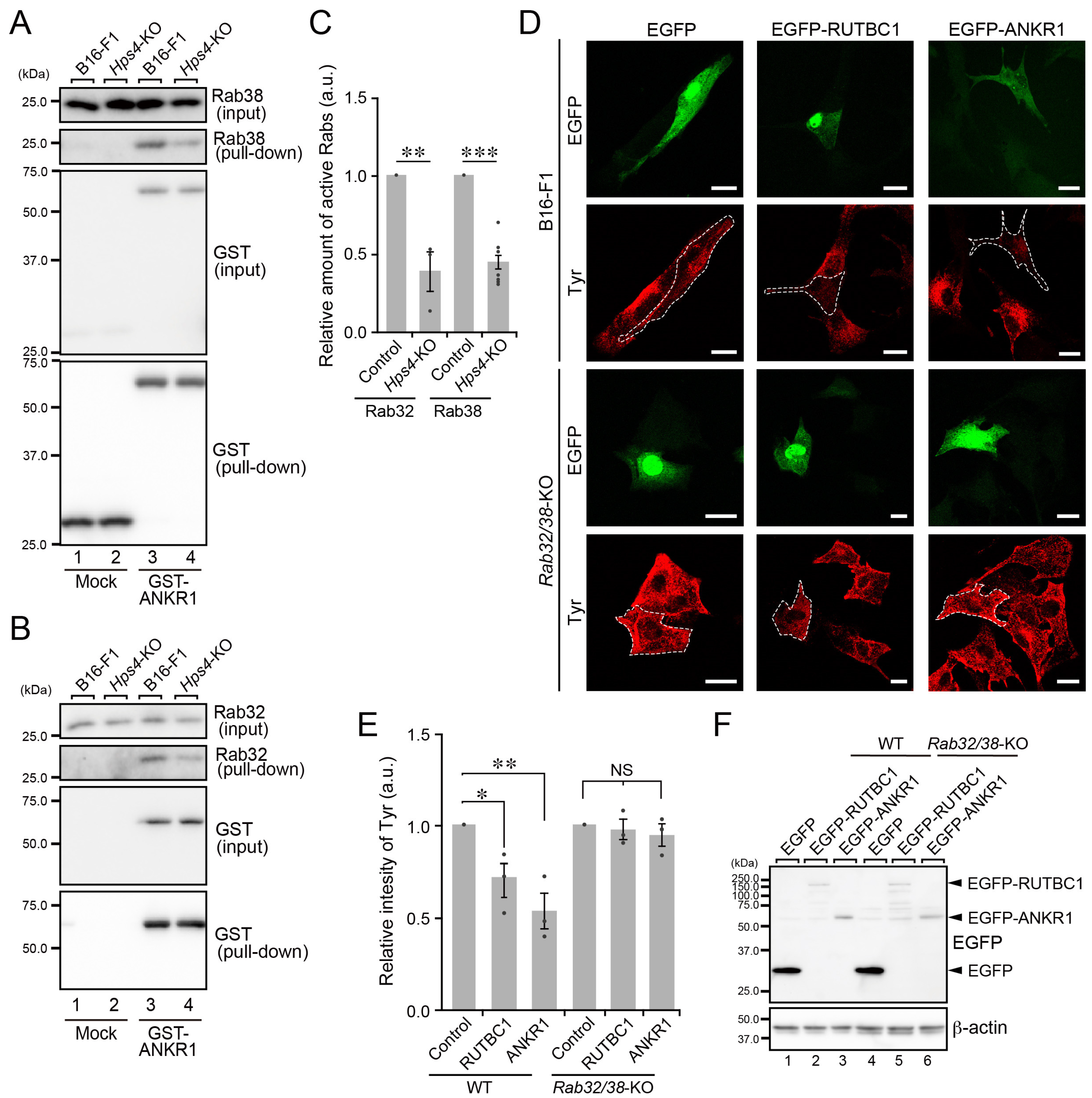
2.2. Normal Melanin Content and Distribution of Melanogenic Enzymes in Hps4-KO B16-F1 Cells
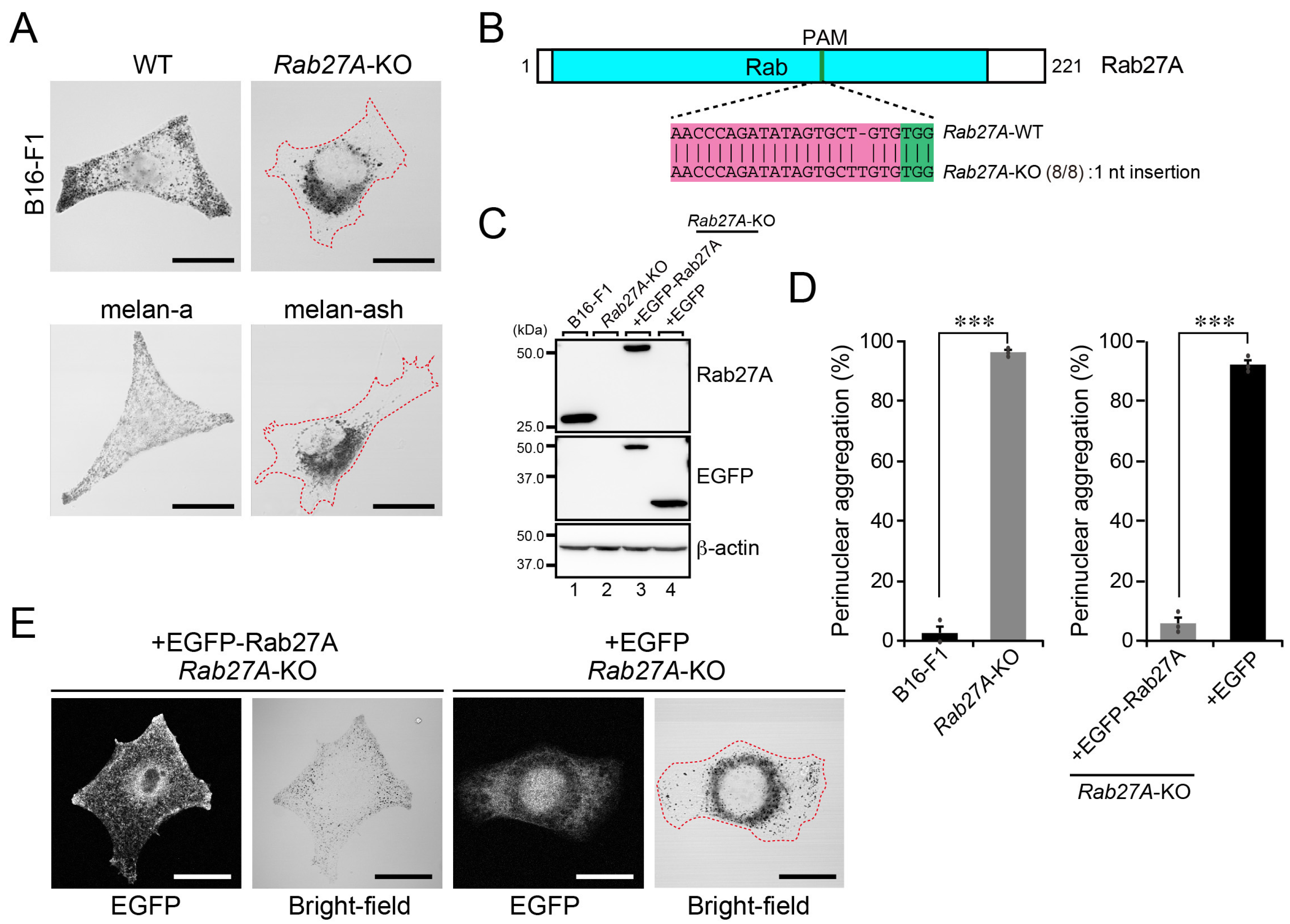
2.3. A Perinuclear Melanosome Aggregation Phenotype of Rab27A-KO B16-F1 Cells
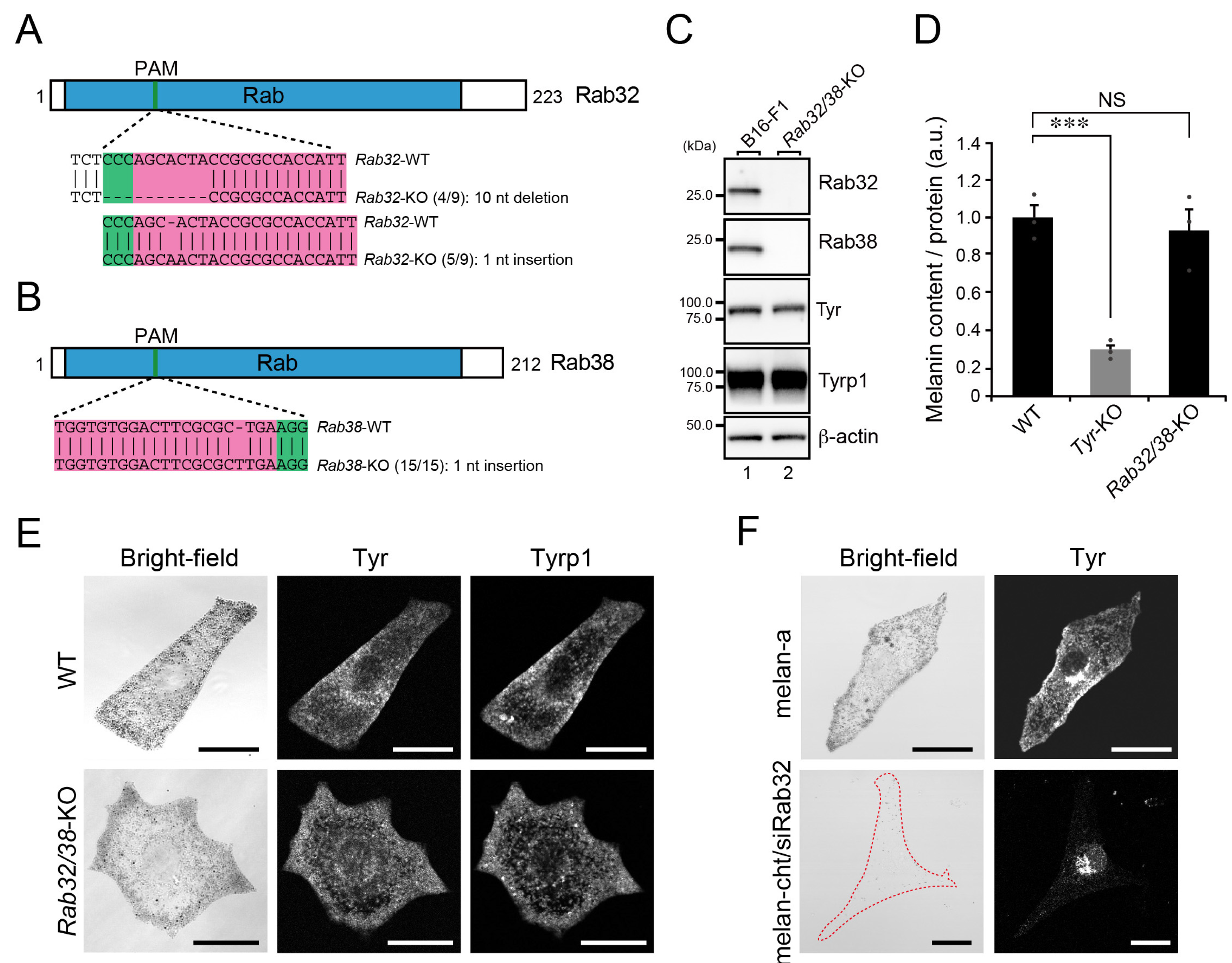
2.4. Rab32/38-Independent Melanogenic Enzyme Transport to Melanosomes in B16-F1 Cells
2.5. Screening for Other Candidate Rabs Involved in Melanogenic Enzyme Transport in B16-F1 Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Cultures and Transfections
4.3. Immunofluorescence Analysis
4.4. Establishment of B16-F1 KO Cells
4.5. GTP-Rab32/38 Pull-Down Assays in B16-F1 Cells
4.6. Immunoblotting
4.7. Melanin Assays
4.8. Reverse-Transcription (RT)-PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANKR1 | ankyrin repeat domain 1 |
| EGFP | enhanced green fluorescent protein |
| GST | glutathione S-transferase |
| HPS | Hermansky–Pudlak syndrome |
| KD | knockdown |
| KO | knockout |
| NS | not significant |
| PAM | protospacer adjacent motif |
| RT | reverse-transcription |
| sgRNA | single guide RNA |
| siRNA | small interfering RNA |
| Tyr | tyrosinase |
| Tyrp1 | tyrosinase-related protein 1 |
| WT | wild-type |
References
- Raposo, G.; Marks, M.S. Melanosomes: Dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007, 8, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Wasmeier, C.; Hume, A.N.; Bolasco, G.; Seabra, M.C. Melanosomes at a glance. J. Cell Sci. 2008, 121, 3995–3999. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hammer, J.A. Melanosome transfer: It is best to give and receive. Curr. Opin. Cell Biol. 2014, 29, 1–7. [Google Scholar] [CrossRef]
- Bowman, S.L.; Bi-Karchin, J.; Le, L.; Marks, M.S. The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic 2019, 20, 404–435. [Google Scholar] [CrossRef]
- Ohbayashi, N.; Fukuda, M. Recent advances in understanding the molecular basis of melanogenesis in melanocytes. F1000 Research 2020, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Tomita, Y.; Suzuki, T. Genetics of pigmentary disorders. Am. J. Med. Genet. C Semin. Med. Genet. 2004, 131C, 75–81. [Google Scholar] [CrossRef]
- Van Gele, M.; Dynoodt, P.; Lambert, J. Griscelli syndrome: A model system to study vesicular trafficking. Pigment Cell Melanoma Res. 2009, 22, 268–282. [Google Scholar] [CrossRef]
- Wei, A.H.; Li, W. Hermansky-Pudlak syndrome: Pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 2013, 26, 176–192. [Google Scholar] [CrossRef]
- Sitaram, A.; Marks, M.S. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology 2012, 27, 85–99. [Google Scholar] [CrossRef]
- Bennett, D.C.; Cooper, P.J.; Dexter, T.J.; Devlin, L.M.; Heasman, J.; Nester, B. Cloned mouse melanocyte lines carrying the germline mutations albino and brown: Complementation in culture. Development 1989, 105, 379–385. [Google Scholar] [CrossRef]
- Suzuki, T.; Li, W.; Zhang, Q.; Karim, A.; Novak, E.K.; Sviderskaya, E.V.; Hill, S.P.; Bennett, D.C.; Levin, A.V.; Nieuwenhuis, H.K.; et al. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat. Genet. 2002, 30, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Ali, B.R.; Wasmeier, C.; Lamoreux, L.; Strom, M.; Seabra, M.C. Multiple regions contribute to membrane targeting of Rab GTPases. J. Cell Sci. 2004, 117, 6401–6412. [Google Scholar] [CrossRef] [PubMed]
- Bennet, D.C.; Cooper, P.I.; Hart, I.R. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int. J. Cancer 1987, 39, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Fidler, I.J. Selection of successive tumour lines for metastasis. Nat. New Biol. 1973, 242, 148–149. [Google Scholar] [CrossRef]
- Gerondopoulos, A.; Langemeyer, L.; Liang, J.R.; Linford, A.; Barr, F.A. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 2012, 22, 2135–2139. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef]
- Pfeffer, S.R. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 2013, 25, 414–419. [Google Scholar] [CrossRef]
- Homma, Y.; Hiragi, S.; Fukuda, M. Rab family of small GTPases: An updated view on their regulation and functions. FEBS J. 2021, 288, 36–55. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, Y.; Kinoshita, R.; Marubashi, S.; Ishida, M.; Fukuda, M. The BLOC-3 subunit HPS4 is required for activation of Rab32/38 GTPases in melanogenesis, but its Rab9 activity is dispensable for melanogenesis. J. Biol. Chem. 2019, 294, 6912–6922. [Google Scholar] [CrossRef] [PubMed]
- Hume, A.N.; Collinson, L.M.; Rapak, A.; Gomes, A.Q.; Hopkins, C.R.; Seabra, M.C. Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J. Cell Biol. 2001, 152, 795–808. [Google Scholar] [CrossRef]
- Bahadoran, P.; Aberdam, E.; Mantoux, F.; Busca, R.; Bille, K.; Yalman, N.; de Saint-Basile, G.; Casaroli-Marano, R.; Ortonne, J.P.; Ballotti, R. Rab27a: A key to melanosome transport in human melanocytes. J. Cell Biol. 2001, 152, 843–850. [Google Scholar] [CrossRef]
- Fukuda, M.; Kuroda, T.S.; Mikoshiba, K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: Implications of a tripartite protein complex for melanosome transport. J. Biol. Chem. 2002, 277, 12432–12436. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.S.; Rao, K.; Zhang, H.; Wang, F.; Sellers, J.R.; Matesic, L.E.; Copeland, N.G.; Jenkins, N.A.; Hammer, J.A., III. Identification of an organelle receptor for myosin-Va. Nat. Cell Biol. 2002, 4, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Loftus, S.K.; Larson, D.M.; Baxter, L.L.; Antonellis, A.; Chen, Y.; Wu, X.; Jiang, Y.; Bittner, M.; Hammer, J.A., III; Pavan, W.J. Mutation of melanosome protein Rab38 in chocolate mice. Proc. Natl. Acad. Sci. USA 2002, 99, 4471–4476. [Google Scholar] [CrossRef]
- Wasmeier, C.; Romao, M.; Plowright, L.; Bannett, D.C.; Raposo, G.; Seabra, M.C. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 2006, 175, 271–281. [Google Scholar] [CrossRef]
- Marubashi, S.; Shimada, H.; Fukuda, M.; Ohbayashi, N. RUTBC1 functions as a GTPase-activating protein for Rab32/38 and regulates melanogenic enzyme trafficking in melanocytes. J. Biol. Chem. 2016, 291, 1427–1440. [Google Scholar] [CrossRef]
- Aguilar, A.; Weber, J.; Boscher, J.; Freund, M.; Ziessel, C.; Eckly, A.; Magnenat, S.; Bourdon, C.; Hechler, B.; Mangin, P.H.; et al. Combined deficiency of RAB32 and RAB38 in the mouse mimics Hermansky-Pudlak syndrome and critically impairs thrombosis. Blood Adv. 2019, 3, 2368–2380. [Google Scholar] [CrossRef]
- Tamura, K.; Ohbayashi, N.; Ishibashi, K.; Fukuda, M. Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J. Biol. Chem. 2011, 286, 7507–7521. [Google Scholar] [CrossRef] [PubMed]
- Nottingham, R.M.; Ganley, I.G.; Barr, F.A.; Lambright, D.G.; Pfeffer, S.R. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J. Biol. Chem. 2011, 286, 33213–33222. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Fukuda, M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013, 14, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Homma, Y.; Kinoshita, R.; Kuchitsu, Y.; Wawro, P.S.; Marubashi, S.; Oguchi, M.E.; Ishida, M.; Fujita, N.; Fukuda, M. Comprehensive knockout analysis of the Rab family GTPases in epithelial cells. J. Cell Biol. 2019, 218, 2035–2050. [Google Scholar] [CrossRef]
- Mahanty, S.; Ravichandran, K.; Chitirala, P.; Prabha, J.; Jani, R.A.; Setty, S.R.G. Rab9A is required for delivery of cargo from recycling endosomes to melanosomes. Pigment Cell Melanoma Res. 2016, 29, 43–59. [Google Scholar] [CrossRef]
- Nag, S.; Rani, S.; Mahanty, S.; Bissig, C.; Arora, P.; Azevedo, C.; Saiardi, A.; van der Sluijs, P.; Delevoye, C.; van Niel, G.; et al. Rab4A organizes endosomal domains for sorting cargo to lysosome-related organelles. J. Cell Sci. 2018, 131, jcs216226. [Google Scholar] [CrossRef]
- Nguyen, T.; Wei, M.L. Hermansky-Pudlak HPS1/pale ear gene regulates epidermal and dermal melanocyte development. J. Investig. Dermatol. 2007, 127, 421–428. [Google Scholar] [CrossRef]
- Schuck, S.; Gerl, M.J.; Ang, A.; Manninen, A.; Keller, P.; Mellman, I.; Simons, K. Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic 2007, 8, 47–60. [Google Scholar] [CrossRef]
- Wang, P.; Liu, H.; Wang, Y.; Liu, O.; Zhang, J.; Gleason, A.; Yang, Z.; Wang, H.; Shi, A.; Grant, B.D. RAB-10 promotes EHBP-1 bridging of filamentous actin and tubular recycling endosomes. PLoS Genet. 2016, 12, e1006093. [Google Scholar] [CrossRef]
- Etoh, K.; Fukuda, M. Rab10 regulates tubular endosome formation through KIF13A and KIF13B motors. J. Cell Sci. 2019, 132, jcs226977. [Google Scholar] [CrossRef]
- Delevoye, C.; Hurbain, I.; Tenza, D.; Sibarita, J.-B.; Uzan-Gafsou, S.; Ohno, H.; Geerts, W.J.; Verkleij, A.J.; Salamero, J.; Marks, M.; et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J. Cell Biol. 2009, 187, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Shakya, S.; Sharma, P.; Bhatt, A.M.; Jani, R.A.; Delevoye, C.; Setty, S.R.G. Rab22A recruits BLOC-1 and BLOC-2 to promote the biogenesis of recycling endosomes. EMBO Rep. 2018, 19, e45918. [Google Scholar] [CrossRef] [PubMed]
- Amaya, C.; Militello, R.D.; Calligaris, S.D.; Colombo, M.I. Rab24 interacts with the Rab7/Rab interacting lysosomal protein complex to regulate endosomal degradation. Traffic 2016, 17, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Munafó, D.B.; Colombo, M.I. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 2002, 3, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Ylä-Anttila, P.; Mikkonen, E.; Happonen, K.E.; Holland, P.; Ueno, T.; Simonsen, A.; Eskelinen, E.L. RAB24 facilitates clearance of autophagic compartments during basal conditions. Autophagy 2015, 11, 1833–1848. [Google Scholar] [CrossRef] [PubMed]
- Kalie, E.; Razi, M.; Tooze, S.A. ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. PLoS ONE 2013, 8, e75313. [Google Scholar] [CrossRef]
- Ho, H.; Kapadia, R.; Al-Tahan, S.; Ahmad, S.; Ganesan, A.K. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J. Biol. Chem. 2011, 286, 12509–12523. [Google Scholar] [CrossRef]
- Kuroda, T.S.; Ariga, H.; Fukuda, M. The actin-binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol. Cell. Biol. 2003, 23, 5245–5255. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishizawa, A.; Maruta, Y.; Fukuda, M. Rab32/38-Dependent and -Independent Transport of Tyrosinase to Melanosomes in B16-F1 Melanoma Cells. Int. J. Mol. Sci. 2022, 23, 14144. https://doi.org/10.3390/ijms232214144
Nishizawa A, Maruta Y, Fukuda M. Rab32/38-Dependent and -Independent Transport of Tyrosinase to Melanosomes in B16-F1 Melanoma Cells. International Journal of Molecular Sciences. 2022; 23(22):14144. https://doi.org/10.3390/ijms232214144
Chicago/Turabian StyleNishizawa, Aya, Yuto Maruta, and Mitsunori Fukuda. 2022. "Rab32/38-Dependent and -Independent Transport of Tyrosinase to Melanosomes in B16-F1 Melanoma Cells" International Journal of Molecular Sciences 23, no. 22: 14144. https://doi.org/10.3390/ijms232214144
APA StyleNishizawa, A., Maruta, Y., & Fukuda, M. (2022). Rab32/38-Dependent and -Independent Transport of Tyrosinase to Melanosomes in B16-F1 Melanoma Cells. International Journal of Molecular Sciences, 23(22), 14144. https://doi.org/10.3390/ijms232214144





