Identification and Analysis of Stress-Associated Proteins (SAPs) Protein Family and Drought Tolerance of ZmSAP8 in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
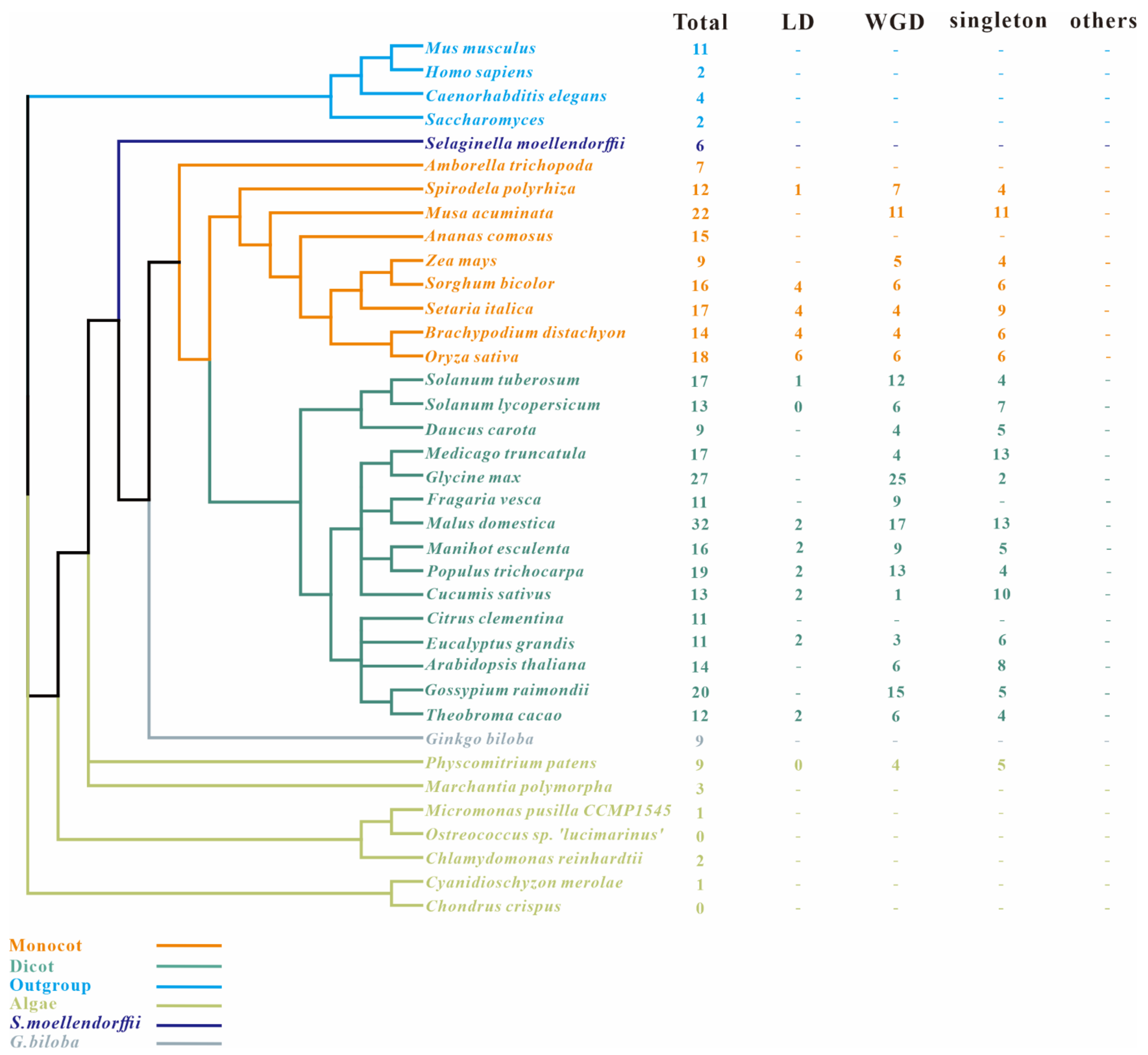
2.1. Identification of Genome-Wide SAP Genes in Maize and 37 Species
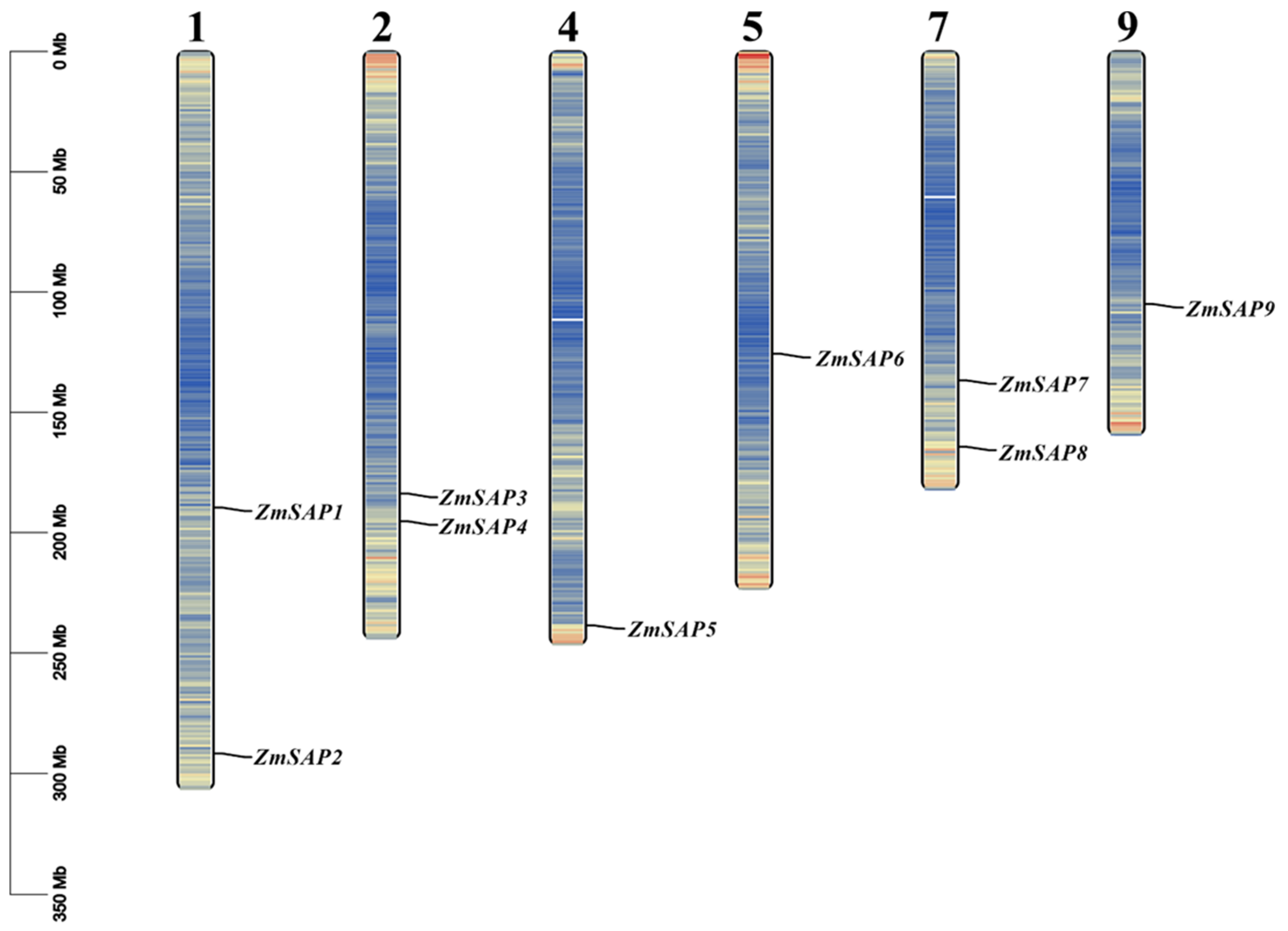
2.2. Identification of Duplication Modes of SAP Members
2.3. Estimation of the Evolutionary Rate of SAPs in Angiosperms
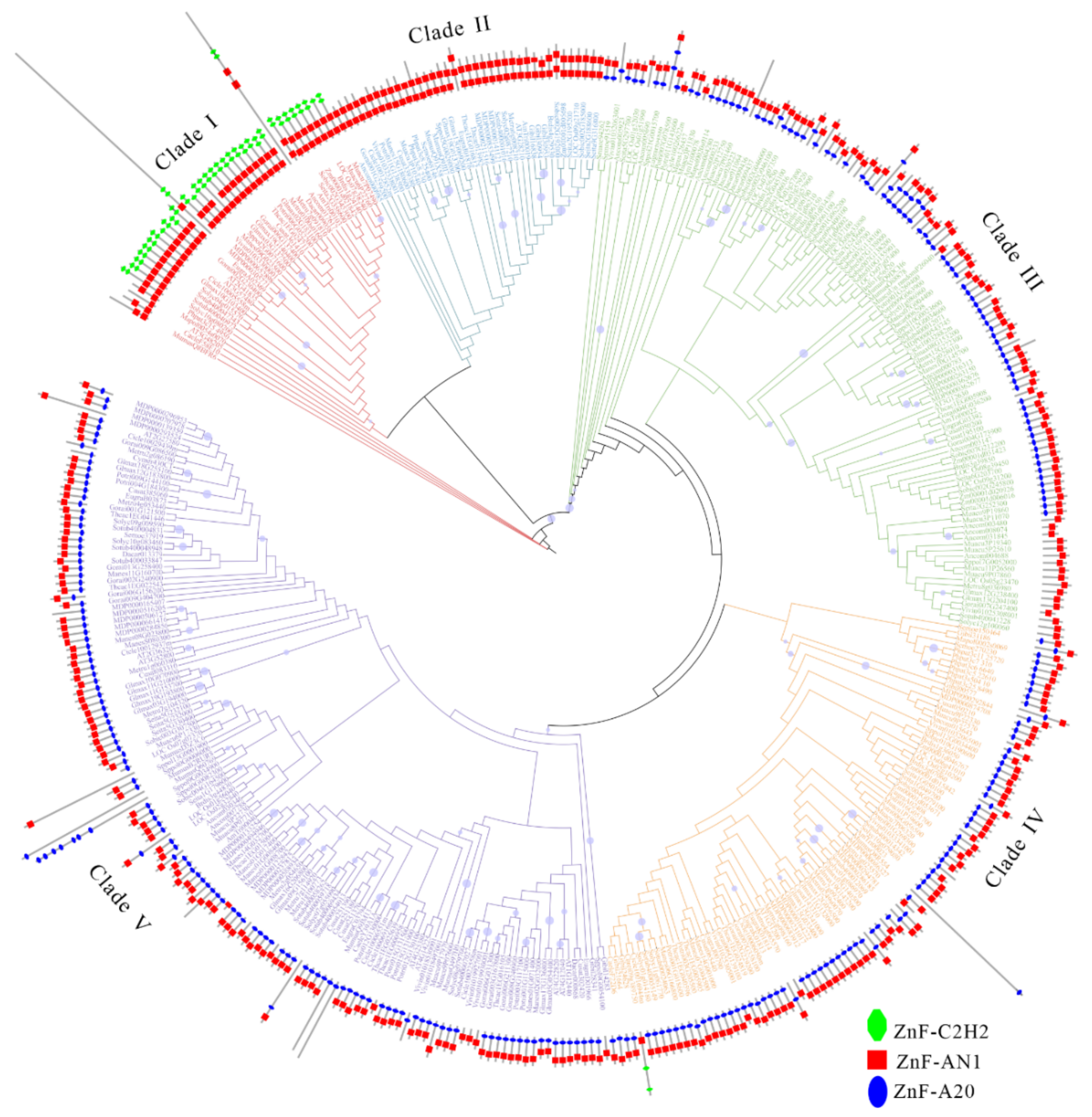
2.4. Phylogenetic Analysis of SAPs in All Studied Species
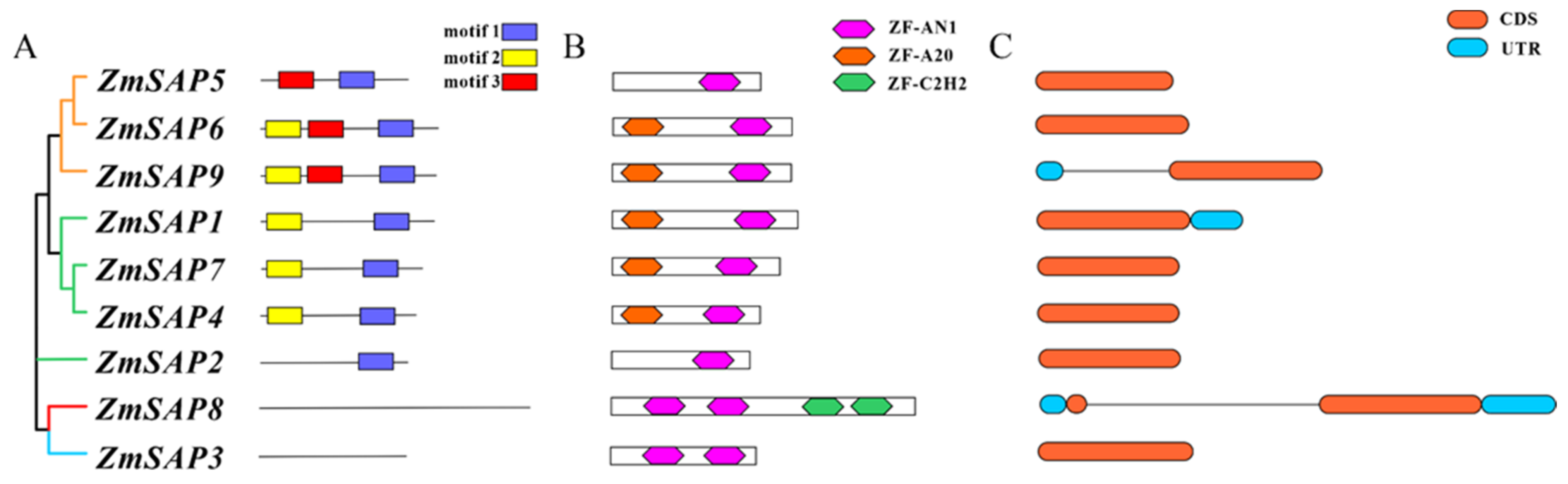
2.5. Gene Structure and Coding Protein Analysis of Maize SAPs
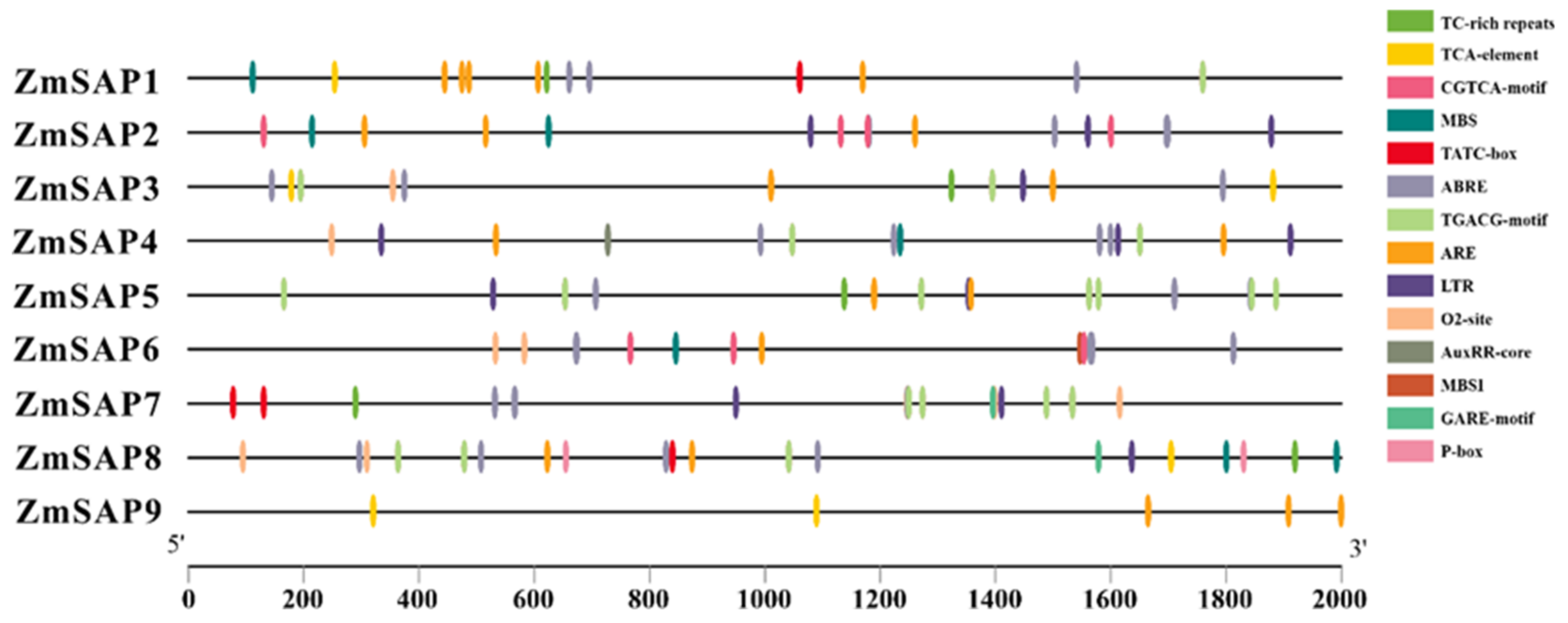
2.6. Cis-Acting Element Analysis of ZmSAPs
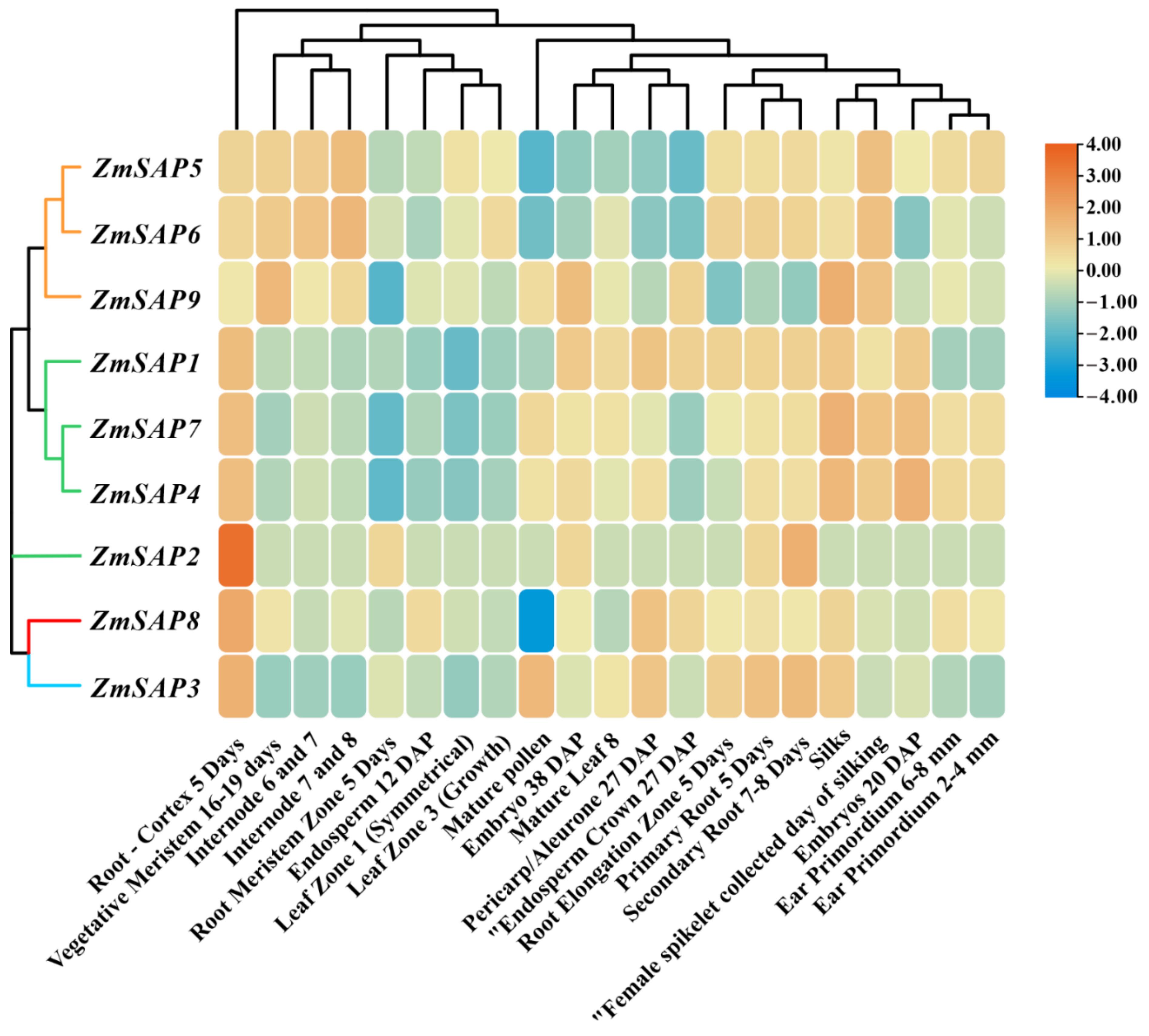
2.7. Expression Patterns of ZmSAPs in Different Tissues
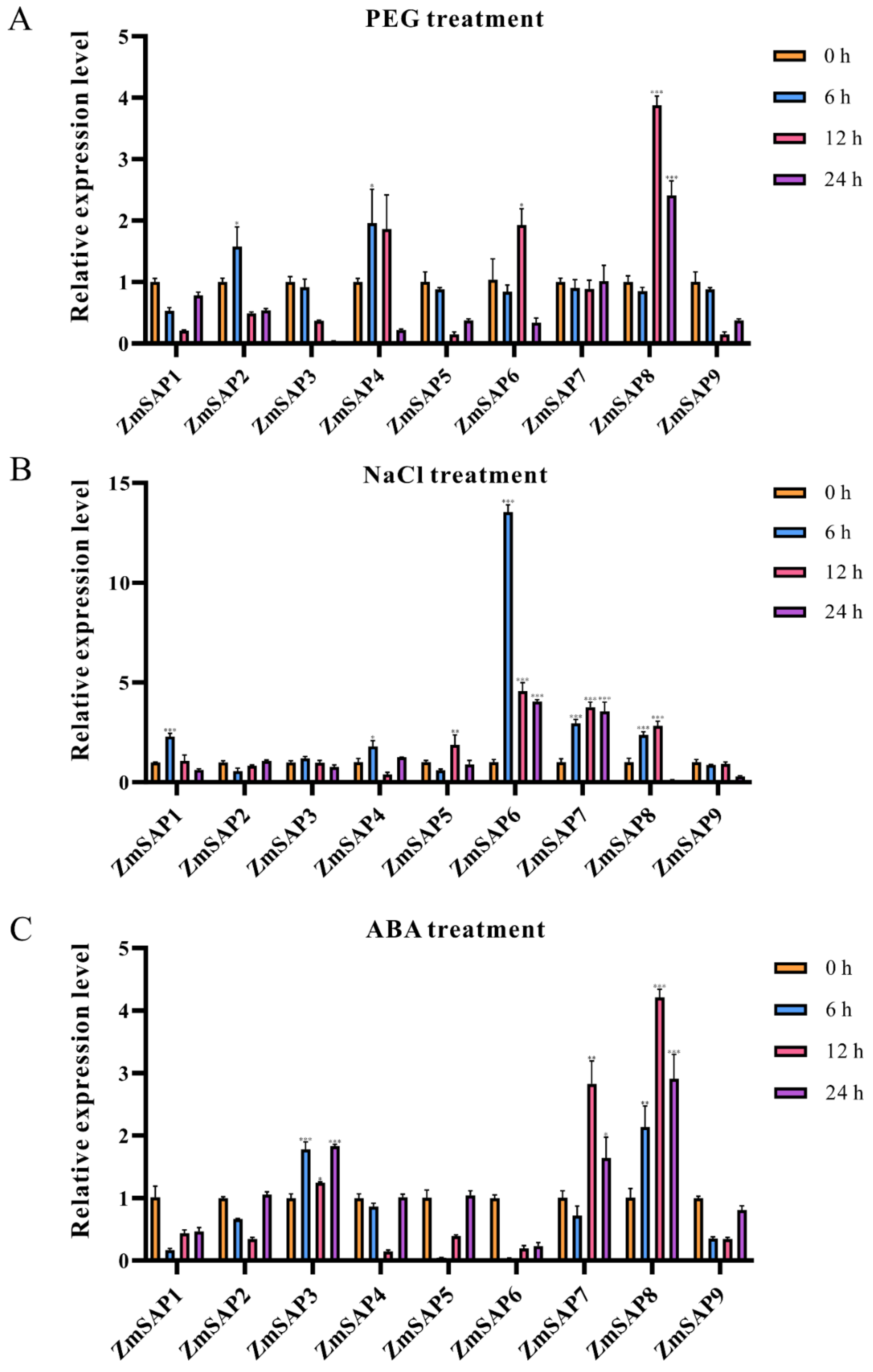
2.8. Effect of Drought Stress Treatment on Relative Gene Expression Levels of ZmSAPs
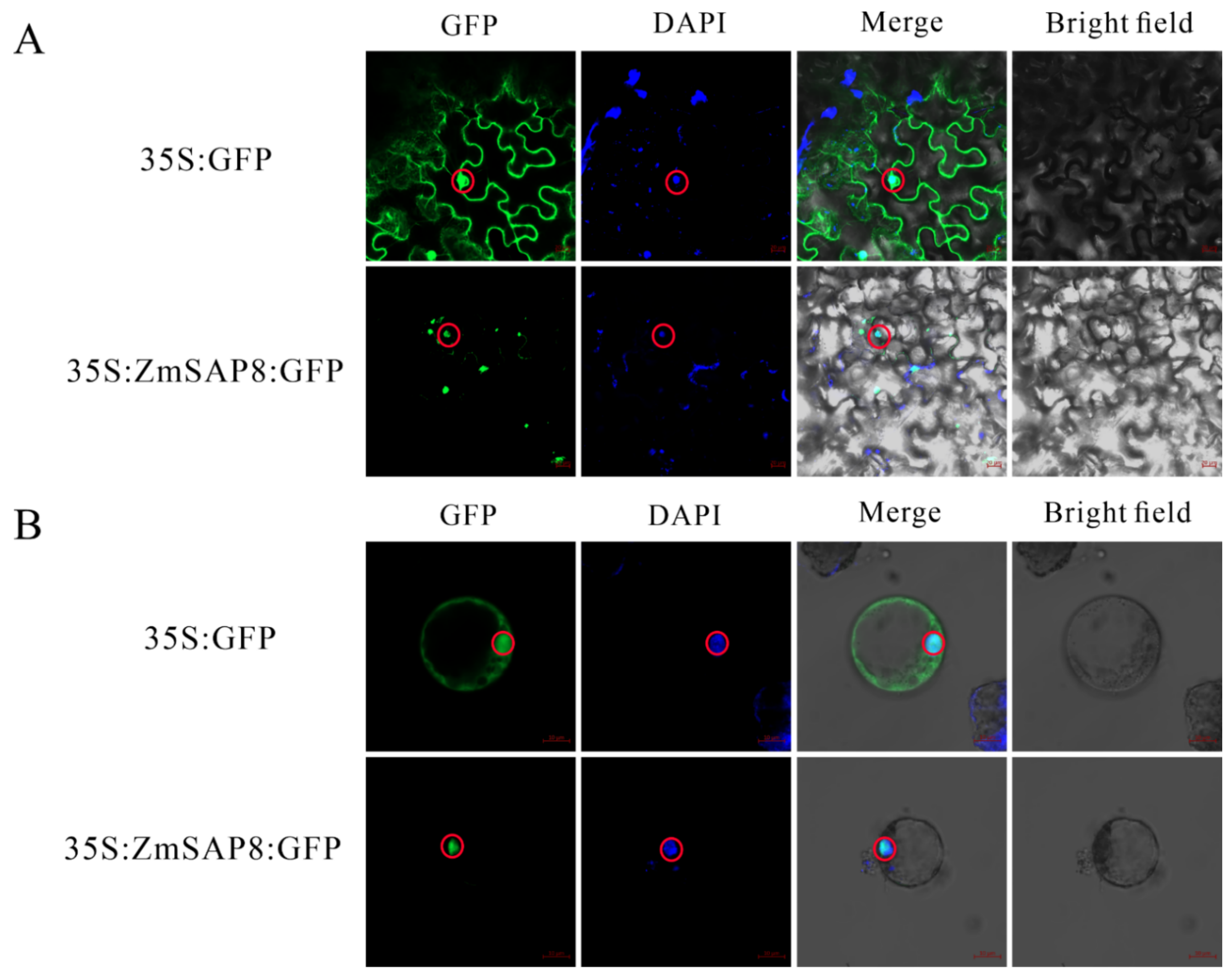
2.9. Subcellular Localization Analysis of ZmSAP8
2.10. Transcriptional Activity Analysis of ZmSAP8 in Yeast Cells
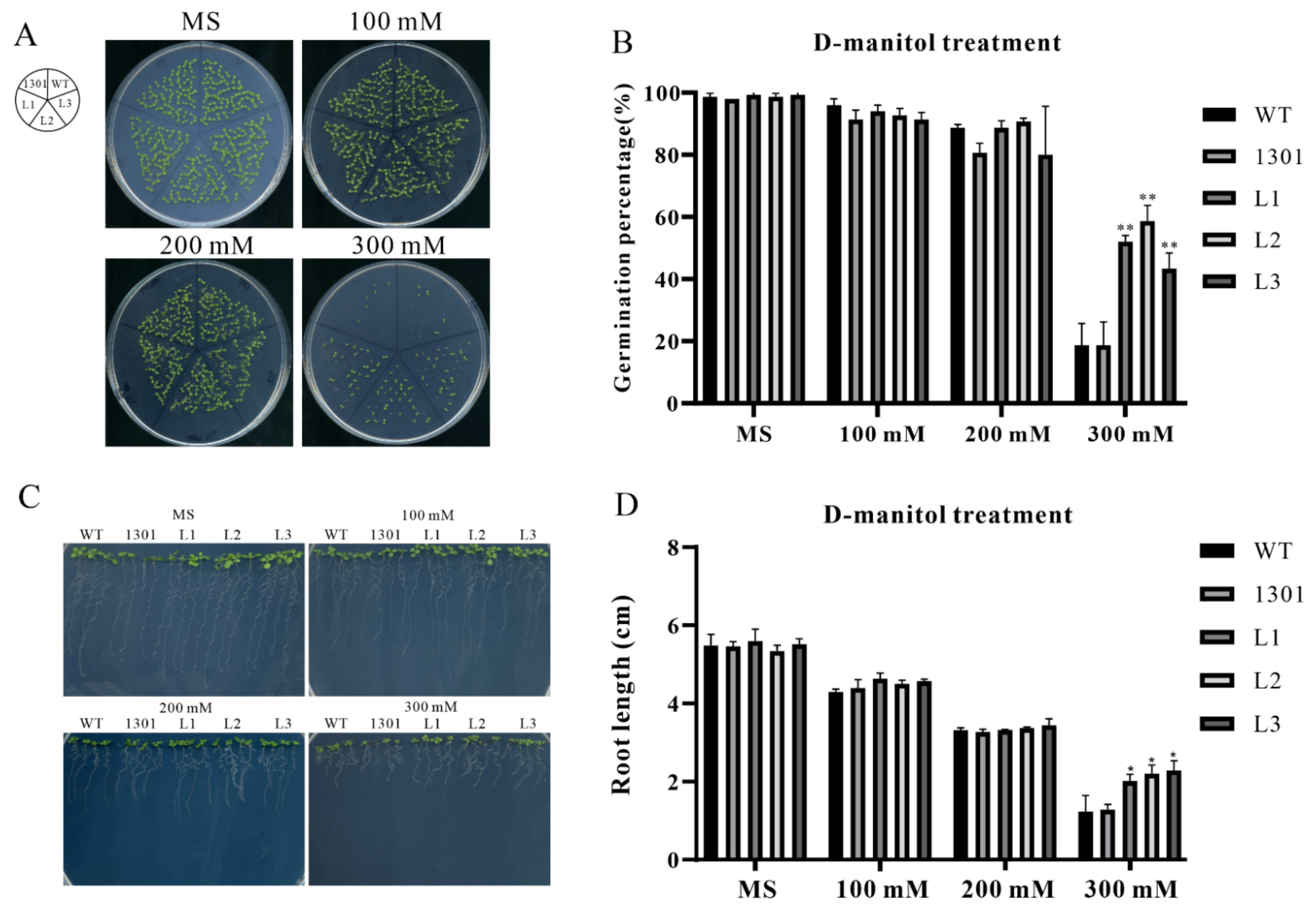
2.11. ZmSAP8 Enhances Drought Resistance in A.thaliana
3. Discussion
4. Method and Materials
4.1. Data Source and Identification of the SAP Gene Family
4.2. Collinearity and Gene Replication Pattern Prediction
4.3. Calculation of the Ratio of Ka to Ks
4.4. Phylogenetic Analysis
4.5. Conserved Motif Analysis
4.6. Expression Analysis of ZmSAPs Genes in Different Tissues
4.7. Plant Material Growth and Stress Treatment
4.8. RNA Extraction and qRT-PCR Analysis
4.9. Subcellular Localization Analysis
4.10. Transcriptional Activation Assay
4.11. Creation of ZmSAP8 Transgenic Arabidopsis
4.12. Statistical Analysis
4.13. Germination Assay
4.14. Root Length Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, L.W.D.; Liu, J.; Dai, L.F.; Deng, Q.W.; Chen, Y.L.; Xie, J.K.; Luo, X.D. Identification and characterisation of cold stress-related proteins in Oryza rufipogon at the seedling stage using label-free quantitative proteomic analysis. Funct. Plant Biol. 2021, 48, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Priya, M.; Dhanker, O.P.; Siddique, K.H.M.; HanumanthaRao, B.; Nair, R.M.; Pandey, S.; Singh, S.; Varshney, R.K.; Prasad, P.V.V.; Nayyar, H. Drought and heat stress-related proteins: An update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 2019, 132, 1607–1638. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J.; Zorb, C.; Geilfus, C.M. Drought and crop yield. Plant Biol. 2021, 23, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ding, Y.F.; Cai, C.; Chen, Z.X.; Zhu, C. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. Plant. 2019, 165, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Gimeno-Gilles, C.; Gervais, M.L.; Planchet, E.; Satour, P.; Limami, A.M.; Lelievre, E. A stress-associated protein containing A20/AN1 zing-finger domains expressed in Medicago truncatula seeds. Plant Physiol. Biochem. 2011, 49, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Sun, P.; Xia, Y.X.; Zheng, G.S.; Sun, J.S.; Jia, H.X. A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress. Int. J. Mol. Sci. 2019, 20, 5782. [Google Scholar] [CrossRef]
- Dong, Q.L.; Duan, D.Y.; Zhao, S.; Xu, B.Y.; Luo, J.W.; Wang, Q.; Huang, D.; Liu, C.H.; Li, C.; Gong, X.Q.; et al. Genome-Wide Analysis and Cloning of the Apple Stress-Associated Protein Gene Family Reveals MdSAP15, Which Confers Tolerance to Drought and Osmotic Stresses in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2478. [Google Scholar] [CrossRef]
- Wang, Z.; Kuang, J.; Han, B.; Chen, S.; Liu, A. Genomic characterization and expression profiles of stress-associated proteins (SAPs) in castor bean (Ricinus communis). Plant Divers. 2021, 43, 152–162. [Google Scholar] [CrossRef]
- Lai, W.; Zhou, Y.; Pan, R.; Liao, L.T.; He, J.C.; Liu, H.J.; Yang, Y.G.; Liu, S.Q. Identification and Expression Analysis of Stress-Associated Proteins (SAPs) Containing A20/AN1 Zinc Finger in Cucumber. Plants 2020, 9, 400. [Google Scholar] [CrossRef]
- Liu, S.X.; Yuan, X.; Wang, Y.Y.; Wang, H.; Wang, J.L.; Shen, Z.H.; Gao, Y.Z.; Cai, J.T.; Li, D.Y.; Song, F.M. Tomato Stress-Associated Protein 4 Contributes Positively to Immunity Against Necrotrophic Fungus Botrytis cinerea. Mol. Plant Microbe 2019, 32, 566–582. [Google Scholar] [CrossRef]
- Vij, S.; Tyagi, A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006, 276, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, P.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Coe, R.A.; Karki, S.; Wanchana, S.; Thakur, V.; Henry, A.; Lin, H.C.; Huang, J.L.; Peng, S.B.; Quick, W.P. Overexpression of OsSAP16 Regulates Photosynthesis and the Expression of a Broad Range of Stress Response Genes in Rice (Oryza sativa L.). PLoS ONE 2016, 11, e0157244. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, H.; He, D.L.; Damaris, R.N.; Yang, P.F. A stress-associated protein OsSAP8 modulates gibberellic acid biosynthesis by reducing the promotive effect of transcription factor OsbZIP58 on OsKO2. J. Exp. Bot. 2022, 73, 2420–2433. [Google Scholar] [CrossRef]
- Giri, J.; Vij, S.; Dansana, P.K.; Tyagi, A.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011, 191, 721–732. [Google Scholar] [CrossRef]
- Kang, M.; Lee, S.; Abdelmageed, H.; Reichert, A.; Lee, H.K.; Fokar, M.; Mysore, K.S.; Allen, R.D. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 2017, 40, 702–716. [Google Scholar] [CrossRef]
- Dixit, A.R.; Dhankher, O.P. A Novel Stress-Associated Protein ‘AtSAP10’ from Arabidopsis thaliana Confers Tolerance to Nickel, Manganese, Zinc, and High Temperature Stress. PLoS ONE 2011, 6, e20921. [Google Scholar] [CrossRef]
- Stroher, E.; Wang, X.J.; Roloff, N.; Klein, P.; Husemann, A.; Dietz, K.J. Redox-Dependent Regulation of the Stress-Induced Zinc-Finger Protein SAP12 in Arabidopsis thaliana. Mol. Plant 2009, 2, 357–367. [Google Scholar] [CrossRef]
- Lloret, A.; Conejero, A.; Leida, C.; Petri, C.; Gil-Munoz, F.; Burgos, L.; Badenes, M.L.; Rios, G. Dual regulation of water retention and cell growth by a stress-associated protein (SAP) gene in Prunus. Sci. Rep. 2017, 7, 332. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, R.; Zhang, Y.; Li, Y.; Yue, Y.Z.; Zhou, T.C.; Wang, C. Comprehensive analysis of the stress associated protein (SAP) gene family in Tamarix hispida and the function of ThSAP6 in salt tolerance. Plant Physiol. Biochem. 2021, 165, 1–9. [Google Scholar] [CrossRef]
- Charrier, A.; Planchet, E.; Cerveau, D.; Gimeno-Gilles, C.; Verdu, I.; Limami, A.M.; Lelievre, E. Overexpression of a Medicago truncatula stress-associated protein gene (MtSAP1) leads to nitric oxide accumulation and confers osmotic and salt stress tolerance in transgenic tobacco. Planta 2012, 236, 567–577. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.P.; Li, J.P.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.Z.; Pan, Q.W.; Wang, X.M.; Chen, Y.; Pan, X.B.; Dong, Y.J. Mutation of the rice AN1-type zinc-finger protein gene ASL4 causes chloroplast development defects and seedling lethality. Plant Biol. 2022, 24, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Hozain, M.; Abdelmageed, H.; Lee, J.; Kang, M.; Fokar, M.; Allen, R.D.; Holaday, A.S. Expression of AtSAP5 in cotton up-regulates putative stress-responsive genes and improves the tolerance to rapidly developing water deficit and moderate heat stress. J. Plant Physiol. 2012, 169, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Han, G.L.; Lu, C.X.; Guo, J.R.; Qiao, Z.Q.; Sui, N.; Qiu, N.W.; Wang, B.S. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Liu, Y.H.; Khan, A.R.; Gan, Y.B. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Yu, X.X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X.H. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Li, Z.Q.; Zhang, J.L.; Li, J.X.; Li, H.J.; Zhang, G.F. The Functional and Regulatory Mechanisms of the Thellungiella salsuginea Ascorbate Peroxidase 6 (TsAPX6) in Response to Salinity and Water Deficit Stresses. PLoS ONE 2016, 11, e0154042. [Google Scholar] [CrossRef]
- Wu, C.H.; Zheng, C.Y.; Ji, G.S.; Jiang, P. Synergistic effects of HSE and LTR elements from hsp70 gene promoter of Ulva prolifera (Ulvophyceae, Chlorophyta) upon temperature induction. J. Phycol. 2019, 55, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, Y.D.; Cai, Y.Y.; Tang, J.J.; Li, Y.; Gai, J.Y. The soybean U-box gene GmPUB6 regulates drought tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 155, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Soboleva, A.; Frolova, N.; Bureiko, K.; Shumilina, J.; Balcke, G.U.; Zhukov, V.A.; Tikhonovich, I.A.; Frolov, A. Dynamics of Reactive Carbonyl Species in Pea Root Nodules in Response to Polyethylene Glycol (PEG)-Induced Osmotic Stress. Int. J. Mol. Sci. 2022, 23, 2726. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Verma, H.; Sarma, R.N. Identification of Markers for Root Traits Related to Drought Tolerance Using Traditional Rice Germplasm. Mol. Biotechnol. 2021, 63, 1280–1292. [Google Scholar] [CrossRef]
- Evans, P.C.; Ovaa, H.; Hamon, M.; Kilshaw, P.J.; Hamm, S.; Bauer, S.; Ploegh, H.L.; Smith, T.S. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 2004, 378, 727–734. [Google Scholar] [CrossRef]
- Giri, J.; Dansana, P.K.; Kothari, K.S.; Sharma, G.; Vij, S.; Tyagi, A.K. SAPs as novel regulators of abiotic stress response in plants. Bioessays 2013, 35, 639–648. [Google Scholar] [CrossRef]
- Ben Saad, R.; Ben Hsouna, A.; Saibi, W.; Ben Hamed, K.; Brini, F.; Ghneim-Herrera, T. A stress-associated protein, LmSAP, from the halophyte Lobularia maritima provides tolerance to heavy metals in tobacco through increased ROS scavenging and metal detoxification processes. J. Plant Physiol. 2018, 231, 234–243. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Jeyasri, R.; Selvaraj, A.; Kalaiyarasi, D.; Aruni, W.; Pandian, S.T.K.; Ramesh, M. Global transcriptome analysis of novel stress associated protein (SAP) genes expression dynamism of combined abiotic stresses in Oryza sativa (L.). J. Biomol. Struct. Dyn. 2021, 39, 2106–2117. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef]
- Schranz, M.E.; Mohammadin, S.; Edger, P.P. Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Ezoe, A.; Shirai, K.; Hanada, K. Degree of Functional Divergence in Duplicates Is Associated with Distinct Roles in Plant Evolution. Mol. Biol. Evol. 2021, 38, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.H.; Yin, H.; Qi, K.J.; Li, L.T.; Wang, R.Z.; Zhang, S.L.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Qin, Q.Q.; Chen, L.; Long, Y.; Song, N.N.; Jiang, H.Y.; Si, W.N. Characterization and phylogenetic analysis of multiple C2 domain and transmembrane region proteins in maize. Bmc Plant Biol. 2022, 22, 388. [Google Scholar] [CrossRef] [PubMed]
- Pophaly, S.D.; Tellier, A. Population Level Purifying Selection and Gene Expression Shape Subgenome Evolution in Maize. Mol. Biol. Evol. 2015, 32, 3226–3235. [Google Scholar] [CrossRef][Green Version]
- Li, J.L.; Han, G.L.; Sun, C.F.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Sheshadri, S.A.; Nishanth, M.J.; Simon, B. Stress-Mediated cis-Element Transcription Factor Interactions Interconnecting Primary and Specialized Metabolism in planta. Front. Plant Sci. 2016, 7, 1725. [Google Scholar] [CrossRef]
- Wang, T.; Guo, J.; Peng, Y.Q.; Lyu, X.G.; Liu, B.; Sun, S.Y.; Wang, X.L. Light-induced mobile factors from shoots regulate rhizobium-triggered soybean root nodulation. Science 2021, 374, 65–71. [Google Scholar] [CrossRef]
- Lin, P.; Dong, T.; Chen, W.L.; Zou, N.X.; Chen, Y.L.; Li, Y.Q.; Chen, K.L.; Wang, M.Y.; Liu, J.F. Expression Analysis of MaTGA8 Transcription Factor in Banana and Its Defence Functional Analysis by Overexpression in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 9344. [Google Scholar] [CrossRef]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Portwood, J.L.; Woodhouse, M.R.; Cannon, E.K.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Walsh, J.R.; Sen, T.Z.; Cho, K.T.; Schott, D.A.; et al. MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Res. 2019, 47, D1146–D1154. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Zhao, Y.P.; Zhang, H.; Fan, G.Y.; Liu, X.; Zhou, W.B.; Shi, C.C.; Wang, J.H.; Liu, W.Q.; Liang, X.M.; et al. Draft genome of the living fossil Ginkgo biloba. Gigascience 2016, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Yau, S.S.T.; Mao, W.G.; Benson, M.; He, R.L. Distinguishing Proteins From Arbitrary Amino Acid Sequences. Sci. Rep.-Uk 2015, 5, 7972. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Mello, B. Estimating TimeTrees with MEGA and the TimeTree Resource. Mol. Biol. Evol. 2018, 35, 2334–2342. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Mitteer, D.R.; Greer, B.D.; Fisher, W.W.; Cohrs, V.L. Teaching behavior technicians to create publication-quality, single-case design graphs in graphpad prism 7. J. Appl. Behav. Anal. 2018, 51, 998–1010. [Google Scholar] [CrossRef]
- Kumari, K.; Rai, M.P.; Bansal, N.; Prashat, G.R.; Kumari, S.; Srivathsa, R.; Dahuja, A.; Sachdev, A.; Praveen, S.; Vinutha, T. Study of subcellular localization of Glycine max γ-tocopherol methyl transferase isoforms in N. benthamiana. 3 Biotech 2020, 10, 110. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Cao, J.; Yao, D.; Lin, F.; Jiang, M. PEG-mediated transient gene expression and silencing system in maize mesophyll protoplasts: A valuable tool for signal transduction study in maize. Acta Physiol. Plant. 2014, 36, 1271–1281. [Google Scholar] [CrossRef]
- Chazotte, B. Labeling nuclear DNA using DAPI. Cold Spring Harb. Protoc. 2011, 2011, pdb.prot5556. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.A.; Ji, W.; Zhu, Y.M.; Gao, P.; Li, Y.; Cai, H.; Bai, X.; Guo, D.J. GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 2010, 61, 2519–2533. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Electroporation. Cold Spring Harb. Protoc. 2019, 2019, pdb.top096271. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.J.; Mott, E.K.; Parsley, K.; Aspinall, S.; Gray, J.C.; Cottage, A. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2006, 2, 19. [Google Scholar] [CrossRef]



| Gene Name | v4 Gene ID | Chromosomal Location | Amino Acid Length | Mw/Da a | pI b | SAP Domain | Duplication Type c | |
|---|---|---|---|---|---|---|---|---|
| AN1 | A20 | |||||||
| ZmSAP1 | Zm00001d031423 | Chr1:189593205-189593937 | 176 | 18,749.01 | 9.12 | 1 | 1 | WGD |
| ZmSAP2 | Zm00001d034389 | Chr1:291685815-291686300 | 161 | 16,678.85 | 9.53 | 1 | 0 | / |
| ZmSAP3 | Zm00001d005698 | Chr2:183790605-183791147 | 180 | 19,399.08 | 9.08 | 2 | 0 | / |
| ZmSAP4 | Zm00001d006016 | Chr2:195298613-195299098 | 161 | 16,782.85 | 9.19 | 1 | 1 | WGD |
| ZmSAP5 | Zm00001d053671 | Chr4:238612589-238613017 | 142 | 15,272.56 | 8.8 | 1 | 0 | WGD |
| ZmSAP6 | Zm00001d015842 | Chr5:125657037-125657552 | 171 | 18,291.03 | 8.28 | 1 | 1 | WGD |
| ZmSAP7 | Zm00001d020926 | Chr7:136725907-136726398 | 163 | 17,195.35 | 9.45 | 1 | 1 | WGD |
| ZmSAP8 | Zm00001d021842 | Chr7:164334705-164337536 | 290 | 32,038.18 | 8.58 | 2 | 0 | / |
| ZmSAP9 | Zm00001d046767 | Chr9:104954887-104960193 | 174 | 18,409.12 | 8.48 | 1 | 1 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, A.; Qin, Q.; Liu, C.; Zhang, J.; Yu, B.; Cheng, Y.; Wang, S.; Tang, J.; Si, W. Identification and Analysis of Stress-Associated Proteins (SAPs) Protein Family and Drought Tolerance of ZmSAP8 in Transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 14109. https://doi.org/10.3390/ijms232214109
Su A, Qin Q, Liu C, Zhang J, Yu B, Cheng Y, Wang S, Tang J, Si W. Identification and Analysis of Stress-Associated Proteins (SAPs) Protein Family and Drought Tolerance of ZmSAP8 in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2022; 23(22):14109. https://doi.org/10.3390/ijms232214109
Chicago/Turabian StyleSu, Anqi, Qianqian Qin, Chao Liu, Jiajun Zhang, Bingxin Yu, Yifeng Cheng, Sijia Wang, Jiawen Tang, and Weina Si. 2022. "Identification and Analysis of Stress-Associated Proteins (SAPs) Protein Family and Drought Tolerance of ZmSAP8 in Transgenic Arabidopsis" International Journal of Molecular Sciences 23, no. 22: 14109. https://doi.org/10.3390/ijms232214109
APA StyleSu, A., Qin, Q., Liu, C., Zhang, J., Yu, B., Cheng, Y., Wang, S., Tang, J., & Si, W. (2022). Identification and Analysis of Stress-Associated Proteins (SAPs) Protein Family and Drought Tolerance of ZmSAP8 in Transgenic Arabidopsis. International Journal of Molecular Sciences, 23(22), 14109. https://doi.org/10.3390/ijms232214109






