Manipulating GA-Related Genes for Cereal Crop Improvement
Abstract
1. Introduction
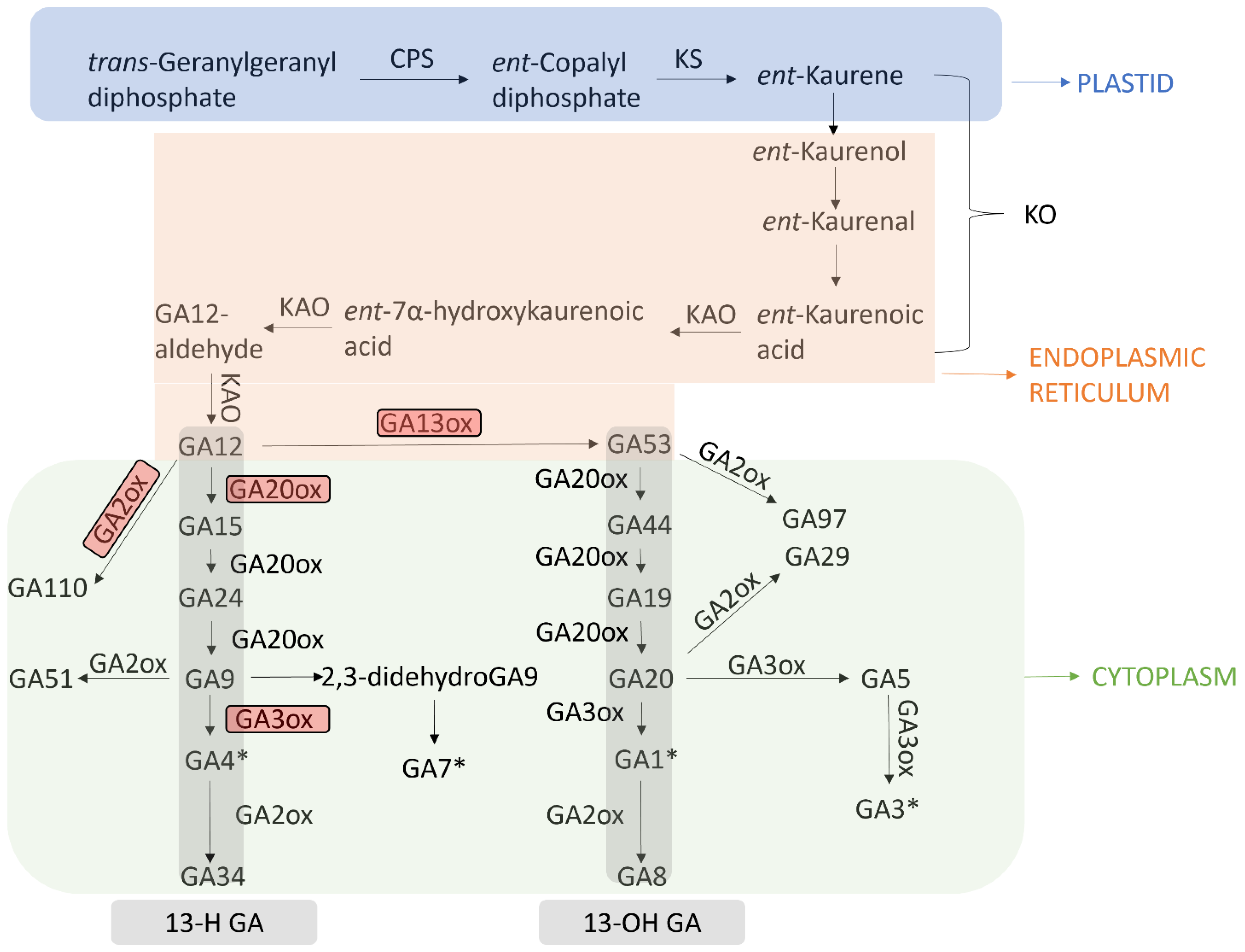
2. Gibberellin Biosynthesis and Deactivation Pathways
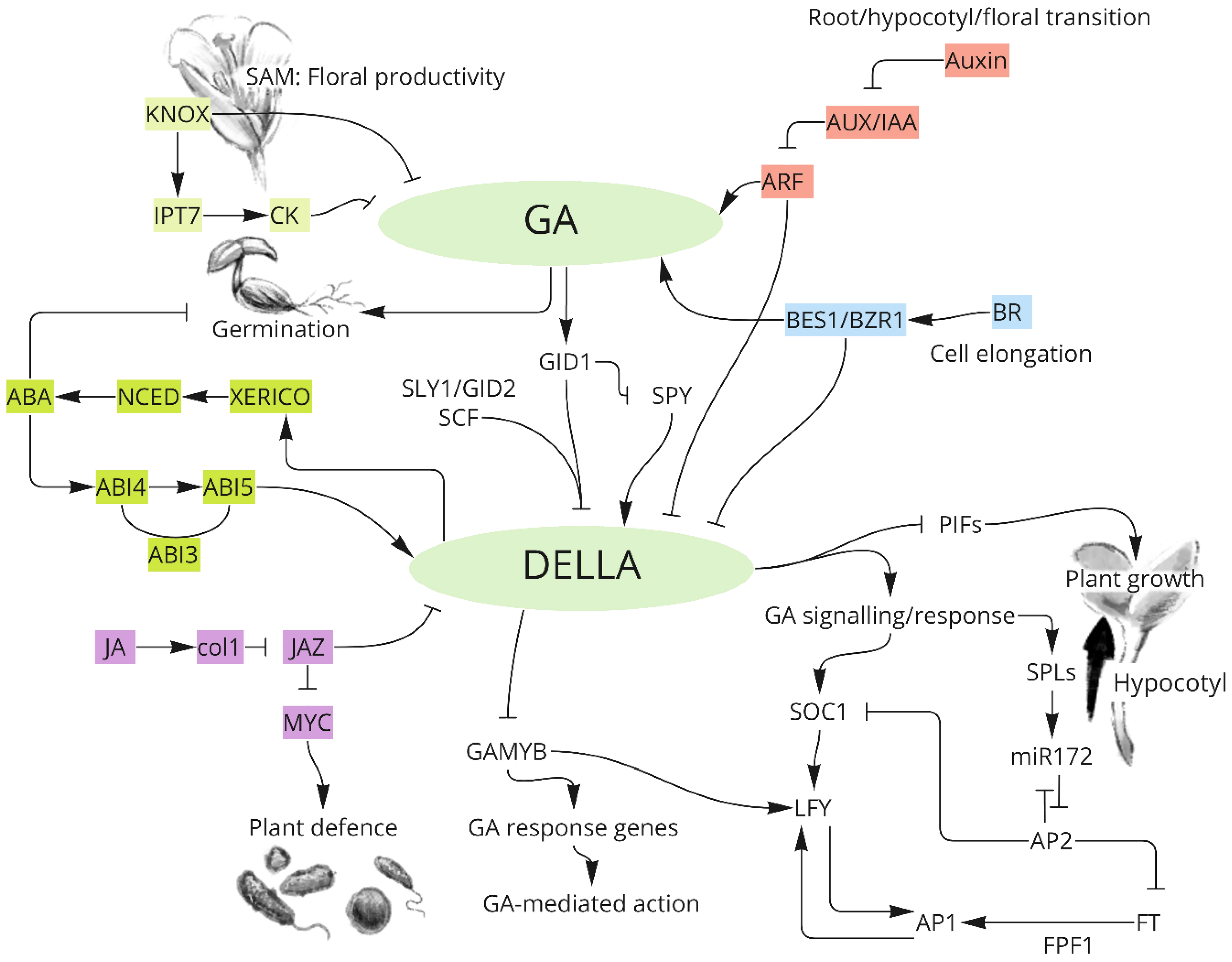
3. GA Signalling Pathway and Network
3.1. GA Signalling Pathway
3.2. Crosstalk between GA and Other Plant Hormones in Plants
3.2.1. Antagonistic Regulation of Phytohormones with GA in Mediating Plant Growth
3.2.2. Synergetic Regulation of Phytohormones with GA in Mediating Plant Growth
4. GA-Related Genes Regulate Agronomic Traits
4.1. Seed Dormancy and Germination
4.2. Plant Stature
4.3. Abiotic Stress Tolerance
4.4. Biotic Stress Tolerance
5. CRISPR/Cas9-Mediated Gene Editing of Gibberellin Genes for Crop Improvement
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ehrlich, P.; Harte, J. Opinion: To feed the world in 2050 will require a global revolution. Proc. Natl. Acad. Sci. USA 2015, 112, 14743–14744. [Google Scholar] [CrossRef]
- Razzaq, A.; Wani, S.H.; Saleem, F.; Yu, M.; Zhou, M.; Shabala, S. Rewilding crops for climate resilience: Economic analysis and de novo domestication strategies. J. Exp. Bot. 2021, 72, 6123–6139. [Google Scholar] [CrossRef]
- Khush, G.S. Green revolution: Preparing for the 21st century. Genome 1999, 42, 646–655. [Google Scholar] [CrossRef]
- Khush, G.S. Modern varieties—Their real contribution to food supply and equity. GeoJournal 1995, 35, 275–284. [Google Scholar] [CrossRef]
- Sakamoto, T.; Matsuoka, M. Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 2004, 15, 144–147. [Google Scholar] [CrossRef]
- Sittampalam, P.; Kuruppu, K. Annual Report for 1965. J. Ceylon Branch R. Asiat. Soc. Great Br. Irel. 1966, 1, 104–105. [Google Scholar]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef]
- Barua, U.M.; Chalmers, K.J.; Thomas, W.T.; Hackett, C.A.; Lea, V.; Jack, P.; Forster, B.P.; Waugh, R.; Powell, W. Molecular mapping of genes determining height, time to heading, and growth habit in barley (Hordeum vulgare). Genome 1993, 36, 1080–1087. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Xu, Y.; Jia, Q.; Zhou, G.; Zhang, X.Q.; Angessa, T.; Broughton, S.; Yan, G.; Zhang, W.; Li, C. Characterization of the sdw1 semi-dwarf gene in barley. BMC Plant Biol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Jia, Q.; Li, C.; Shang, Y.; Zhu, J.; Hua, W.; Wang, J.; Yang, J.; Zhang, G. Molecular characterization and functional analysis of barley semi-dwarf mutant Riso no. 9265. BMC Genom. 2015, 16, 927. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Steber, C.M. GA signaling is essential for the embryo-to-seedling transition during Arabidopsis seed germination, a ghost story. Plant Signal. Behav. 2020, 15, 1705028. [Google Scholar] [CrossRef]
- Smith, B.R.; Njardarson, J.T. [2.2.2]- to [3.2.1]-Bicycle Skeletal Rearrangement Approach to the Gibberellin Family of Natural Products. Org. Lett. 2018, 20, 2993–2996. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Kamiya, Y. Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000, 41, 251–257. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Kasahara, H.; Hanada, A.; Kuzuyama, T.; Takagi, M.; Kamiya, Y.; Yamaguchi, S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 2002, 277, 45188–45194. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Koornneef, M.; van der Veen, J.H. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1980, 58, 257–263. [Google Scholar] [CrossRef]
- Keeling, C.I.; Dullat, H.K.; Yuen, M.; Ralph, S.G.; Jancsik, S.; Bohlmann, J. Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol. 2010, 152, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef]
- Xu, M.; Ross Wilderman, P.; Morrone, D.; Xu, J.; Roy, A.; Margis-Pinheiro, M.; Upadhyaya, N.M.; Coates, R.M.; Peters, R.J. Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 2007, 68, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Regnault, T.; Davière, J.M.; Achard, P. Long-distance transport of endogenous gibberellins in Arabidopsis. Plant Signal. Behav. 2016, 11, e1110661. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mizutani, M.; Ohta, D. Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 2010, 61, 291–315. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Sullivan, J.A.; Mould, R.M.; Gray, J.C.; Peacock, W.J.; Dennis, E.S. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001, 28, 201–208. [Google Scholar] [CrossRef]
- Morrone, D.; Chen, X.; Coates, R.M.; Peters, R.J. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J. 2010, 431, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.W.; Lin, F.; Katsumata, T.; Sugai, Y.; Miyazaki, S.; Kawaide, H.; Okada, K.; Nojiri, H.; Yamane, H. Functional identification of a rice ent-kaurene oxidase, OsKO2, using the Pichia pastoris expression system. Biosci. Biotechnol. Biochem. 2008, 72, 3285–3288. [Google Scholar] [CrossRef]
- Hedden, P. The oxidases of gibberellin biosynthesis: Their function and mechanism. Physiol. Plant. 1997, 101, 709–719. [Google Scholar] [CrossRef]
- Davidson, S.E.; Elliott, R.C.; Helliwell, C.A.; Poole, A.T.; Reid, J.B. The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol. 2003, 131, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Fambrini, M.; Mariotti, L.; Parlanti, S.; Picciarelli, P.; Salvini, M.; Ceccarelli, N.; Pugliesi, C. The extreme dwarf phenotype of the GA-sensitive mutant of sunflower, dwarf2, is generated by a deletion in the ent-kaurenoic acid oxidase1 (HaKAO1) gene sequence. Plant Mol. Biol. 2011, 75, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Igielski, R.; Kępczyńska, E. Gene expression and metabolite profiling of gibberellin biosynthesis during induction of somatic embryogenesis in Medicago truncatula Gaertn. PLoS ONE 2017, 12, e0182055. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A Century of gibberellin research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B.; Rojas, M.C.; Gaskin, P.; Hedden, P. The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J. Biol. Chem. 2002, 277, 21246–21253. [Google Scholar] [CrossRef] [PubMed]
- Lester, D.; Ross, J.; Davies, P.; Reid, J. Mendel’s stem length gene (Le) encodes a gibberellin 3b-hydroxylase. Plant Cell 1997, 9, 1435–1443. [Google Scholar] [CrossRef]
- Pitel, D.W.; Vining, L.C.; Arsenault, G.P. Biosynthesis of gibberellins in Gibberella fujikuroi. The sequence after gibberellin A4. Can. J. Biochem. 1971, 49, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Appleford, N.; Evans, D.; Lenton, J.; Gaskin, P.; Croker, S.; Devos, K.; Phillips, A.; Hedden, P. Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta 2006, 223, 568–582. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Ubeda-Tomás, S.; Gisbert, C.; García-Martínez, J.L.; Moritz, T.; López-Díaz, I. Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant Cell Physiol. 2008, 49, 679–690. [Google Scholar] [CrossRef]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef]
- Elliott, R.C.; Ross, J.J.; Smith, J.J.; Lester, D.R.; Reid, J.B. Feed-forward regulation of gibberellin deactivation in pea. J. Plant Growth Regul. 2001, 20, 87–94. [Google Scholar] [CrossRef]
- Serrani, J.C.; Sanjuán, R.; Ruiz-Rivero, O.; Fos, M.; García-Martínez, J.L. Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 2007, 145, 246–257. [Google Scholar] [CrossRef]
- Han, F.; Zhu, B. Evolutionary analysis of three gibberellin oxidase genes in rice, Arabidopsis, and soybean. Gene 2011, 473, 23–35. [Google Scholar] [CrossRef]
- Zhu, Y.; Nomura, T.; Xu, Y.; Zhang, Y.; Peng, Y.; Mao, B.; Hanada, A.; Zhou, H.; Wang, R.; Li, P.; et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 2006, 18, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; Conde, E.S.N.; Kreis, M.; Deveaux, Y. WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, N.; Girin, T.; Sorefan, K.; Fuentes, S.; Wood, T.A.; Lawrenson, T.; Sablowski, R.; Østergaard, L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010, 24, 2127–2132. [Google Scholar] [CrossRef] [PubMed]
- de Lucas, M.; Davière, J.M.; Rodríguez-Falcón, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blázquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Su, D.; Li, J.; Ying, S.; Deng, H.; He, X.; Zhu, Y.; Li, Y.; Chen, Y.; Pirrello, J.; et al. Overexpression of bHLH95, a basic helix-loop-helix transcription factor family member, impacts trichome formation via regulating gibberellin biosynthesis in tomato. J. Exp. Bot. 2020, 71, 3450–3462. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Digby, J.; Thomas, T.H.; Wareing, P.F. Promotion of cell division in tissue cultures by gibberellic acid. Nature 1964, 203, 547–548. [Google Scholar] [CrossRef]
- MacMillan, J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 2001, 20, 387–442. [Google Scholar] [CrossRef]
- Sachs, R.M.; Bretz, C.; Lang, A. Cell division and gibberellic acid. Exp. Cell Res. 1959, 18, 230–244. [Google Scholar] [CrossRef]
- Hedden, P.; Kamiya, Y. GIBBERELLIN BIOSYNTHESIS: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 431–460. [Google Scholar] [CrossRef] [PubMed]
- Harberd, N.P. Botany. Relieving DELLA restraint. Science 2003, 299, 1853–1854. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.P.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Asano, K.; Tsuji, H.; Kawamura, M.; Mori, H.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 2010, 22, 2680–2696. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Thomas, S.G.; Hu, J.; Steber, C.M.; Sun, T.P. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Jung, H.S.; Dill, A.; Kawaide, H.; Kamiya, Y.; Sun, T.P. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 2001, 13, 1555–1566. [Google Scholar] [CrossRef]
- Lee, S.; Soh, M.-S. How plants make and sense changes in their levels of Gibberellin. J. Plant Biol. 2007, 50, 90–97. [Google Scholar] [CrossRef]
- Li, K.; Yu, R.; Fan, L.M.; Wei, N.; Chen, H.; Deng, X.W. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 2016, 7, 11868. [Google Scholar] [CrossRef]
- Gocal, G.F.; Sheldon, C.C.; Gubler, F.; Moritz, T.; Bagnall, D.J.; MacMillan, C.P.; Li, S.F.; Parish, R.W.; Dennis, E.S.; Weigel, D.; et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 2001, 127, 1682–1693. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Conti, L. Hormonal control of the floral transition: Can one catch them all? Dev. Biol. 2017, 430, 288–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009, 7, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arab. Book 2008, 6, e0103. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Okada, K.; Fukazawa, J.; Takahashi, Y. DELLA-dependent and -independent gibberellin signaling. Plant Signal. Behav. 2018, 13, e1445933. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Binkowski, K.A.; Olszewski, N.E. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 1996, 93, 9292–9296. [Google Scholar] [CrossRef]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N.; et al. ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef]
- Li, Z.; Luo, X.; Wang, L.; Shu, K. ABSCISIC ACID INSENSITIVE 5 mediates light-ABA/gibberellin crosstalk networks during seed germination. J. Exp. Bot. 2022, 73, 4674–4682. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed dormancy and germination-emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, S.; Tang, S.; Yang, W.; Xie, Q. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J. Exp. Bot. 2016, 67, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, S.; Lu, M.; Zhang, Y.; Li, J.; Wang, W.; Wang, P.; Zhang, J.; Hu, Z.; Li, L.; et al. Biosynthesis of DHGA(12) and its roles in Arabidopsis seedling establishment. Nat. Commun. 2019, 10, 1768. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef]
- Boden, S.A.; Weiss, D.; Ross, J.J.; Davies, N.W.; Trevaskis, B.; Chandler, P.M.; Swain, S.M. EARLY FLOWERING3 Regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 2014, 26, 1557–1569. [Google Scholar] [CrossRef]
- Liao, X.; Li, M.; Liu, B.; Yan, M.; Yu, X.; Zi, H.; Liu, R.; Yamamuro, C. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. USA 2018, 115, E11542–E11550. [Google Scholar] [CrossRef]
- Youssef, H.M.; Hansson, M. Crosstalk among hormones in barley spike contributes to the yield. Plant Cell Rep. 2019, 38, 1013–1016. [Google Scholar] [CrossRef]
- Buhrow, L.M.; Cram, D.; Tulpan, D.; Foroud, N.A.; Loewen, M.C. Exogenous abscisic acid and gibberellic acid elicit opposing effects on fusarium graminearum infection in wheat. Phytopathology 2016, 106, 986–996. [Google Scholar] [CrossRef]
- Buhrow, L.M.; Liu, Z.; Cram, D.; Sharma, T.; Foroud, N.A.; Pan, Y.; Loewen, M.C. Wheat transcriptome profiling reveals abscisic and gibberellic acid treatments regulate early-stage phytohormone defense signaling, cell wall fortification, and metabolic switches following Fusarium graminearum-challenge. BMC Genom. 2021, 22, 798. [Google Scholar] [CrossRef]
- Kramell, R.; Atzorn, R.; Schneider, G.; Miersch, O.; Brückner, C.; Schmidt, J.; Sembdner, G.; Parthier, B. Occurrence and identification of jasmonic acid and its amino acid conjugates induced by osmotic stress in barley leaf tissue. J. Plant Growth Regul. 1995, 14, 29. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Jang, G.; Yoon, Y.; Choi, Y.D. Crosstalk with jasmonic acid integrates multiple responses in plant development. Int. J. Mol. Sci. 2020, 21, 305. [Google Scholar] [CrossRef]
- Greenboim-Wainberg, Y.; Maymon, I.; Borochov, R.; Alvarez, J.; Olszewski, N.; Ori, N.; Eshed, Y.; Weiss, D. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 2005, 17, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, N.C.S.; Dornelas, M.C. The roles of gibberellins and cytokinins in plant phase transitions. Trop. Plant Biol. 2021, 14, 11–21. [Google Scholar] [CrossRef]
- Subbaraj, A.K.; Funnell, K.A.; Woolley, D.J. Dormancy and Flowering Are Regulated by the Reciprocal Interaction Between Cytokinin and Gibberellin in Zantedeschia. J. Plant Growth Regul. 2010, 29, 487–499. [Google Scholar] [CrossRef]
- Sato, Y.; Fukuda, Y.; Hirano, H.-Y. Mutations that cause amino acid substitutions at the invariant positions in homeodomain of OSH3 KNOX protein suggest artificial selection during rice domestication. Genes Genet. Syst. 2001, 76, 381–392. [Google Scholar] [CrossRef][Green Version]
- Sakamoto, T.; Sakakibara, H.; Kojima, M.; Yamamoto, Y.; Nagasaki, H.; Inukai, Y.; Sato, Y.; Matsuoka, M. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 2006, 142, 54–62. [Google Scholar] [CrossRef]
- Takei, K.; Sakakibara, H.; Sugiyama, T. Identification of Genes Encoding Adenylate Isopentenyltransferase, a Cytokinin Biosynthesis Enzyme, inArabidopsis thaliana. J. Biol. Chem. 2001, 276, 26405–26410. [Google Scholar] [CrossRef]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Frigerio, M.; Alabadí, D.; Pérez-Gómez, J.; García-Cárcel, L.; Phillips, A.L.; Hedden, P.; Blázquez, M.A. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 2006, 142, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Harberd, N.P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Ben-Targem, M.; Ripper, D.; Bayer, M.; Ragni, L. Auxin and gibberellin signaling cross-talk promotes hypocotyl xylem expansion and cambium homeostasis. J. Exp. Bot. 2021, 72, 3647–3660. [Google Scholar] [CrossRef]
- Yin, C.; Gan, L.; Ng, D.; Zhou, X.; Xia, K. Decreased panicle-derived indole-3-acetic acid reduces gibberellin A1 level in the uppermost internode, causing panicle enclosure in male sterile rice Zhenshan 97A. J. Exp. Bot. 2007, 58, 2441–2449. [Google Scholar] [CrossRef]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Peng, S. Heat-induced phytohormone changes are associated with disrupted early reproductive development and reduced yield in rice. Sci. Rep. 2016, 6, 34978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bai, M.Y.; Chong, K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef]
- Gallego-Bartolomé, J.; Minguet, E.G.; Grau-Enguix, F.; Abbas, M.; Locascio, A.; Thomas, S.G.; Alabadí, D.; Blázquez, M.A. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 13446–13451. [Google Scholar] [CrossRef]
- Li, Q.F.; Wang, C.; Jiang, L.; Li, S.; Sun, S.S.; He, J.X. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 2012, 5, ra72. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Rozhon, W.; Poppenberger, B. Reply: Interaction between Brassinosteroids and Gibberellins: Synthesis or Signaling? In Arabidopsis, Both! Plant Cell 2016, 28, 836–839. [Google Scholar] [CrossRef]
- Zentella, R.; Hu, J.; Hsieh, W.P.; Matsumoto, P.A.; Dawdy, A.; Barnhill, B.; Oldenhof, H.; Hartweck, L.M.; Maitra, S.; Thomas, S.G.; et al. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016, 30, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zheng, J.; Huang, R.; Huang, Y.; Wang, H.; Jiang, L.; Fang, X. Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep. 2016, 35, 2423–2433. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Ueguchi-Tanaka, M.; Sakamoto, T.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Sazuka, T.; Ashikari, M.; Matsuoka, M. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006, 48, 390–402. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Fujisawa, Y.; Kobayashi, M.; Ashikari, M.; Iwasaki, Y.; Kitano, H.; Matsuoka, M. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 2000, 97, 11638–11643. [Google Scholar] [CrossRef]
- Davière, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef]
- Richards, D.E.; King, K.E.; Ait-Ali, T.; Harberd, N.P. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 67–88. [Google Scholar] [CrossRef]
- He, J.; Chen, Q.; Xin, P.; Yuan, J.; Ma, Y.; Wang, X.; Xu, M.; Chu, J.; Peters, R.J.; Wang, G. CYP72A enzymes catalyse 13-hydrolyzation of gibberellins. Nat. Plants 2019, 5, 1057–1065. [Google Scholar] [CrossRef]
- Cao, D.; Hussain, A.; Cheng, H.; Peng, J. Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 2005, 223, 105–113. [Google Scholar] [CrossRef]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002, 16, 646–658. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Olszewski, N.E. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 1993, 5, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef]
- Lange, M.J.; Lange, T. Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nat. Plants 2015, 1, 14025. [Google Scholar] [CrossRef]
- Li, Q.F.; Zhou, Y.; Xiong, M.; Ren, X.Y.; Han, L.; Wang, J.D.; Zhang, C.Q.; Fan, X.L.; Liu, Q.Q. Gibberellin recovers seed germination in rice with impaired brassinosteroid signalling. Plant Sci. 2020, 293, 110435. [Google Scholar] [CrossRef]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Lo, S.F.; Ho, T.D.; Liu, Y.L.; Jiang, M.J.; Hsieh, K.T.; Chen, K.T.; Yu, L.C.; Lee, M.H.; Chen, C.Y.; Huang, T.P.; et al. Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice. Plant Biotechnol. J. 2017, 15, 850–864. [Google Scholar] [CrossRef]
- Wang, Y.; Du, F.; Wang, J.; Li, Y.; Zhang, Y.; Zhao, X.; Zheng, T.; Li, Z.; Xu, J.; Wang, W.; et al. Molecular dissection of the gene OsGA2ox8 conferring osmotic stress tolerance in rice. Int. J. Mol. Sci. 2021, 22, 9107. [Google Scholar] [CrossRef]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Liu, J.H.; Zhao, W.S.; Chen, X.J.; Guo, Z.J.; Peng, Y.L. Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol. Plant Microbe Interact. 2013, 26, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Matsuoka, M.; Kitano, H.; Asano, T.; Kaku, H.; Komatsu, S. gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant Cell Environ. 2006, 29, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Itoh, H.; Shimada, A.; Ueguchi-Tanaka, M.; Kamiya, N.; Hasegawa, Y.; Ashikari, M.; Matsuoka, M. Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J. 2005, 44, 669–679. [Google Scholar] [CrossRef]
- Alghabari, F.; Ihsan, M.Z.; Khaliq, A.; Hussain, S.; Daur, I.; Fahad, S.; Nasim, W. Gibberellin-sensitive Rht alleles confer tolerance to heat and drought stresses in wheat at booting stage. J. Cereal Sci. 2016, 70, 72–78. [Google Scholar] [CrossRef]
- Alghabari, F.; Ihsan, M.Z.; Hussain, S.; Aishia, G.; Daur, I. Effect of Rht alleles on wheat grain yield and quality under high temperature and drought stress during booting and anthesis. Environ. Sci. Pollut. Res. 2015, 22, 15506–15515. [Google Scholar] [CrossRef]
- Tian, X.; Xia, X.; Xu, D.; Liu, Y.; Xie, L.; Hassan, M.A.; Song, J.; Li, F.; Wang, D.; Zhang, Y.; et al. Rht24b, an ancient variation of TaGA2ox-A9, reduces plant height without yield penalty in wheat. New Phytol. 2022, 233, 738–750. [Google Scholar] [CrossRef]
- Filardo, F.; Robertson, M.; Singh, D.P.; Parish, R.W.; Swain, S.M. Functional analysis of HvSPY, a negative regulator of GA response, in barley aleurone cells and Arabidopsis. Planta 2009, 229, 523–537. [Google Scholar] [CrossRef]
- Chandler, P.M. Hormonal regulation of gene expression in the “slender” mutant of barley (Hordeum vulgare L.). Planta 1988, 175, 115–120. [Google Scholar] [CrossRef]
- Chandler, P.M.; Marion-Poll, A.; Ellis, M.; Gubler, F. Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 2002, 129, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Chandler, P.M.; Harding, C.A. ‘Overgrowth’ mutants in barley and wheat: New alleles and phenotypes of the ‘Green Revolution’ DELLA gene. J. Exp. Bot. 2013, 64, 1603–1613. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Zhang, J.; Westcott, S.; Zhang, X.Q.; Bellgard, M.; Lance, R.; Li, C. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct. Integr. Genom. 2009, 9, 255–262. [Google Scholar] [CrossRef]
- Mikołajczak, K.; Kuczyńska, A.; Ogrodowicz, P.; Kiełbowicz-Matuk, A.; Ćwiek-Kupczyńska, H.; Daszkowska-Golec, A.; Szarejko, I.; Surma, M.; Krajewski, P. High-throughput sequencing data revealed genotype-specific changes evoked by heat stress in crown tissue of barley sdw1 near-isogenic lines. BMC Genom. 2022, 23, 177. [Google Scholar] [CrossRef] [PubMed]
- Wiegmann, M.; Maurer, A.; Pham, A.; March, T.J.; Al-Abdallat, A.; Thomas, W.T.B.; Bull, H.J.; Shahid, M.; Eglinton, J.; Baum, M.; et al. Barley yield formation under abiotic stress depends on the interplay between flowering time genes and environmental cues. Sci. Rep. 2019, 9, 6397. [Google Scholar] [CrossRef]
- Agata, A.; Ando, K.; Ota, S.; Kojima, M.; Takebayashi, Y.; Takehara, S.; Doi, K.; Ueguchi-Tanaka, M.; Suzuki, T.; Sakakibara, H.; et al. Diverse panicle architecture results from various combinations of Prl5/GA20ox4 and Pbl6/APO1 alleles. Commun. Biol. 2020, 3, 302. [Google Scholar] [CrossRef]
- Robertson, M.; Swain, S.M.; Chandler, P.M.; Olszewski, N.E. Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 1998, 10, 995–1007. [Google Scholar] [CrossRef][Green Version]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef]
- Zentella, R.; Yamauchi, D.; Ho, T.H. Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 2002, 14, 2289–2301. [Google Scholar] [CrossRef]
- Ogawa, M.; Kusano, T.; Katsumi, M.; Sano, H. Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 2000, 245, 21–29. [Google Scholar] [CrossRef]
- Tian, X.; Wen, W.; Xie, L.; Fu, L.; Xu, D.; Fu, C.; Wang, D.; Chen, X.; Xia, X.; Chen, Q.; et al. Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front. Plant Sci. 2017, 8, 1379. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tian, X.; Xue, L.; Zhang, X.; Yang, S.; Traw, M.B.; Huang, J. CRISPR-based assessment of gene specialization in the gibberellin metabolic pathway in rice. Plant Physiol. 2019, 180, 2091–2105. [Google Scholar] [CrossRef] [PubMed]
- Trethowan, R.M.; Singh, R.P.; Huerta-Espino, J.; Crossa, J.; van Ginkel, M. Coleoptile length variation of near-isogenic Rht lines of modern CIMMYT bread and durum wheats. Field Crop. Res. 2001, 70, 167–176. [Google Scholar] [CrossRef]
- Mickelson, H.R.; Rasmusson, D.C. Genes for short stature in barley. Crop Sci. 1994, 34, 1180–1183. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Verbyla, A.P.; Verbyla, K.L.; Morell, M.K.; Cavanagh, C.R. Use of a large multiparent wheat mapping population in genomic dissection of coleoptile and seedling growth. Plant Biotechnol. J. 2014, 12, 219–230. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, E.; Kirkegaard, J.A.; Rebetzke, G.J. Novel wheat varieties facilitate deep sowing to beat the heat of changing climates. Nat. Clim. Chang. 2022, 12, 291–296. [Google Scholar] [CrossRef]
- Kandemir, N.; Saygili, İ.; Sönmezoğlu, Ö.A.; Yildirim, A. Evaluation of barley semi-dwarf allele sdw1.d in a near isogenic line. Euphytica 2022, 218, 31. [Google Scholar] [CrossRef]
- Tong, C.; Hill, C.B.; Zhou, G.; Zhang, X.Q.; Jia, Y.; Li, C. Opportunities for improving waterlogging tolerance in cereal crops-physiological traits and genetic mechanisms. Plants 2021, 10, 1560. [Google Scholar] [CrossRef]
- Hill, C.B.; Li, C. Genetic architecture of flowering phenology in cereals and opportunities for crop improvement. Front. Plant Sci. 2016, 7, 1906. [Google Scholar] [CrossRef]
- Alagoz, Y.; Gurkok, T.; Zhang, B.; Unver, T. Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci. Rep. 2016, 6, 30910. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, S.; Xu, J.; Sui, C.; Wei, J. Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B 2017, 7, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, L.; Yang, Y.; Emenecker, R.; Smoker, M.; Taylor, J.; Perkins, S.; Smith, J.; MacLean, D.; Olszewski, N.E.; Jones, J.D.G. Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 2019, 17, 132–140. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Chen, R.; Yang, L.; Fan, K.; Liu, Y.; Wang, G.; Ren, Z.; Liu, Y. Generation of transgene-free semidwarf maize plants by gene editing of Gibberellin-Oxidase20-3 using CRISPR/Cas9. Front. Plant Sci. 2020, 11, 1048. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Wu, S.; Dou, T.; Zhu, H.; Hu, C.; Huo, H.; He, W.; Deng, G.; Sheng, O.; Bi, F.; et al. Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 2020, 18, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Kawai, K.; Takehara, S.; Kashio, T.; Morii, M.; Sugihara, A.; Yoshimura, H.; Ito, A.; Hattori, M.; Toda, Y.; Kojima, M.; et al. Evolutionary alterations in gene expression and enzymatic activities of gibberellin 3-oxidase 1 in Oryza. Commun. Biol. 2022, 5, 67. [Google Scholar] [CrossRef]
- Karunarathne, S.D.; Han, Y.; Zhang, X.Q.; Li, C. CRISPR/Cas9 gene editing and natural variation analysis demonstrate the potential for HvARE1 in improvement of nitrogen use efficiency in barley. J. Integr. Plant Biol. 2022, 64, 756–770. [Google Scholar] [CrossRef]
| Gene Name (Abbreviation) | Plant Species | Annotation | Function | Reference |
|---|---|---|---|---|
| CYTOCHROME P450, FAMILY 72, SUBFAMILY A, POLUPEPTIDE 9 (CYP72A9) | Arabidopsis thaliana | Encode gibberellin 13-oxidase | Negatively regulate bioactive GA4 to maintain seed dormancy. | [108] |
| RGA-like2 (RGL2) | Arabidopsis thaliana | Encode a DELLA protein | Negatively regulate GA signalling response to decrease seed germination rates. | [109,110] |
| SPINDLY (SPY) | Arabidopsis thaliana | An O-linked N-acetylglucosamine (GlcNAc) transferase (OGT); negative regulator of the GA signaling pathway; | Negative regulator of GA biosynthesis pathway to inhibit seed germination frequency. | [67,111] |
| REPRESSOR OF GA (RGA) | Arabidopsis thaliana | Encode a DELLA protein; repressor of GA signalling pathway | Negatively regulate GA signalling response to negatively mediate stem elongation. | [112] |
| GA INSENSITIVE (GAI) | Arabidopsis thaliana | Encode a DELLA protein; a repressor of GA response | Negatively regulate GA signalling response to inhibit stem elongation. | [113] |
| GA2-oxidase 7 (GA2ox7) | Arabidopsis thaliana | Encode a gibberellin catabolic enzyme gibberellin 2-oxidase that acts specifically on C-20 gibberellins | Reduce endogenous GA biosynthesis to improve salinity tolerance and resistance to pathogens. | [114,115] |
| Late Embryogenesis Abundant 33 (LEA33) | Oryza sativa | Regulate OsGA20ox1 to mediate gibberellin biosynthesis | Negative regulator of GA biosynthesis to negatively regulate grain size and seed germination rates. | [116] |
| SLENDER RICE1 (SLR1) | Oryza sativa | Encode a DELLA protein; an O-linked N-acetylglucosamine transferase; negative regulator of GA response | Negatively regulate GA signalling pathway to inhibit stem elongation. | [117] |
| GA20-oxidase2 (GA20ox2) | Oryza sativa | Encode a gibberellin biosynthetic enzyme gibberellin 20-oxidase | Enhance GA biosynthesis to positively regulate stem elongation. | [118] |
| GA2-oxidase6 (GA2ox6) | Oryza sativa | Encode a gibberellin catabolic enzyme gibberellin 2-oxidase | Reduce endogenous GA biosynthesis to decrease plant height, increase tiller number, and modify crop architecture to enhance drought stress tolerance. | [119] |
| GA2-oxidase8 (GA2ox8) | Oryza sativa | Encode a gibberellin catabolic enzyme gibberellin 2-oxidase | Positively regulate osmotic regulators and antioxidase to enhance osmotic stress tolerance. | [120] |
| Semi-dwarf1 (SD1) | Oryza sativa | Encode a gibberellin biosynthetic enzyme gibberellin 20-oxidase | Enhance endogenous GA biosynthesis to promote stem elongation to escape waterlogging. | [121] |
| Submergence 1A (Sub1A) | Oryza sativa | An ethylene response factor; promote GA signalling repressors, SLR1 and SLRL1, activities | Negatively regulate GA signalling to limit shoot elongation to escape submergence stress. | [122] |
| GA20-oxidase3 (GA20ox3) | Oryza sativa | Encode a gibberellin biosynthetic enzyme gibberellin 20-oxidase | Knockout reduces the endogenous GA to inhibit stem elongation and improve resistance to pathogens. | [123] |
| Gibberellin-insensitive dwarf1 (gid1) | Oryza sativa | A soluble gibberellic acid receptor | A recessive GA-insensitive severe dwarf mutant and positively regulate resistance to rice blast fungus. | [124] |
| GA20-oxidase1 (GA20ox1) | Triticum aestivum | Encode a gibberellin biosynthesis enzyme gibberellin 20-oxidase | Enhance endogenous GA biosynthesis to positively regulate seed dormancy breakage and seed germination. | [35,125] |
| GA3-oxidase2 (GA3ox2) | Triticum aestivum | Encode a gibberellin biosynthesis enzyme gibberellin 3-oxidase | Enhance endogenous GA biosynthesis to positively regulate seed dormancy breakage and seed germination. | [35,125] |
| REDUCED HEIGHT-B1b/D1b (RhtB1b/D1b) | Triticum aestivum | Encode a DELLA protein | Negatively regulate GA signalling response to reduce stem elongation and enhance resistance to high temperature and drought stress. | [126,127] |
| REDUCED HEIGHT 12 (Rht12) | Triticum aestivum | Encode a DELLA protein | Negatively regulate GA signalling response to inhibit stem elongation and enhance resistance to high temperature and drought stress. | [128] |
| GA2-oxidaseA9 (GA2oxA9) | Triticum aestivum | Encode a gibberellin catabolic enzyme gibberellin 2-oxidase | Reduce endogenous GA biosynthesis to negatively regulate stem elongation without yield loss. | [129] |
| SPINDLY (SPY) | Hordeum vulgare | A negative regulator of GA response | Negatively regulate GA signalling to suppress the expression of α-amylase induced by GA and maintain seed dormancy. | [130] |
| SLENDER1 (SLN1) | Hordeum vulgare | Encode a DELLA protein; negatively regulate GA signalling | Negatively regulate GA signalling response to suppress the expression of α-amylase and maintain seed dormancy, as well as negatively regulate stem elongation. | [131,132,133] |
| GA20-oxidase2/Sdw1/denso (GA20ox2) | Hordeum vulgare | Encode a gibberellin biosynthetic enzyme gibberellin 20-oxidase | Enhance endogenous GA biosynthesis to positively regulate stem elongation and enhance resistance to drought stress. | [134,135,136] |
| PANICLE RACHIS LENGTH 5 (Prl5) | Oryza sativa | Encode a gibberellin biosynthesis enzyme gibberellin 20-oxidase4 | Positively regulate endogenous GA biosynthesis to increase panicle rachis elongation. | [137] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Hill, C.B.; Shabala, S.; Li, C.; Zhou, M. Manipulating GA-Related Genes for Cereal Crop Improvement. Int. J. Mol. Sci. 2022, 23, 14046. https://doi.org/10.3390/ijms232214046
Cheng J, Hill CB, Shabala S, Li C, Zhou M. Manipulating GA-Related Genes for Cereal Crop Improvement. International Journal of Molecular Sciences. 2022; 23(22):14046. https://doi.org/10.3390/ijms232214046
Chicago/Turabian StyleCheng, Jingye, Camilla Beate Hill, Sergey Shabala, Chengdao Li, and Meixue Zhou. 2022. "Manipulating GA-Related Genes for Cereal Crop Improvement" International Journal of Molecular Sciences 23, no. 22: 14046. https://doi.org/10.3390/ijms232214046
APA StyleCheng, J., Hill, C. B., Shabala, S., Li, C., & Zhou, M. (2022). Manipulating GA-Related Genes for Cereal Crop Improvement. International Journal of Molecular Sciences, 23(22), 14046. https://doi.org/10.3390/ijms232214046










