The Shared Proteome of the Apomictic Fern Dryopteris affinis ssp. affinis and Its Sexual Relative Dryopteris oreades
Abstract
1. Introduction
2. Results
2.1. Identification of Proteins by LC-MS/MS
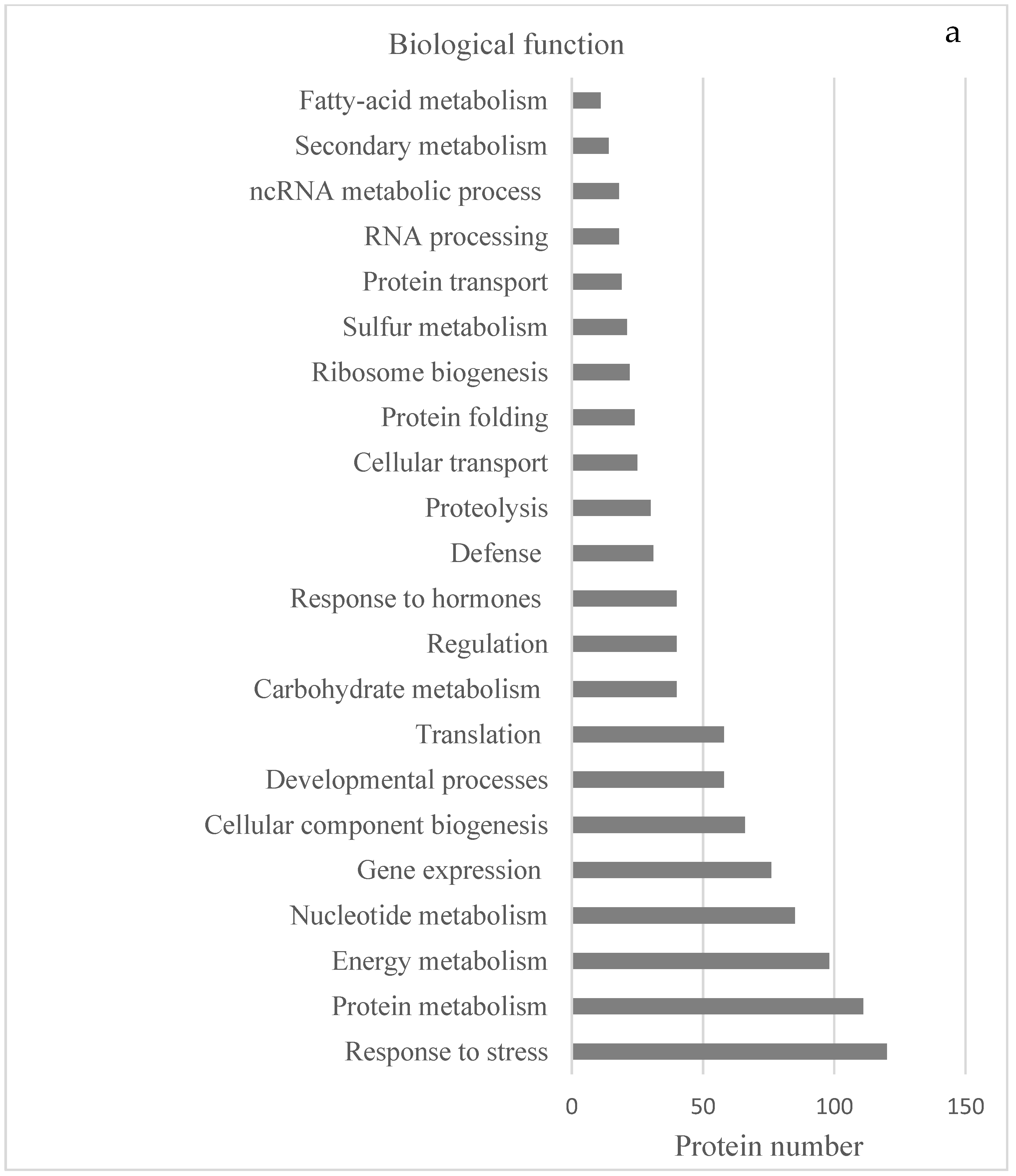
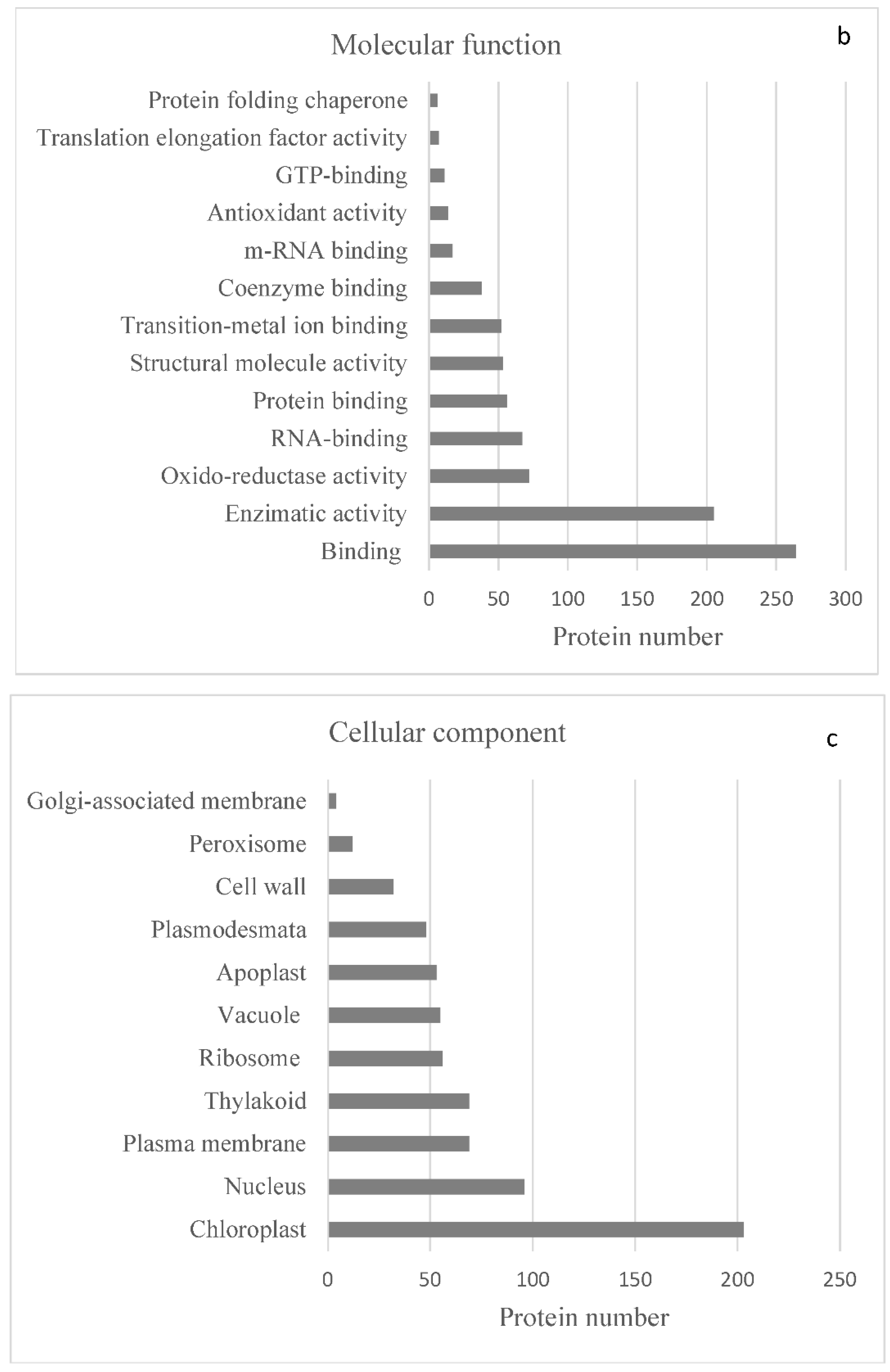
2.2. Protein Analyses in Fern Gametophytes
3. Discussion
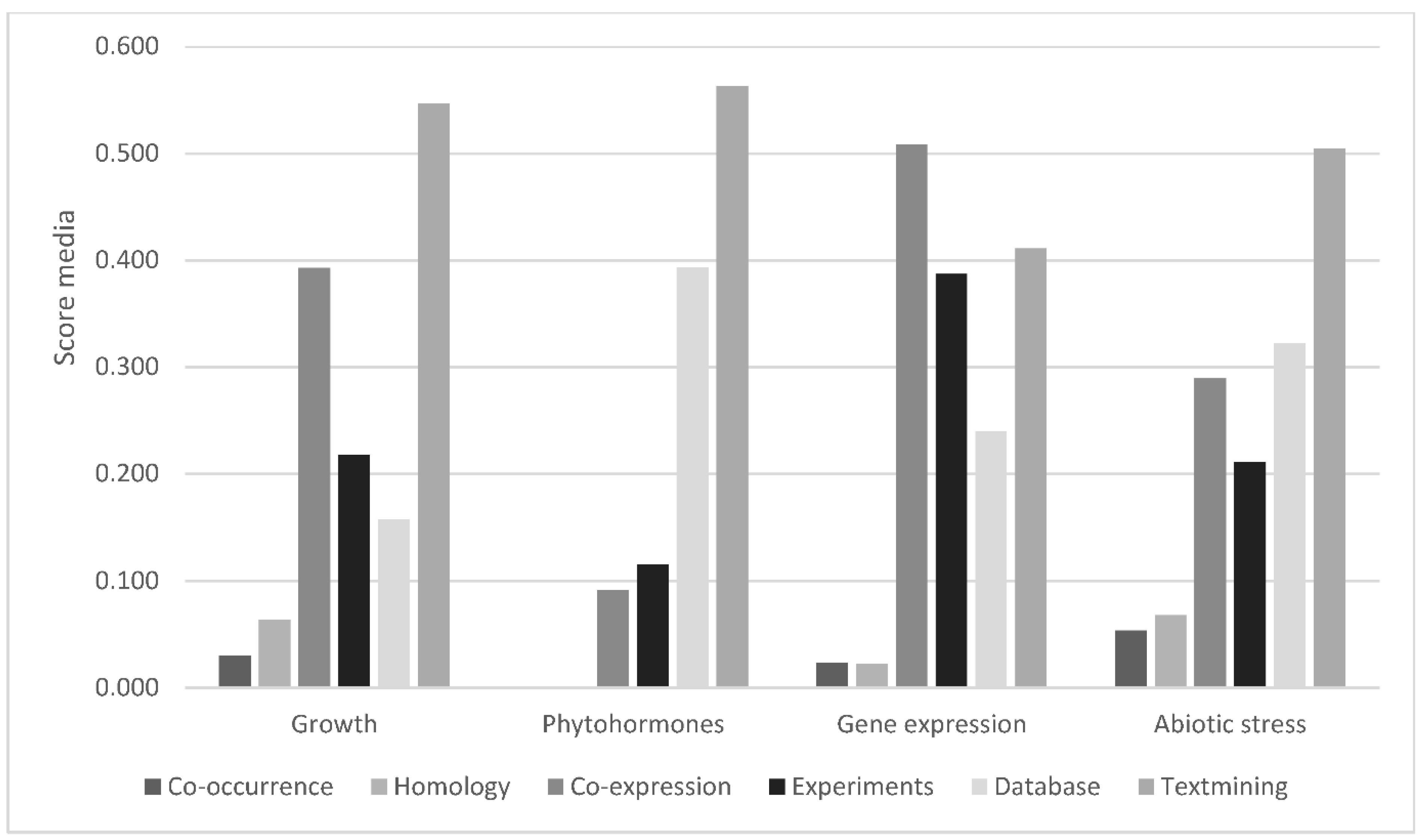
3.1. Growth
3.2. Reproduction
3.3. Phytohormone Signaling and Biosynthesis
3.4. Gene Expression
3.5. Biotic Stress
3.6. Abiotic Stress
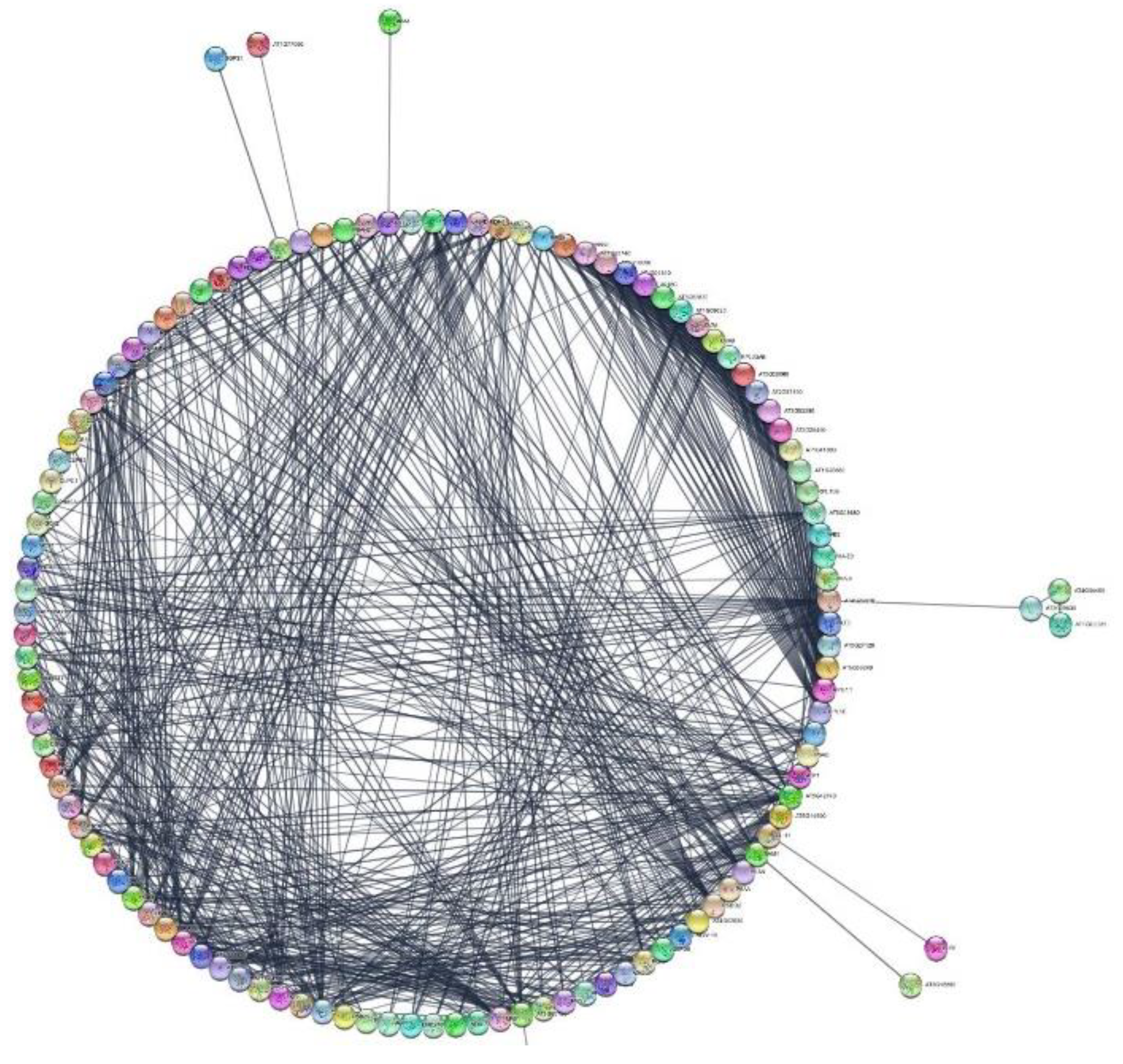
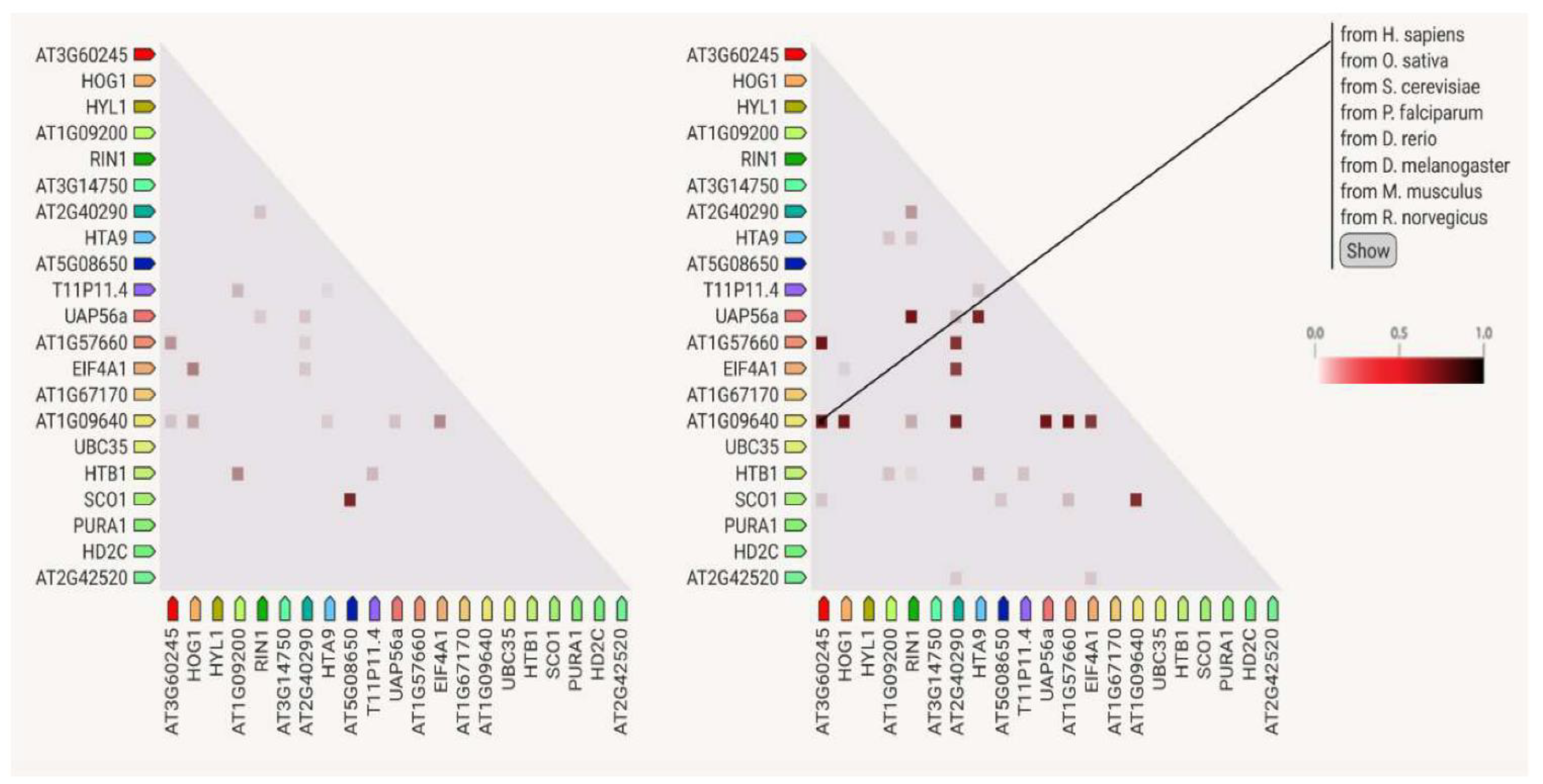
3.7. Protein–Protein Interactions
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Protein Extraction
4.3. Protein Separation and In-Gel Digestion
4.4. Mass Spectrometry and Peptide Identification (Orbitrap XL)
4.5. Protein Identification, Verification, and Bioinformatic Downstream Analyses
4.6. Protein Analysis Using the STRING Platform
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wada, M. The fern as a model system to study photomorphogenesis. J. Plant Res. 2007, 120, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Salmi, M.L.; Bushart, T.; Stout, S.; Roux, S. Profile and analysis of gene expression changes during early development in germinating spores of Ceratopteris richardii. Plant Physiol. 2005, 138, 1734–1745. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Zhao, Q.; Zhang, Z.; Chen, S.; Cao, J.; Liu, G.; Wei, X.; Wang, T.; Yang, C.; Dai, S. Cytological and proteomic analyses of Osmunda cinnamomea germinating spores reveal characteristics of fern spore germination and rhizoid tip growth. Mol. Cell Proteomics. 2015, 14, 2510–2534. [Google Scholar] [CrossRef] [PubMed]
- Salmi, M.L.; Morris, K.E.; Roux, S.J.; Porterfield, D.M. Nitric oxide and cGMP signaling in calcium-dependent development of cell polarity in Ceratopteris richardii. Plant Physiol. 2007, 144, 94–104. [Google Scholar] [CrossRef]
- Eeckhout, S.; Leroux, O.; Willats, W.G.T.; Popper, Z.A.; Viane, R.L.L. Comparative glycan profiling of Ceratopteris richardii “C-Fern” gametophytes and sporophytes links cell-wall composition to functional specialization. Ann Bot. 2014, 114, 1295–1307. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rivera, A.; Cañal, M.J.; Grossniklauss, U.; Fernández, H. The gametophyte of fern: Born to reproducing. In Recent Advances in Fern Research; Helena, F., Ed.; Springer: New York, NY, USA, 2018; pp. 3–19. [Google Scholar]
- Rathinasabapathi, B. Ferns represent an untapped biodiversity for improving crops for environmental stress tolerance. New Phytol. 2006, 172, 385–390. [Google Scholar] [CrossRef]
- Femi-Adepoju, A.G.; Dada, A.O.; Otun, K.O.; Adepoju, A.O.; Fatoba, O.P. Green synthesis of silver nanoparticles using terrestrial fern Gleichenia pectinata (Willd.) C. Presl.: Characterization and antimicrobial studies. Heliyon 2019, 5, e01543. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Nogler, G.A.; van Dijk, P.J. How to avoid Sex: The genetic control of developmental aspects. Plant Cell. 2001, 13, 1491–1497. [Google Scholar]
- Spillane, C.; Curtis, M.D.; Grossniklaus, U. Apomixis technology development—Virgin births in farmers’ fields? Nat. Biotechnol. 2004, 22, 687–691. [Google Scholar] [CrossRef]
- Underwood, C.J.; Mercier, R. Engineering apomixis: Clonal seeds approaching the fields. Annu. Rev. Plant Biol. 2022, 73, 201–225. [Google Scholar] [CrossRef]
- Dyer, R.J.; Savolainen, V.; Schneider, H. Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Ann. Bot. 2012, 110, 1515–1529. [Google Scholar] [CrossRef] [PubMed]
- Fraser-Jenkins, C.R. Dryopteris affinis: A new treatment for a complex species in the European Pteridophyte flora. Willdenowia 1980, 10, 107–115. [Google Scholar]
- Quintanilla, L.G.; Escudero, A. Spore fitness components do not differ between diploid and allotetraploid species of Dryopteris (Dryopteridaceae). Ann. Bot. 2006, 98, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Thagela, P.; Yadav, R.K.; Dahuja, A.; Singh, P.K.; Abraham, G. Physiological and proteomic changes in Azolla microphylla roots upon exposure to salinity. Indian J. Biotechnol. 2016, 15, 101–106. [Google Scholar]
- Cordle, A.; Irish, E.; Cheng, C.l.L. Gene expression associated with apogamy commitment in Ceratopteris richardii. Sex Plant Reprod. 2012, 25, 293–304. [Google Scholar] [CrossRef]
- Domżalska, L.; Kędracka-Krok, S.; Jankowska, U.; Grzyb, M.; Sobczak, M.; Rybczyński, J.J.; Mikuła, A. Proteomic analysis of stipe explants reveals differentially expressed proteins involved in early direct somatic embryogenesis of the tree fern Cyathea delgadii Sternb. Plant Sci. 2017, 258, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Sareen, B.; Thapa, P.; Joshi, R.; Bhattacharya, A. Proteome analysis of the gametophytes of a Western Himalayan fern Diplazium maximum reveals their adaptive responses to changes in their micro-environment. Front. Plant Sci. 2019, 10, 1623. [Google Scholar] [CrossRef]
- Aya, K.; Kobayashi, M.; Tanaka, J.; Ohyanagi, H.; Suzuki, T.; Yano, K.; Takano, T.; Yano, K.; Matsuoka, M. De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database, Ljtrans DB. Plant Cell Physiol. 2015, 56, e5. [Google Scholar] [CrossRef]
- Gill, A.T.; Farrant, J.M.; Rafudeen, M.S. Characterisation of the heat stable proteome of the desiccation tolerant form of the resurrection fern Mohria caffrorum. S. Afr. J. Bot. 2009, 75, 433. [Google Scholar] [CrossRef]
- Bona, E.; Cattaneo, C.; Cesaro, P.; Marsano, F.; Lingua, G.; Cavaletto, M.; Berta, G. Proteomic analysis of Pteris vittata fronds: Two arbuscular mycorrhizal fungi differentially modulate protein expression under arsenic contamination. Proteomics 2010, 10, 3811–3834. [Google Scholar] [CrossRef]
- Der, J.P.; Barker, M.S.; Wickett, N.J.; dePamphilis, C.W.; Wolf, P.G. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genom. 2011, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.S.; Zhang, Y.; Li, J.; Wang, L.B.; Yu, Z.F. Proteomic changes in the stem of wild Pteridium aquilinum during development. J. Plant Growth Regul. 2016, 35, 504–517. [Google Scholar] [CrossRef]
- Valledor, L.; Menéndez, V.; Canal, M.J.; Revilla, A.; Fernández, H. Proteomic approaches to sexual development mediated by antheridiogen in the fern Blechnum spicant L. Proteomics 2014, 14, 2061–2071. [Google Scholar] [CrossRef]
- Grossmann, J.; Fernández, H.; Chaubey, P.M.; Valdés, A.E.; Gagliardini, V.; Canal-Villanueva, M.J.F.; Russo, G.; Grossniklaus, U. Proteogenomic analysis greatly expands the identification of proteins related to reproduction in the apogamous fern Dryopteris affinis ssp. affinis. Front. Plant Sci. 2017, 8, 336. [Google Scholar] [CrossRef]
- Wyder, S.; Rivera, A.; Valdés, A.E.; Cañal, M.J.; Gagliardini, V.; Fernández, H.; Grossniklaus, U. Plant physiology and biochemistry differential gene expression profiling of one- and two-dimensional apogamous gametophytes of the fern Dryopteris affinis ssp. affinis. Plant Physiol. Biochem. 2020, 148, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Fernández, H.; Grossmann, J.; Gagliardini, V.; Feito, I.; Rivera, A.; Rodríguez, L.; Quintanilla, L.G.; Quesada, V.; Cañal, M.J.; Grossniklaus, U. Sexual and apogamous species of woodferns show different protein and phytohormone profiles. Front. Plant Sci. 2021, 12, 2326. [Google Scholar] [CrossRef]
- Ishiguro, S.; Watanabe, Y.; Ito, N.; Nonaka, H.; Takeda, N.; Sakai, T.; Kanaya, H.; Okada, K. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 2002, 21, 898–908. [Google Scholar] [CrossRef]
- Park, S.; Rancour, D.M.; Bednarek, S.Y. In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol. 2008, 148, 246–258. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Gilliland, L.U.; McKinney, E.C.; Meagher, R.B. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell. 2001, 13, 1541. [Google Scholar] [CrossRef]
- Lee, S.A.; Yoon, E.K.; Heo, J.O.; Lee, M.H.; Hwang, I.; Cheong, H.; Sung Lee, W.; Hwang, Y.-S.; Lim, J. Analysis of Arabidopsis glucose-insensitive growth mutants reveals the involvement of the plastidial copper transporter PAA1 in glucose-induced intracellular signaling. Plant Physiol. 2012, 159, 1001–1012. [Google Scholar] [CrossRef]
- Yoder, D.W.; Kadirjan-Kalbach, D.; Olson, B.J.S.C.; Miyagishima, S.-Y.; DeBlasio, S.L.; Hangarter, R.P.; Osteryoung, K.W. Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 2007, 48, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, V.; Löttgert, T.; Gigolashvili, T.; Bell, K.; Flügge, U.-I.; Häusler, R.E. Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett. 2009, 583, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Hargett, S.R.; Frankel, L.K.; Bricker, T.M. The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J. Biol. Chem. 2006, 281, 26260–26267. [Google Scholar] [CrossRef]
- Chen, I.-C.; Huang, I.-C.; Liu, M.-J.; Wang, Z.-G.; Chung, S.-S.; Hsieh, H.-L. Glutathione S-transferase interacting with FAR-RED INSENSITIVE219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol. 2007, 143, 1189–1202. [Google Scholar] [CrossRef]
- Schenck, C.A.; Nadella, V.; Clay, S.L.; Lindner, J.; Abrams, Z.; Wyatt, S.E. A proteomics approach identifies novel proteins involved in gravitropic signal transduction. Am. J. Bot. 2013, 100, 194–202. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Ho, S.-S.; Kuo, T.-Y.; Hsieh, H.-L.; Cheng, Y.-S. Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E1815–E1824. [Google Scholar] [CrossRef]
- Lucas, W.J.; Bouché-Pillon, S.; Jackson, D.P.; Nguyen, L.; Baker, L.; Ding, B.; Hake, S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 1995, 270, 1980–1983. [Google Scholar] [CrossRef] [PubMed]
- Hassidim, M.; Yakir, E.; Fradkin, D.; Hilman, D.; Kron, I.; Keren, N.; Harir, Y.; Yerushalmi, S.; Green, R.M. Mutations in CHLOROPLAST RNA BINDING provide evidence for the involvement of the chloroplast in the regulation of the circadian clock in Arabidopsis. Plant J. 2007, 51, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Berardini, T.Z.; Bollman, K.; Sun, H.; Poethig, R.S.; Scott Poethig, R. Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 2001, 291, 2405–2407. [Google Scholar] [CrossRef]
- Berg, M.; Rogers, R.; Muralla, R.; Meinke, D. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 2005, 44, 866–878. [Google Scholar] [CrossRef]
- Feng, J.; Fan, P.; Jiang, P.; Lv, S.; Chen, X.; Li, Y. Chloroplast-targeted Hsp90 plays essential roles in plastid development and embryogenesis in Arabidopsis possibly linking with VIPP1. Physiol. Plant 2014, 150, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Grebe, M.; Gadea, J.; Steinmann, T.; Kientz, M.; Rahfeld, J.-U.; Salchert, K.; Koncz, C.; Jürgensa, G. A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 2000, 12, 343. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.C.; Yu, H.-J.; Ngo, Q.A.; Rajani, S.; Mayalagu, S.; Johnson, C.S.; Capron, A.; Xie, L.-F.; Ye, D.; Sundaresan, V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 2005, 132, 603–614. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Michaels, S.D.; Amasino, R.M. Regulation of flowering time by histone acetylation in Arabidopsis. Science 2003, 302, 1751–1754. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, S.; Raitskin, O.; Wu, Z.; Liu, F.; Sun, Q.; Dean, C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 2014, 54, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.S.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef]
- Wiszniewski, A.A.G.; Zhou, W.; Smith, S.M.; Bussell, J.D. Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol. Biol. 2009, 69, 503–515. [Google Scholar] [CrossRef]
- Jurkuta, R.J.; Kaplinsky, N.J.; Spindel, J.E.; Kathryn Barton, M.; Barton, M.K. Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 2009, 21, 1957–1971. [Google Scholar] [CrossRef][Green Version]
- Kitakura, S.; Vanneste, S.; Robert, S.; Löfke, C.; Teichmann, T.; Tanaka, H.; Friml, J. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 2011, 23, 1920–1931. [Google Scholar] [CrossRef]
- Gebbie, L.K.; Burn, J.E.; Hocart, C.H.; Williamson, R.E. Genes encoding ADP-ribosylation factors in Arabidopsis thaliana L. Heyn.; genome analysis and antisense suppression. J. Exp. Bot. 2005, 56, 1079–1091. [Google Scholar] [CrossRef]
- Buer, C.S.; Sukumar, P.; Muday, G.K. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 2006, 140, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Lee, H.; Ishitani, M.; Zhu, J.K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 2002, 277, 8588–8596. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.M.; Piqueras, P.; González-Guzmán, M.; Serrano, R.; Rodríguez, P.L.; Ponce, M.R.; Micol, J.L. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 2005, 56, 2071–2083. [Google Scholar] [CrossRef] [PubMed]
- Rabert, C.; Inostroza, K.; Bravo, S.; Sepúlveda, N.; Bravo, L.A. Exploratory study of fatty acid profile in two filmy ferns with contrasting desiccation tolerance reveal the production of very long chain polyunsaturated omega-3 fatty acids. Plants 2020, 9, 1431. [Google Scholar] [CrossRef]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef]
- Palanivelu, R.; Brass, L.; Edlund, A.F.; Preuss, D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 2003, 114, 47–59. [Google Scholar] [CrossRef]
- Ramos-Ruiz, R.; Martinez, F.; Knauf-Beiter, G. The effects of GABA in plants. Tejada Moral, M., editor. Cogent Food Agric. 2019, 5, 1670553. [Google Scholar] [CrossRef]
- Lu, C.; Fedoroff, N. A Mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 2000, 12, 2351. [Google Scholar] [CrossRef]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004, 14, 346–351. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Z.; Lu, F.; Dong, A.; Huang, H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006, 47, 841–850. [Google Scholar] [CrossRef]
- Sridha, S.; Wu, K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006, 46, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Schmidt, W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 2010, 62, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Betsuyaku, S.; Peart, J.; Takahashi, A.; Noel, L.; Sadanandom, A.; Casais, C.; Parker, J.; Shirasu, K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006, 25, 2007–2016. [Google Scholar] [CrossRef]
- Tor, M.; Gordon, P.; Cuzick, A.; Eulgem, T.; Sinapidou, E.; Mert-Türk, F.; Can, C.; Dangl, J.L.; Holub, E.B. Arabidopsis SGT1b Is Required for Defense Signaling Conferred by Several Downy Mildew Resistance Genes. Plant Cell 2002, 14, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A Downstream Mediator in the Growth Repression Limb of the Jasmonate Pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef]
- Kachroo, P.; Venugopal, S.C.; Navarre, D.A.; Lapchyk, L.; Kachroo, A. Role of Salicylic Acid and Fatty Acid Desaturation Pathways in ssi2-Mediated Signaling. Plant Physiol. 2005, 139, 1717–1735. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Gao, Q.-M.; Yu, K.; Lapchyk, L.; Navarre, D.; Hildebrand, D.; Kachroo, A.; Kachroo, P. An Intact Cuticle in Distal Tissues Is Essential for the Induction of Systemic Acquired Resistance in Plants. Cell Host Microbe 2009, 5, 151–165. [Google Scholar] [CrossRef]
- Wang, C.; Zien, C.A.; Afitlhile, M.; Welti, R.; Hildebrand, D.F.; Wang, X. Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 2000, 12, 2237. [Google Scholar] [CrossRef]
- Rajashekar, C.; Zhou, H.-E.; Zhang, Y.; Li, W.; Wang, X. Suppression of phospholipase Dα1 induces freezing tolerance in Arabidopsis: Response of cold-responsive genes and osmolyte accumulation. J. Plant Physiol. 2006, 163, 916–926. [Google Scholar] [CrossRef]
- Uraji, M.; Katagiri, T.; Okuma, E.; Ye, W.; Hossain, M.A.; Masuda, C.; Miura, A.; Nakamura, Y.; Mori, I.C.; Shinozaki, K.; et al. Cooperative Function of PLDδ and PLDα1 in Abscisic Acid-Induced Stomatal Closure in Arabidopsis. Plant Physiol. 2012, 159, 450–460. [Google Scholar] [CrossRef]
- Afitlhile, M.M.; Fukushige, H.; Nishimura, M.; Hildebrand, D.F. A defect in glyoxysomal fatty acid beta-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Van Hulten, M.; Zhang, Y.; Berkowitz, O.; López, A.; Pétriacq, P.; Sellwood, M.A.; Chen, B.; Burrell, M.; van de Meene, A.; et al. Plant perception of β-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat. Chem. Biol. 2014, 10, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Mustilli, A.-C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 Protein Kinase Mediates the Regulation of Stomatal Aperture by Abscisic Acid and Acts Upstream of Reactive Oxygen Species Production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef]
- Lu, M.; Tang, X.; Zhou, J. Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 2001, 13, 437–447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreau, M.; Westlake, T.; Zampogna, G.; Popescu, G.; Tian, M.; Noutsos, C.; Popescu, S. The Arabidopsis oligopeptidases TOP1 and TOP2 are salicylic acid targets that modulate SA-mediated signaling and the immune response. Plant J. 2013, 76, 603–614. [Google Scholar] [CrossRef]
- Moreno, J.I.; Martín, R.; Castresana, C. Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 2004, 41, 451–463. [Google Scholar] [CrossRef]
- Yang, Y.; Sulpice, R.; Himmelbach, A.; Meinhard, M.; Christmann, A.; Grill, E. Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. USA 2006, 103, 6061–6066. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Pezza, A.; Biarc, J.; Burlingame, A.L.; Casati, P. Plant L10 Ribosomal Proteins Have Different Roles during Development and Translation under Ultraviolet-B Stress. Plant Physiol. 2010, 153, 1878–1894. [Google Scholar] [CrossRef]
- Barroso, C.; Romero, L.C.; Cejudo, F.J.; Vega, J.M.; Gotor, C. Salt-specific regulation of the cytosolic O-acetylserine(thiol)lyase gene from Arabidopsis thaliana is dependent on abscisic acid. Plant Mol. Biol. 1999, 40, 729–736. [Google Scholar] [CrossRef]
- Shin, J.S.; So, W.M.; Kim, S.Y.; Noh, M.; Hyoung, S.; Yoo, K.S.; Shin, J.S. CBSX3-Trxo-2 regulates ROS generation of mitochondrial complex II (succinate dehydrogenase) in Arabidopsis. Plant Sci. 2020, 294, 110458. [Google Scholar] [CrossRef]
- Popova, A.V.; Rausch, S.; Hundertmark, M.; Gibon, Y.; Hincha, D.K. The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim. Biophys. Acta—Proteins Proteom. 2015, 1854, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Lee, H.; Xiong, L.; Jagendorf, A.; Stevenson, B.; Zhu, J.-K. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 2002, 99, 11507–11512. [Google Scholar] [CrossRef] [PubMed]
- Myouga, F.; Motohashi, R.; Kuromori, T.; Nagata, N.; Shinozaki, K. An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. Plant J. 2006, 48, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-C.; Chen, L.-J.; Su, P.-H.; Li, H.-M. Evolution of Chloroplast J Proteins. PLoS ONE 2013, 8, e70384. [Google Scholar] [CrossRef] [PubMed]
- Kupsch, C.; Ruwe, H.; Gusewski, S.; Tillich, M.; Small, I.; Schmitz-Linneweber, C. Arabidopsis Chloroplast RNA Binding Proteins CP31A and CP29A Associate with Large Transcript Pools and Confer Cold Stress Tolerance by Influencing Multiple Chloroplast RNA Processing Steps. Plant Cell 2012, 24, 4266–4280. [Google Scholar] [CrossRef] [PubMed]
- Yano, R.; Nakamura, M.; Yoneyama, T.; Nishida, I. Starch-Related α-Glucan/Water Dikinase Is Involved in the Cold-Induced Development of Freezing Tolerance in Arabidopsis. Plant Physiol. 2005, 138, 837–846. [Google Scholar] [CrossRef]
- Titiz, O.; Tambasco-Studart, M.; Warzych, E.; Apel, K.; Amrhein, N.; Laloi, C.; Fitzpatrick, T.B. PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 2006, 48, 933–946. [Google Scholar] [CrossRef]
- Machado, C.R.; Costa de Oliveira, R.L.; Boiteux, S.; Praekelt, U.M.; Meacock, P.A.; Menck, C.F.M. THI1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Mol Biol. 1996, 31, 585–593. [Google Scholar] [CrossRef]
- Rapala-Kozik, M.; Wolak, N.; Kujda, M.; Banas, A.K. The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol. 2012, 12, 2. [Google Scholar] [CrossRef]
- Mao, G.; Wang, R.; Guan, Y.; Liu, Y.; Zhang, S. Sulfurtransferases 1 and 2 Play Essential Roles in Embryo and Seed Development in Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 7548–7557. [Google Scholar] [CrossRef]
- Ma, F.; Liu, Z.; Wang, T.-W.; Hopkins, M.T.; Peterson, C.A.; Thompson, J.E. Arabidopsis eIF5A3 influences growth and the response to osmotic and nutrient stress. Plant Cell Environ. 2010, 33, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Scheibe, R. Pollen tube growth: Where does the energy come from? Plant Signal Behav. 2014, 9, e977200. [Google Scholar] [CrossRef] [PubMed]
- Thierry-Mieg, D.; Thierry-Mieg, J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006, 7, S12. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-S.; Li, Z.-Y.; Chen, Y.; Chen, M.; Li, L.-C.; Ma, Y.-Z. Heat Shock Protein 90 in Plants: Molecular Mechanisms and Roles in Stress Responses. Int. J. Mol. Sci. 2012, 13, 15706–15723. [Google Scholar] [CrossRef]
- Damodaran, S.; Strader, L.C. Indole 3-butyric acid metabolism and transport in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 851. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, Y.; Huang, J.; Hu, Y.; Zhang, E.; Xie, Z.; Ma, S.; Gao, Y.; Song, S.; Xu, C.; et al. Ancient horizontal transfer of transaldolase-like protein gene and its role in plant vascular development. New Phytol. 2015, 206, 807–816. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Baerenfaller, K.; Grossmann, J.; Grobei, M.A.; Hull, R.; Hirsch-Hoffmann, M.; Yalovsky, S.; Zimmermann, P.; Grossniklaus, U.; Gruissem, W.; Baginsky, S. Genome-Scale Proteomics Reveals Arabidopsis thaliana Gene Models and Proteome Dynamics. Science 2008, 320, 938–941. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]

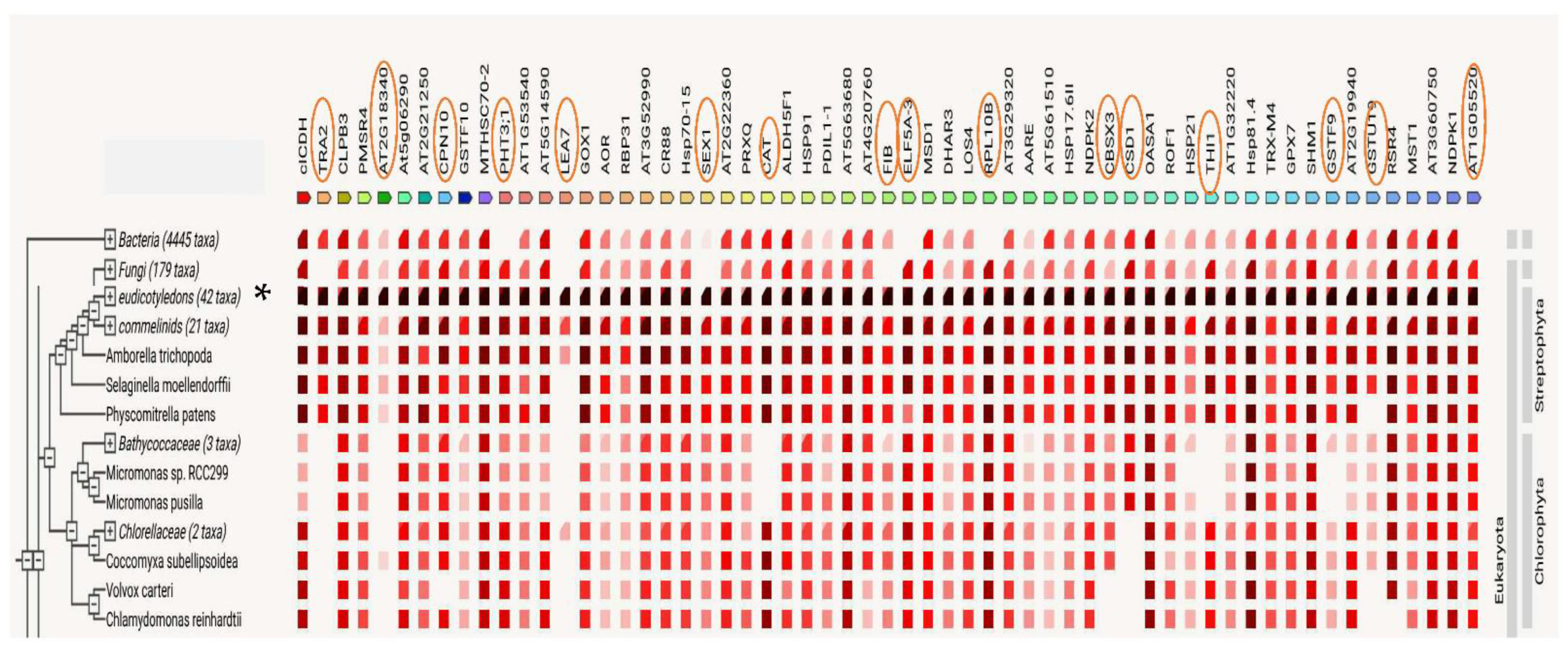
| Category | Accession Number | UniProtKB/Swiss-Prot | Gene Name | Protein Name | MW (kDa) | Amino Acids | Probability |
|---|---|---|---|---|---|---|---|
| Growth | tr|A7YAU9|A7YAU9_PINTA | Q9STX5 | SHD | Endoplasmin homolog | 95 | 1.030 | 0 |
| Growth | 173702-163_1_ORF1 | Q9LZF6 | CDC48A | Cell division control protein 48 homolog E | 65 | 894 | 0 |
| Growth | 149166-199_4_ORF2 | A0A1P8B649 | HMA6 | Copper-transporting ATPase PAA1, chloroplastic | 25 | 246 | 1.62 × 10−22 |
| Growth | tr|F2DY57|F2DY57_HORVD | Q9C9C4 | ENO1 | Enolase 1, chloroplastic | 51 | 477 | 0 |
| Growth | tr|Q6AV23|Q6AV23_ORYSJ | Q56XV8 | CPN60A2 | Chaperonin 60 subunit alpha 1, chloroplastic | 57 | 811 | 0 |
| Growth | 3278-1178_2_ORF2 | Q9C566 | CYP40 | Peptidyl-prolyl cis-trans isomerase CYP40 | 40 | 541 | 0 |
| Growth | tr|O82565|O82565_ANEPH | P53492 | ACT7 | Actin-7 | 42 | 377 | 0 |
| Growth | 152409-192_5_ORF1 | Q42545 | FTSZ1 | Cell division protein FtsZ homolog 1, chloroplastic | 47 | 433 | 2.99 × 10−165 |
| Growth | tr|A9NV11|A9NV11_PICSI | Q94K05 | CCT8 | T-complex protein 1 subunit theta | 59 | 549 | 0 |
| Growth | 195-2609_5_ORF2 | Q41932 | PSBQ2 | Oxygen-evolving enhancer protein 3-2, chloroplastic | 24 | 230 | 3.05 × 10−77 |
| Reproduction | 229414-114_3_ORF1 | O49485 | EDA9 | D-3-phosphoglycerate dehydrogenase 1, chloroplastic | 68 | 918 | 0 |
| Reproduction | 155382-187_1_ORF2 | F4I6W4 | PGM2 | Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | 68 | 915 | 0 |
| Reproduction | 40155-402_5_ORF3 | Q38905 | PRF5 | Profilin-5 | 14 | 62 | 8.5 × 10−59 |
| Reproduction | 65126-313_6_ORF2 | O81644 | VLN2 | Villin-2 | 107 | 1.055 | 0 |
| Reproduction | 88715-266_1_ORF2 | Q8RXE9 | OVA4 | Tryptophan--tRNA ligase, chloroplastic/mitochondrial | 45 | 975 | 0 |
| Reproduction | 12555-680_1_ORF2 | Q8W4F3 | OVA9 | Glutamine--tRNA ligase, cytoplasmic | 80 | 1.021 | 1 × 10−175 |
| Reproduction | 135880-210_3_ORF2 | Q8LD94 | CYP5 | Glucan endo-1,3-beta-glucosidase, acidic isoform | 25 | 251 | 0 |
| Reproduction | 152971-191_2_ORF2 | Q9SIE1 | PAT | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase | 55 | 778 | 0 |
| Reproduction | 116951-229_1_ORF2 | F4IFC5 | EMB2761 | Threonine--tRNA ligase, chloroplastic/mitochondrial 2 | 45 | 625 | 0 |
| Reproduction | 76060-289_2_ORF2 | Q9CAE3 | FLD | Protein flowering locus D | 52 | 742 | 0 |
| Reproduction | tr|D7MAI5|D7MAI5_ARALL | Q9SSD2 | EMB14 | Pre-mRNA processing-splicing factor 8A | 252 | 1.121 | 0 |
| Reproduction | 1422-1543_1_ORF2 | O23654 | VHA-A | V-type proton ATPase catalytic subunit A | 69 | 623 | 0 |
| Reproduction | 50471-358_5_ORF2 | Q9SAJ6 | GAPCP1 | Glyceraldehyde-3-phosphate dehydrogenase GAPCP1, chloroplastic | 51 | 422 | 0 |
| Reproduction | 35041-428_5_ORF2 | Q9M8D3 | PUR4 | Probable phosphoribosylformylglycinamidine synthase, chloroplastic/mitochondrial | 156 | 1.407 | 0 |
| Reproduction | tr|C5XKU8|C5XKU8_SORBI | Q9SRT9 | RGP1 | UDP-arabinopyranose mutase 1 | 40 | 357 | 0 |
| Reproduction | 186611-150_1_ORF2 | Q9SIF2 | CR88 | Heat shock protein 90-5, chloroplastic | 88 | 780 | 0 |
| Reproduction | 135880-210_3_ORF2 | Q8LDP4 | CYP19-4 | Peptidyl-prolyl cis-trans isomerase CYP19-4 | 25 | 201 | 9.98 × 10−112 |
| Hormones | 184149-152_1_ORF2 | Q9S9W2 | SDRA | Short-chain dehydrogenase/reductase SDRA | 193 | 254 | 1.35 × 10−115 |
| Hormones | 167580-170_3_ORF1 | Q9LV09 | BOB1 | Protein BOBBER 1 | 20 | 304 | 4.20 × 10−87 |
| Hormones | 177400-159_3_ORF2 | Q0WLB5 | CHC2 | Clathrin heavy chain 2 | 56 | 1.112 | 0 |
| Hormones | tr|Q9ZPJ2|Q9ZPJ2_SOYBN | Q6ID97 | ARFA1F | ADP-ribosylation factor A1F | 46 | 153 | 1.95 × 10−132 |
| Hormones | 246374-102_1_ORF1 | Q9SK82 | UGT85A1 | UDP-glycosyltransferase 85A1 | 14 | 791 | 1.2 × 10−90 |
| Hormones | 10257-745_6_ORF2 | Q9FGC7 | ABA1 | Zeaxanthin epoxidase, chloroplastic | 46 | 667 | 2.62 × 10−26 |
| Hormones | 118026-228_6_ORF2 | Q9LLR6 | LTP4 | Non-specific lipid-transferprotein 4 | 14 | 56 | 2.04 × 10−90 |
| Hormones | tr|G7JG11|G7JG11_MEDTR | Q27GK7 | TPR4 | Topless-related protein 4 | 88 | 1.001 | 0 |
| Hormones | 359046-40_1_ORF1 | Q9FK25 | OMT1 | Flavone 3’-O-methyltransferase 1 | 42 | 562 | 1.75 × 10−102 |
| Hormones | 294468-71_2_ORF2 | Q42521 | GAD1 | Glutamate descarboxylase 1 | 61 | 851 | 0 |
| Hormones | 53416-347_4_ORF2 | A1A6K6 | AT1G76940 | Nuclear speckle RNA-binding protein A | 29 | 233 | 3.86 × 10−28 |
| Hormones | 114827-231_2_ORF3 | P13114 | CHS | Chalcone synthase | 51 | 395 | 4.93 × 10−124 |
| Gene expression | 118026-228_6_ORF2 | A0A1P8AQD8 | HYL1 | dsRNA-binding domain-like superfamily protein | 31 | 367 | 3.83 × 10−43 |
| Gene expression | tr|G7JG11|G7JG11_MEDTR | Q56XG6 | UAP56A | DEAD-box ATP-dependent RNA helicase 15 | 49 | 688 | 0 |
| Gene expression | 36369-420_5_ORF2 | Q9LZR5 | HD2C | Histone deacetylase HDT3 | 36 | 478 | 3.02 × 10−25 |
| Gene expression | 57659-333_6_ORF2 | Q9SKZ1 | PURA1 | Transcription factor Pur-alpha 1 | 33 | 422 | 1.37 × 10−90 |
| Gene expression | 135393-211_5_ORF1 | Q94A97 | UBC35 | Ubiquitin-conjugating enzyme E2 35 | 22 | 153 | 2.33 × 10−107 |
| Gene expression | tr|A8J568|A8J568_CHLRE | Q9FMR9 | RIN1 | RuvB-like protein 1 | 164 | 1.104 | 0 |
| Gene expression | 239719-107_1_ORF2 | O23255 | HOG1 | Adenosylhomocysteinase 1 | 48 | 485 | 0 |
| Gene expression | tr|B9RUZ2|B9RUZ2_RICCO | Q9C944 | HTA9 | Histone H2A.Z variant | 14 | 134 | 6.67 × 10−76 |
| Biotic stress | 24640-505_5_ORF2 | Q940H6 | OST1 | Serine/threonine-protein kinase SRK2E | 41 | 362 | 0 |
| Biotic stress | 59878-327_6_ORF2 | Q9SW21 | ACP4 | Acyl carrier protein 4, chloroplastic | 14 | 137 | 4.47 × 10−29 |
| Biotic stress | 23981-512_3_ORF1 | Q38882 | PLDALPHA1 | Phospholipase D alpha 1 | 91 | 810 | 0 |
| Biotic stress | 9434-771_1_ORF2 | Q9SUT5 | SGT1B | Protein SGT1 homolog B | 39 | 358 | 8.74 × 10−118 |
| Biotic stress | 30827-456_4_ORF1 | P29976 | DHS1 | Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplastic | 57 | 525 | 0 |
| Biotic stress | tr|D7MBC8|D7MBC8_ARALL | Q9M084 | IBI1 | Aspartate-tRNA ligase 2, cytoplasmic | 62 | 558 | 0 |
| Biotic stress | 3234-1184_1_ORF1 | Q9M8L4 | ACP4 | Glycerol kinase | 56 | 522 | 0 |
| Biotic stress | 284049-77_6_ORF2 | Q94AM1 | OOP | Organellar oligopeptidase A, chloroplastic/mitochondrial | 88 | 791 | 0 |
| Abiotic stress | 177876-158_1_ORF2 | Q8LE52 | DHAR3 | Glutathione S-transferase DHAR3, chloroplastic | 28 | 258 | 3.08 10−103 |
| Abiotic stress | 10787-728_3_ORF2 | P24704 | CSD1 | Superoxide dismutase[Cu-Zn] 1 | 15 | 152 | 2.09 × 10−77 |
| Abiotic stress | 180631-155_1_ORF1 | O81235 | MSD1 | Superoxide dismutase[Mn] 1, mitochondrial | 25 | 231 | 1.51 × 10−104 |
| Abiotic stress | 393073-25_4_ORF2 | Q9SZJ5 | SHM1 | Serine hydroxymethyltransferase 1, mitochondrial | 57 | 517 | 0 |
| Abiotic stress | 372464-34_1_ORF1 | Q9LU86 | PRXQ | Peroxiredoxine Q, chloroplastic | 23 | 216 | 2.11 × 10−94 |
| Abiotic stress | 139597-207_6_ORF1 | Q9FMU6 | PHT3;1 | Mitochondrial phosphate carrier protein 3, mitochondrial | 40 | 375 | 4.08 × 10−136 |
| Abiotic stress | 133552-212_2_ORF2 | Q96270 | LEA7 | Late embryogenesis abundant protein 7 | 18 | 169 | 4.76 × 10−5 |
| Abiotic stress | 145003-202_6_ORF2 | Q93ZG7 | LOS4 | DEAD-box ATP-dependent RNA helicase 38 | 55 | 496 | 3.52 × 10−177 |
| Abiotic stress | 396305-24_6_ORF2 | Q9C5R8 | 2CPB | 2-Cys peroxiredoxin BAS1-like, chloroplastic | 31 | 273 | 1.94 × 10−129 |
| Abiotic Stress | 167311-170_6_ORF2 | Q9LEV3 | CBSX3 | CBS domain-containing protein CBSX3, mitochondrial | 23 | 206 | 1.18 × 10−104 |
| Abiotic stress | 289581-74_6_ORF2 | Q9ZRW8 | GSTU19 | Glutathione S-transferase U19 | 26 | 219 | 2.18 × 10−54 |
| Abiotic stress | 14237-641_3_ORF2 | Q38931 | FKBP62 | Peptidyl-prolyl cis-trans isomerase FKBP62 | 67 | 551 | 0 |
| Abiotic stress | 14173-642_3_ORF2 | P34893 | CPN10 | 10 kDa chaperonin, mitochondrial | 11 | 98 | 1.4 × 10−41 |
| Abiotic stress | 64-3566_3_ORF2 | Q9LF37 | CLPB3 | Chaperone protein ClpB3, chloroplastic | 114 | 968 | 0 |
| Abiotic stress | 4712-1020_6_ORF2 | Q04836 | RBP31 | 31 kDa ribonucleoprotein, chloroplastic | 83 | 329 | 2.38 × 10−17 |
| Abiotic stress | 4641-1023_4_ORF2 | Q38814 | THI1 | Thiamine thiazole synthase, chloroplastic | 42 | 349 | 0 |
| Abiotic stress | 102649-246_6_ORF1 | O64530 | STR1 | Thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial | 43 | 379 | 2.01 × 10−141 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojosnegros, S.; Alvarez, J.M.; Grossmann, J.; Gagliardini, V.; Quintanilla, L.G.; Grossniklaus, U.; Fernández, H. The Shared Proteome of the Apomictic Fern Dryopteris affinis ssp. affinis and Its Sexual Relative Dryopteris oreades. Int. J. Mol. Sci. 2022, 23, 14027. https://doi.org/10.3390/ijms232214027
Ojosnegros S, Alvarez JM, Grossmann J, Gagliardini V, Quintanilla LG, Grossniklaus U, Fernández H. The Shared Proteome of the Apomictic Fern Dryopteris affinis ssp. affinis and Its Sexual Relative Dryopteris oreades. International Journal of Molecular Sciences. 2022; 23(22):14027. https://doi.org/10.3390/ijms232214027
Chicago/Turabian StyleOjosnegros, Sara, José Manuel Alvarez, Jonas Grossmann, Valeria Gagliardini, Luis G. Quintanilla, Ueli Grossniklaus, and Helena Fernández. 2022. "The Shared Proteome of the Apomictic Fern Dryopteris affinis ssp. affinis and Its Sexual Relative Dryopteris oreades" International Journal of Molecular Sciences 23, no. 22: 14027. https://doi.org/10.3390/ijms232214027
APA StyleOjosnegros, S., Alvarez, J. M., Grossmann, J., Gagliardini, V., Quintanilla, L. G., Grossniklaus, U., & Fernández, H. (2022). The Shared Proteome of the Apomictic Fern Dryopteris affinis ssp. affinis and Its Sexual Relative Dryopteris oreades. International Journal of Molecular Sciences, 23(22), 14027. https://doi.org/10.3390/ijms232214027








