Human Vascular Endothelial Cells Promote the Secretion of Vascularization Factors and Migration of Human Skin Fibroblasts under Co-Culture and Its Preliminary Application
Abstract
1. Introduction
2. Results
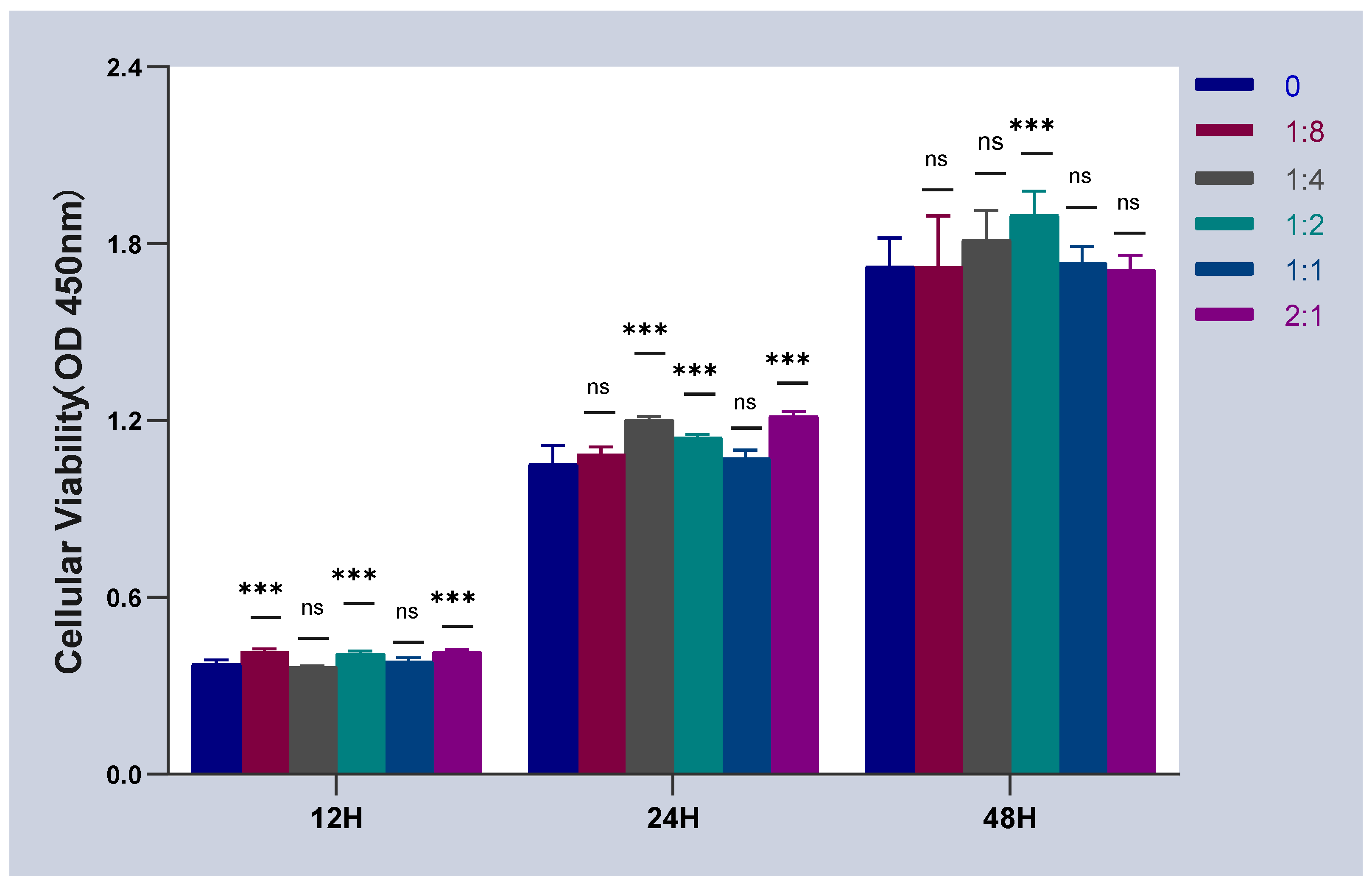
2.1. Effect of Cell Co-Culture on the Activity of HSFs
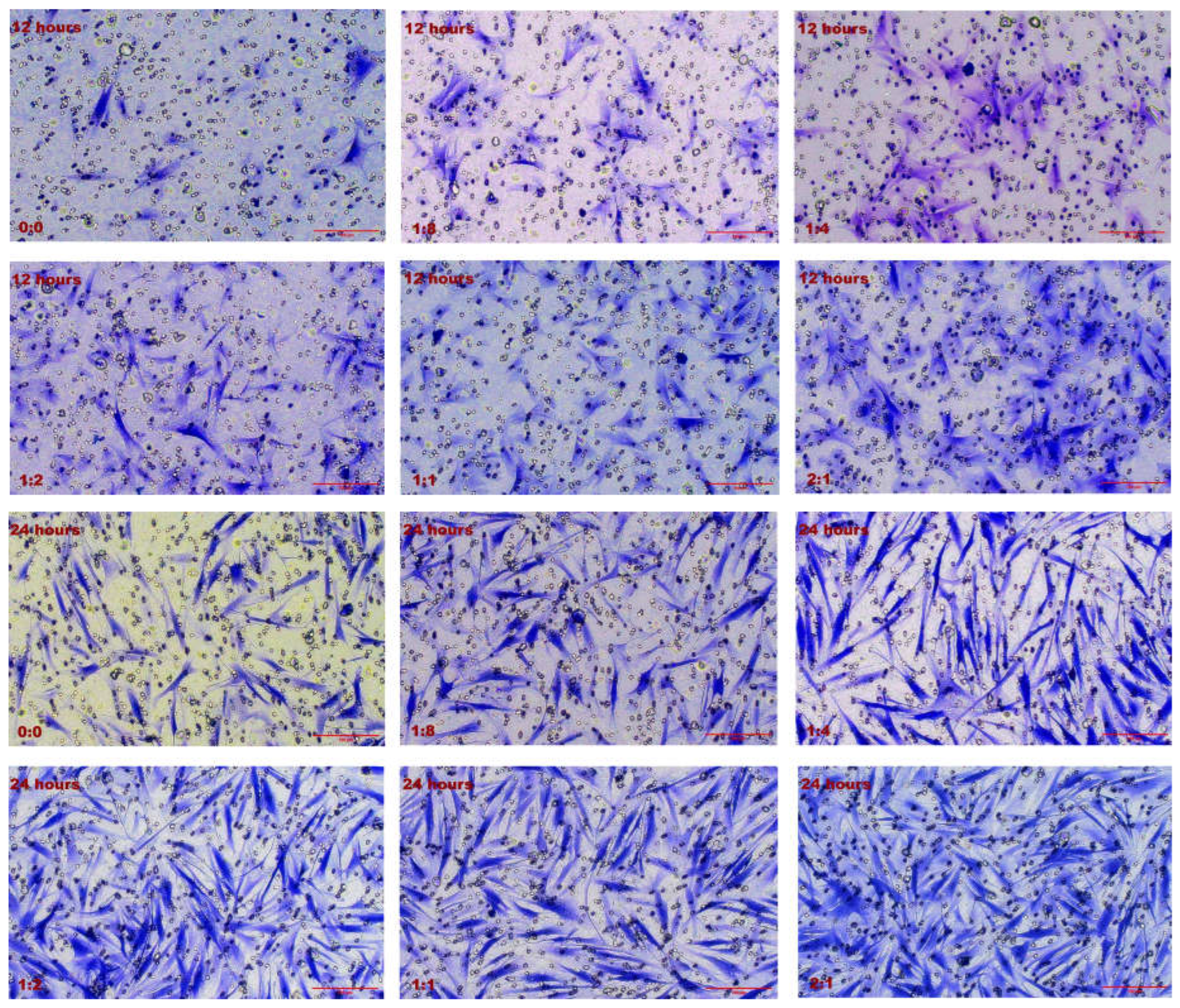
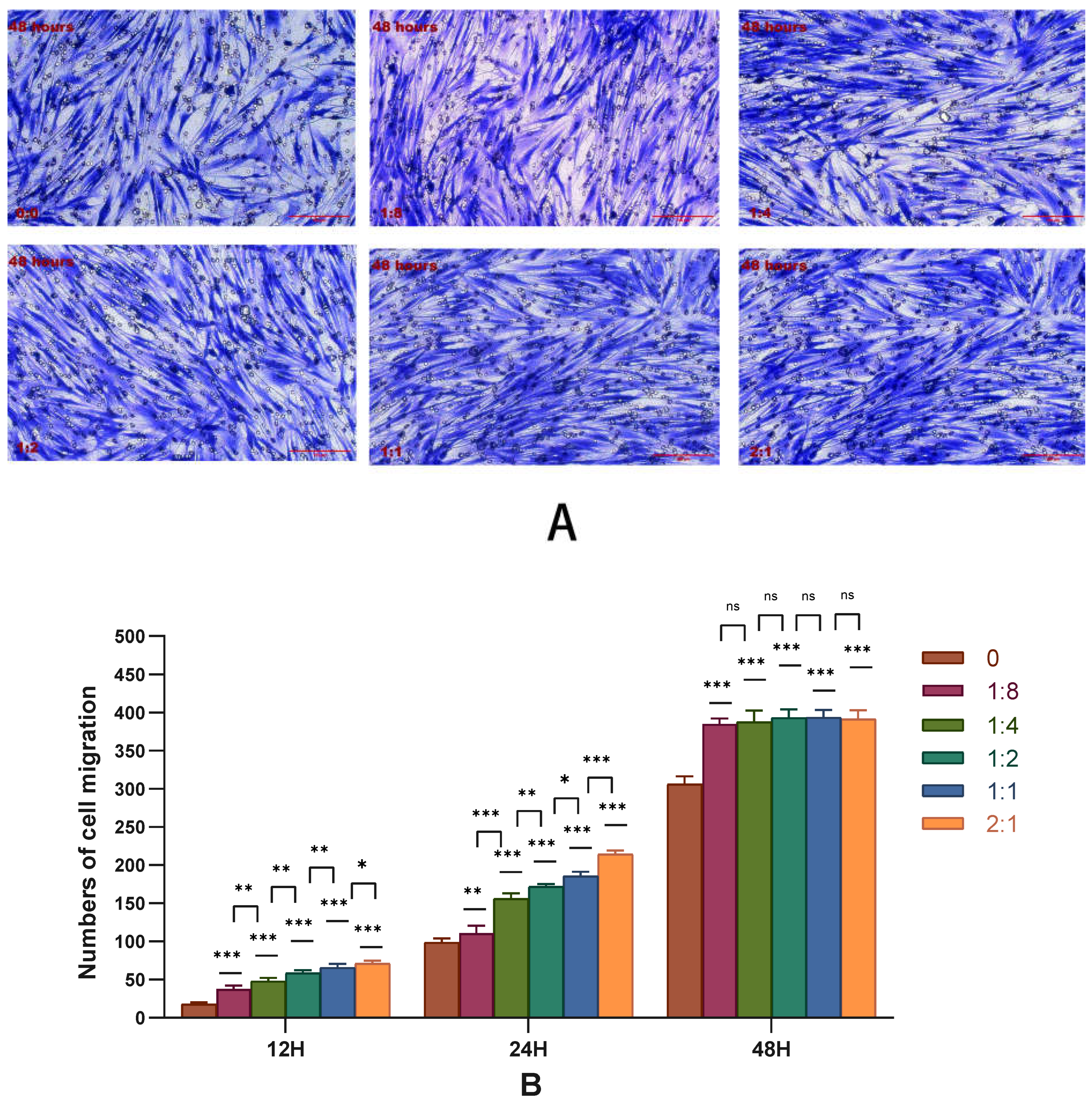
2.2. Effect of Cell Co-Culture on the Migration Ability of HSFs
2.2.1. Results of Transwell Cell Migration Experiment
2.2.2. Migration Factor-HMGB1 Was Detected by RT-qPCR
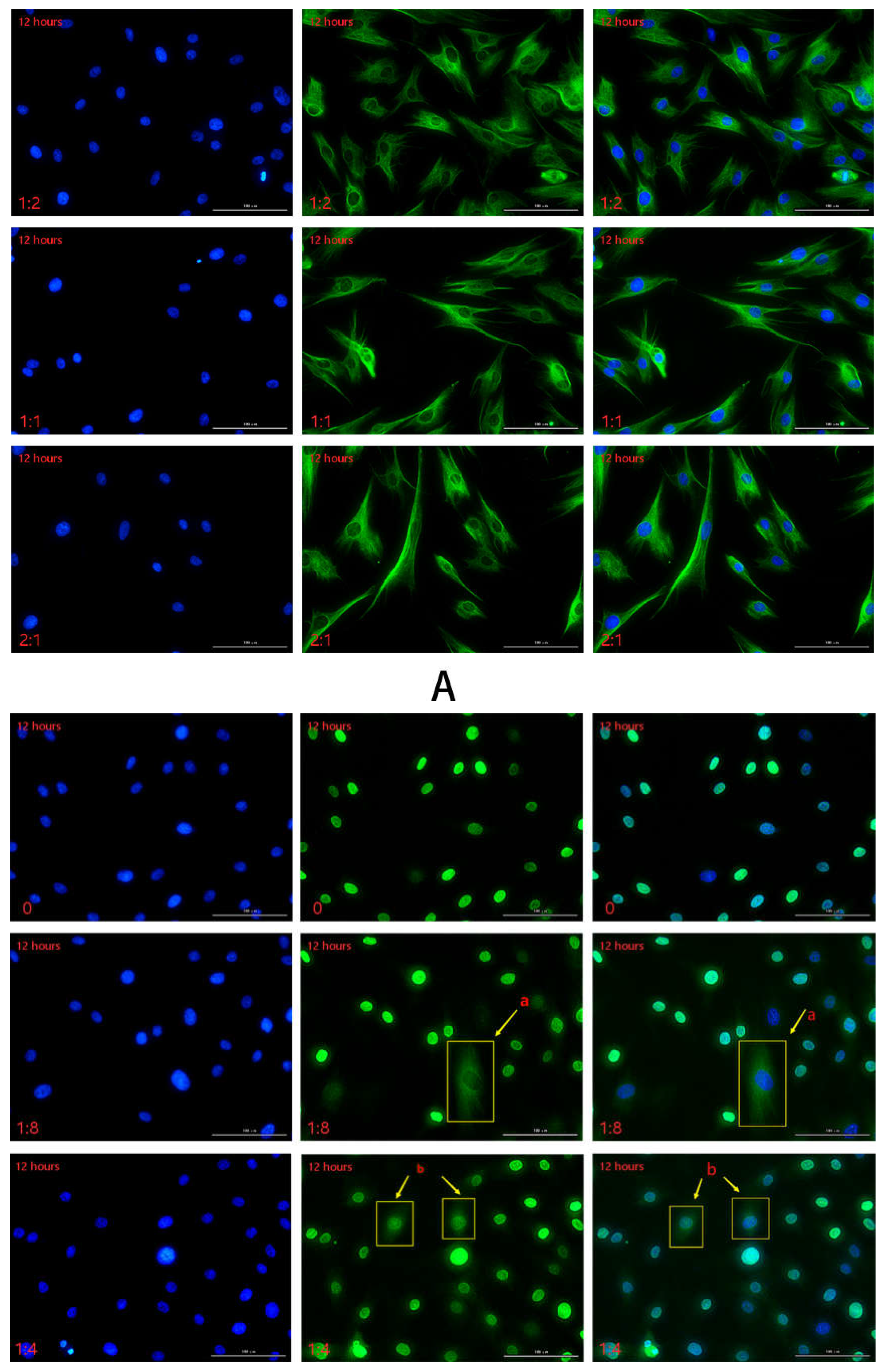
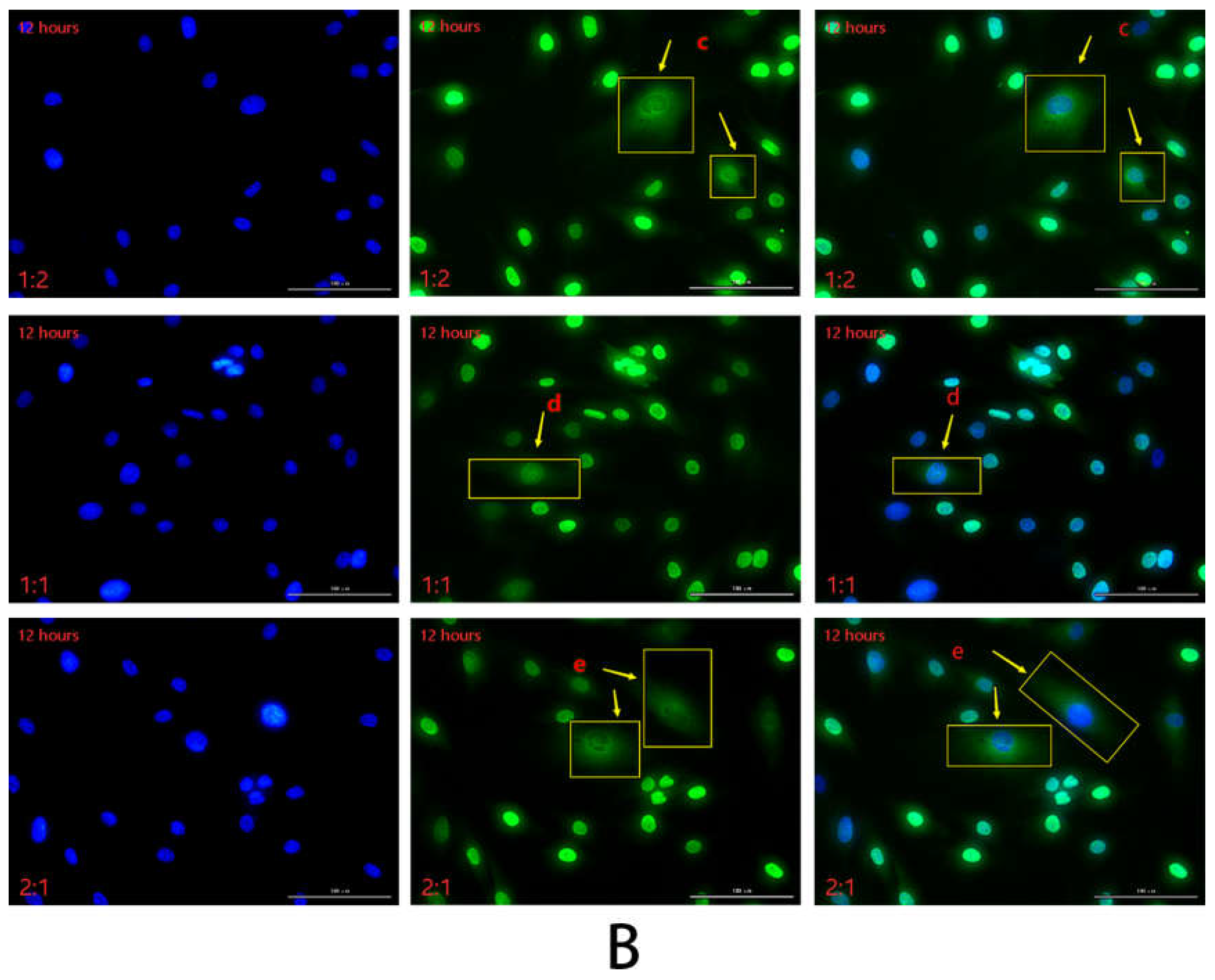
2.2.3. Detection of HSFs Migration Factor HMGB1 by Immunofluorescence
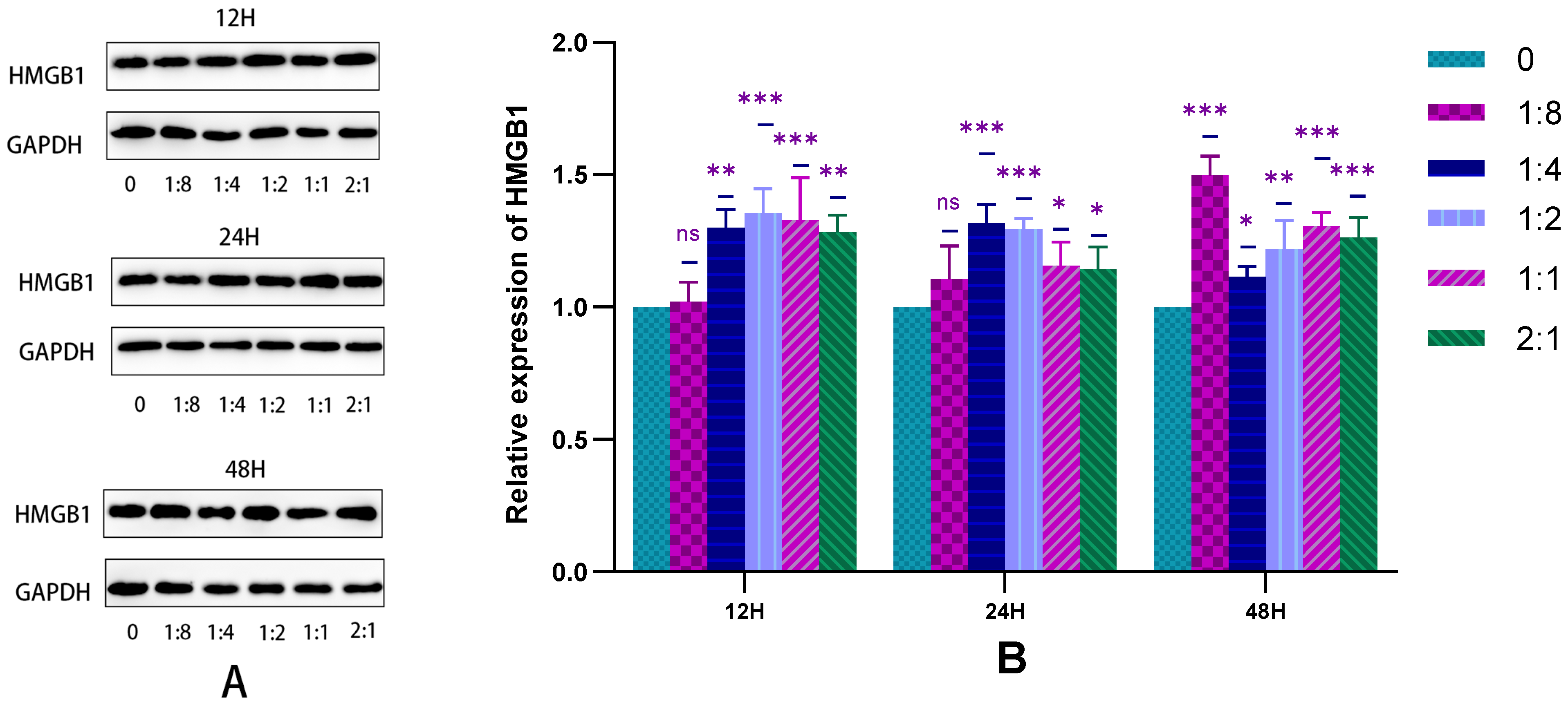
2.2.4. Detection of HSFs Migration Factor HMGB1 by WB
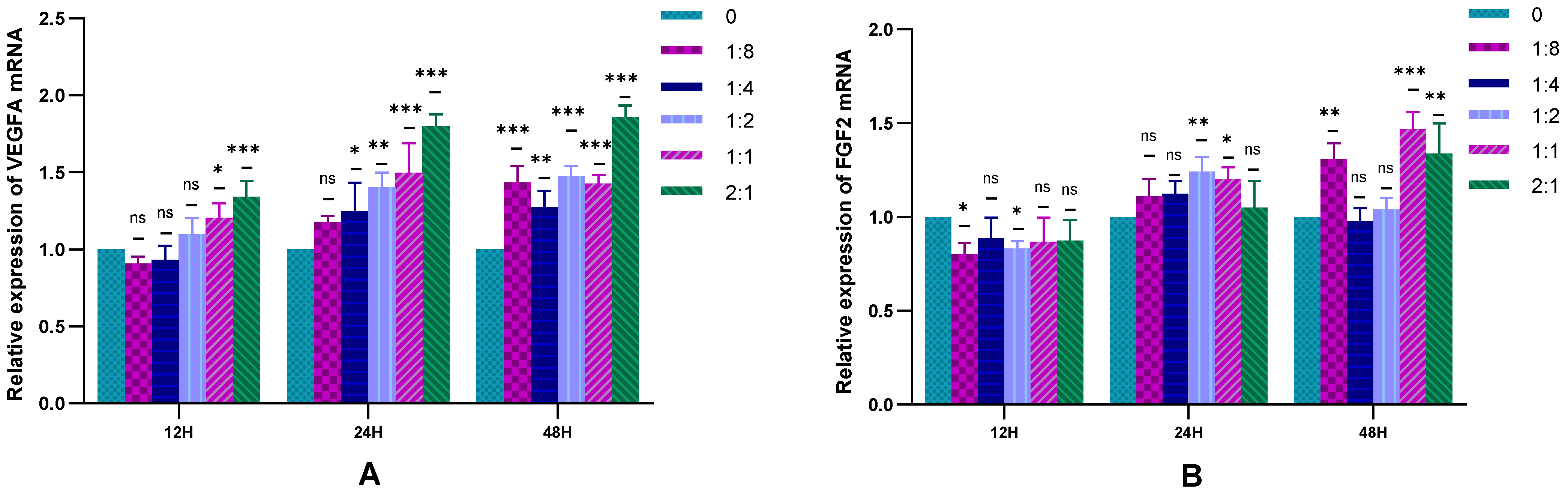
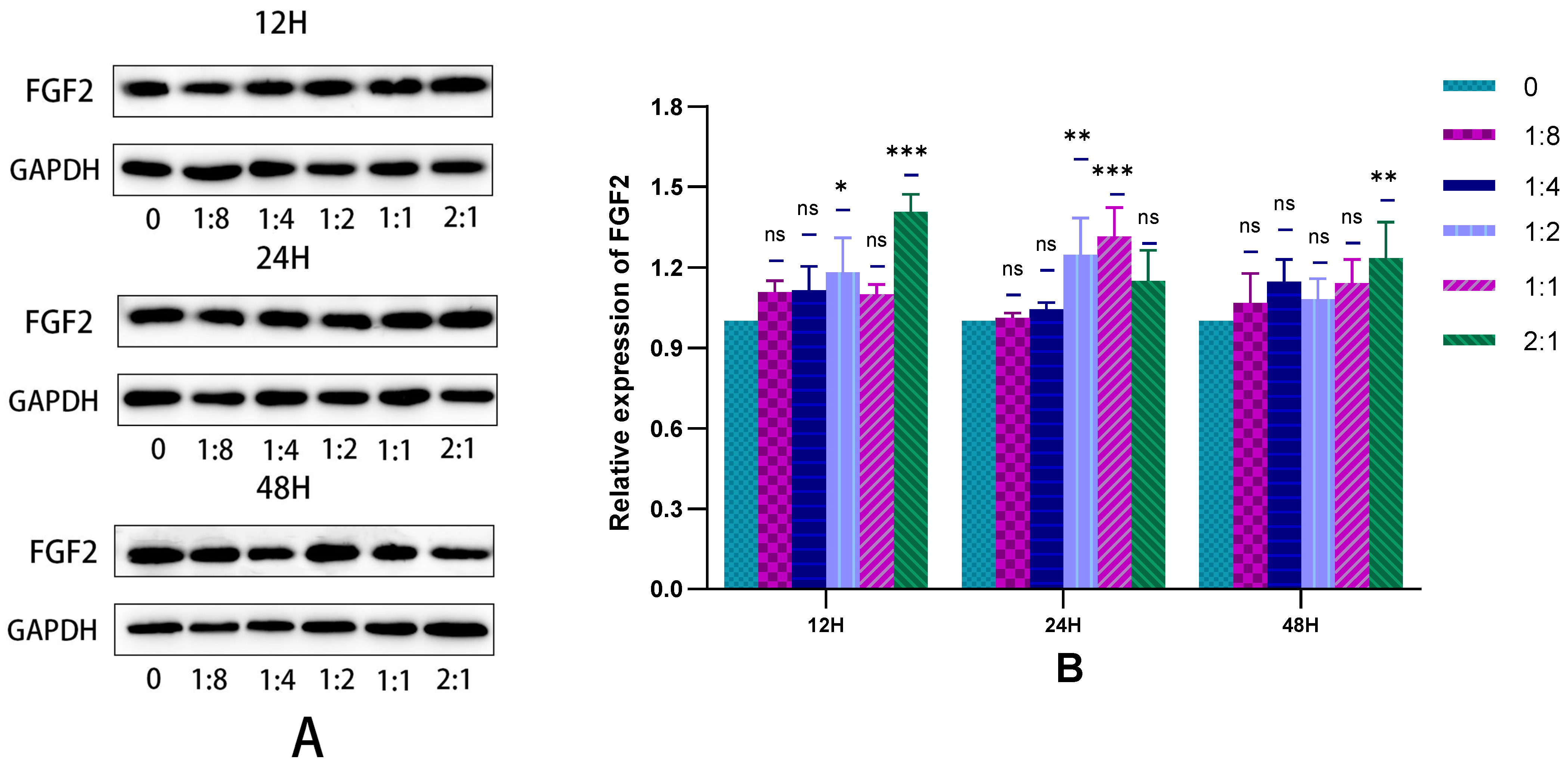
2.3. Detection of HSFs Vascularization Factor-VEGFA and FGF2
2.3.1. The Expression of HSFs VEGFA and FGF2 Detected by RT-qPCR
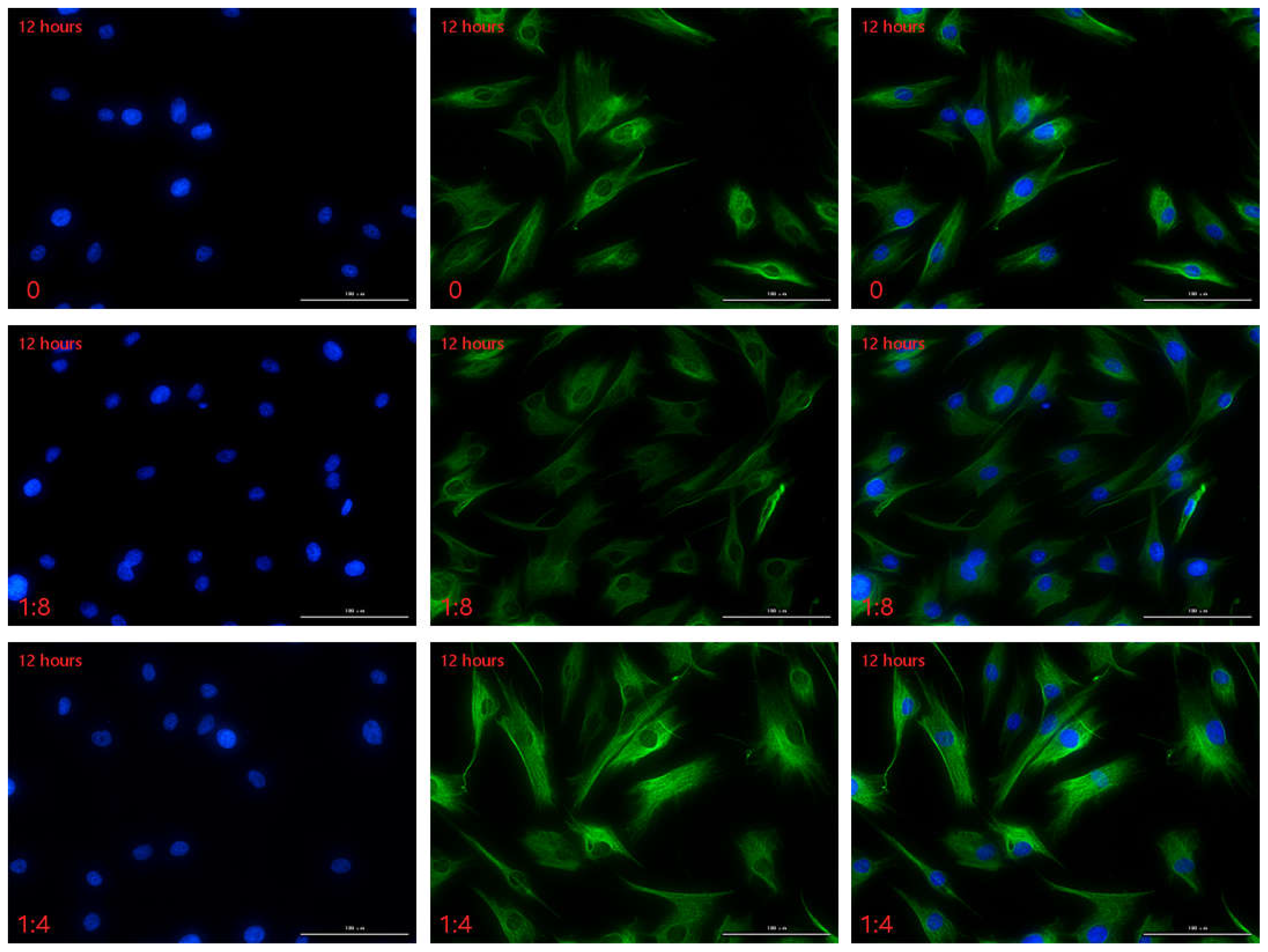
2.3.2. The Secretion of HSFs Proteins VEGFA and FGF2 Detected by Immunofluorescence
2.3.3. The Secretion of HSFs Proteins VEGFA and FGF2 Detected by WB
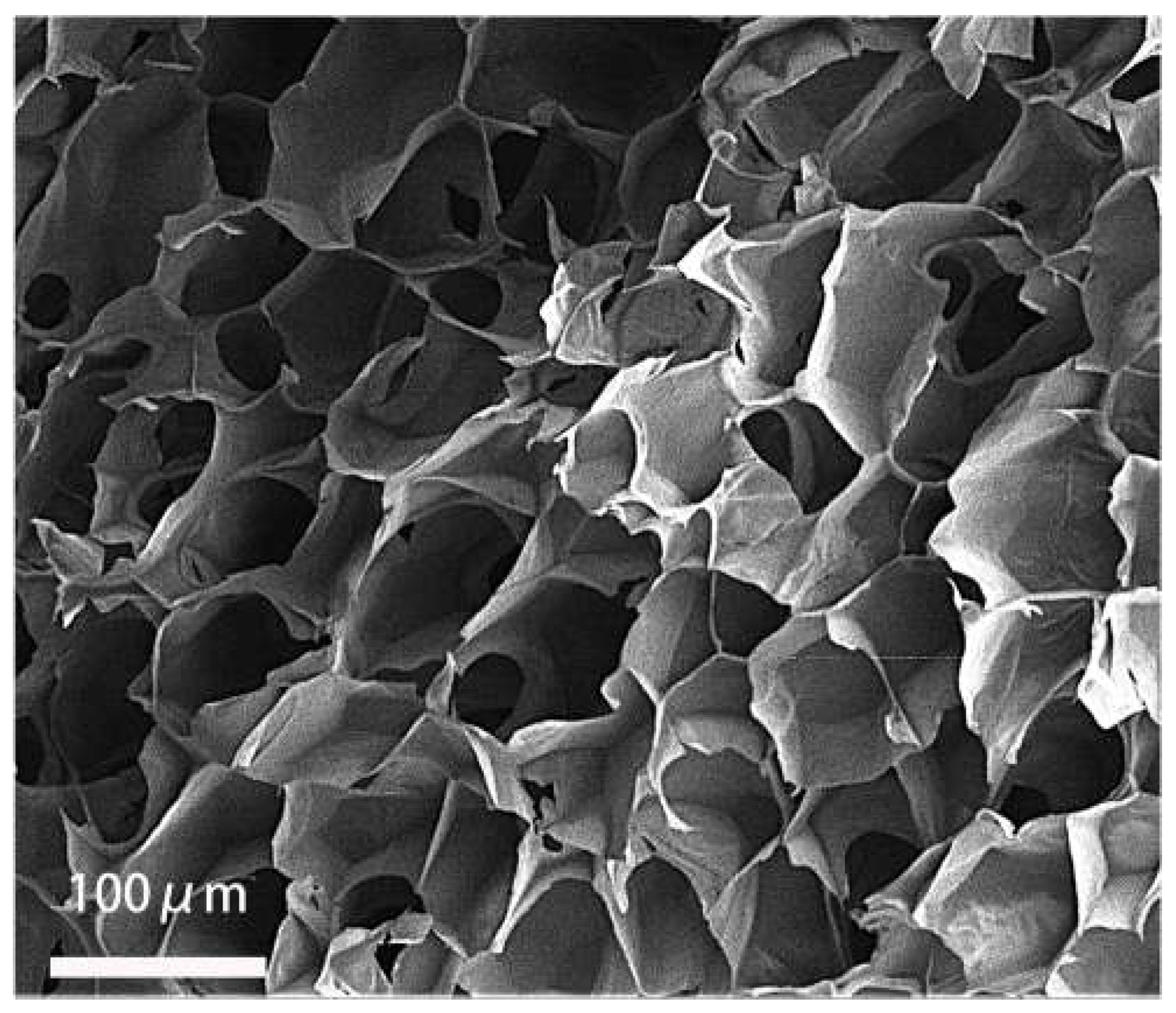
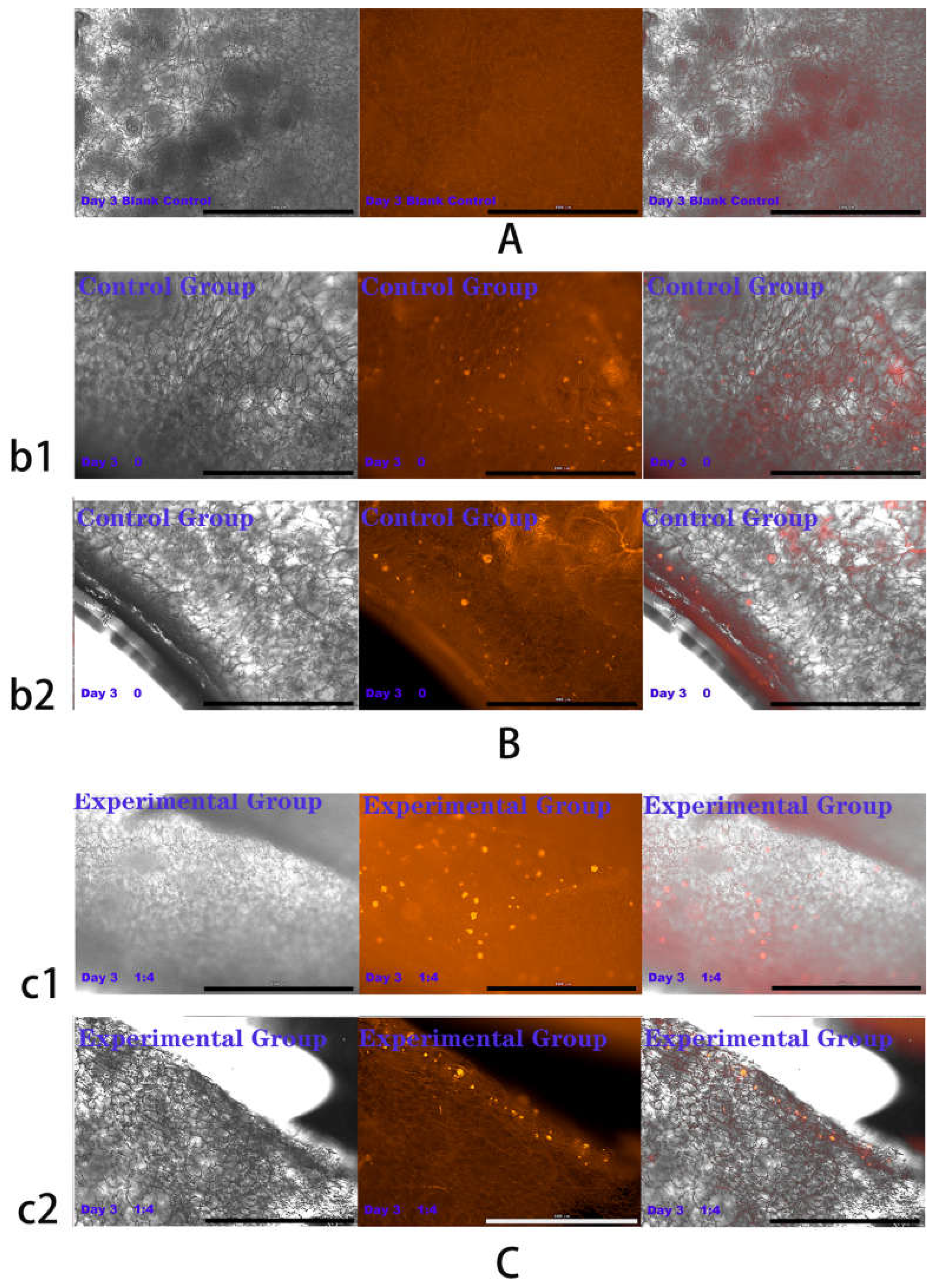
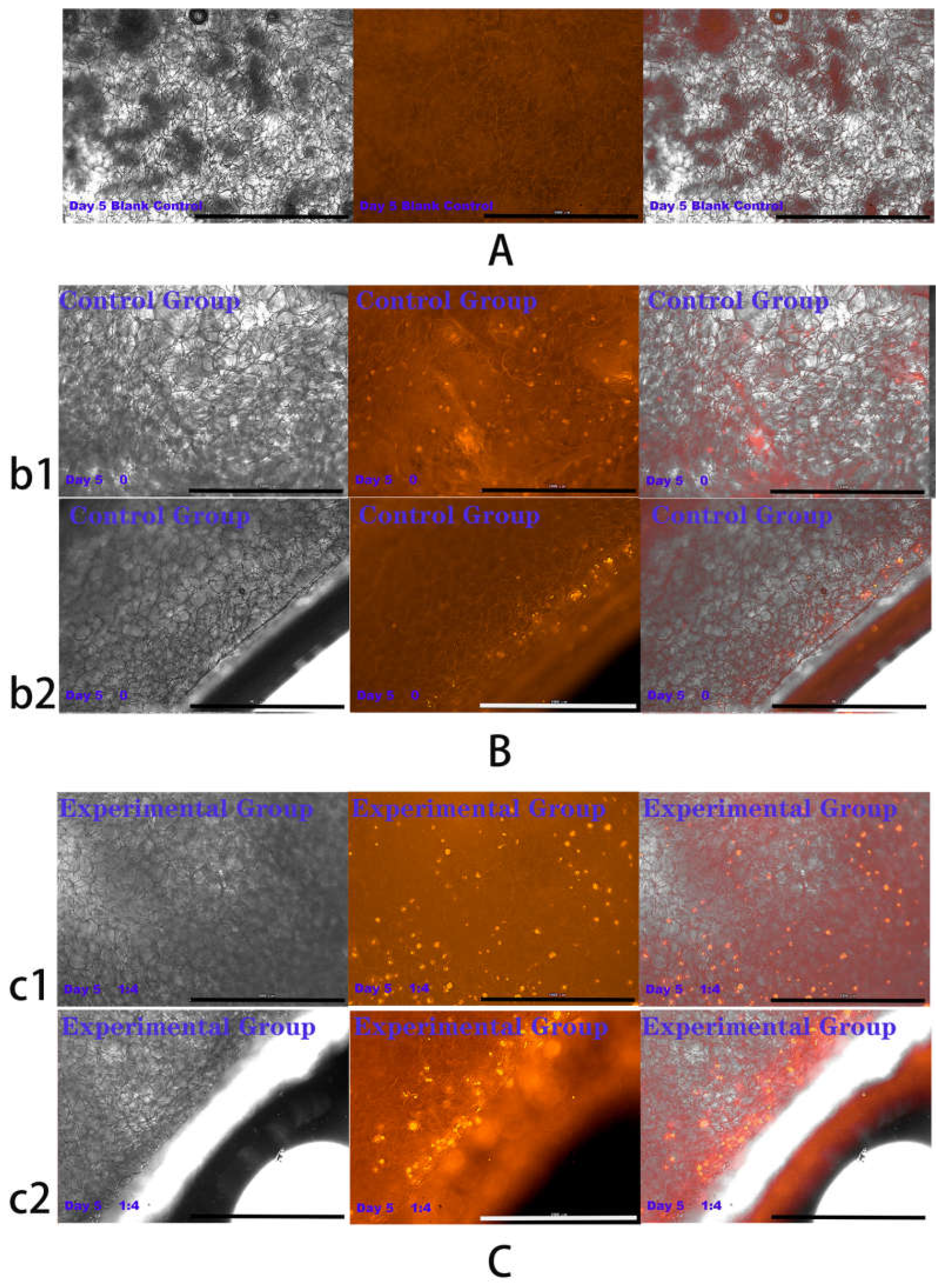
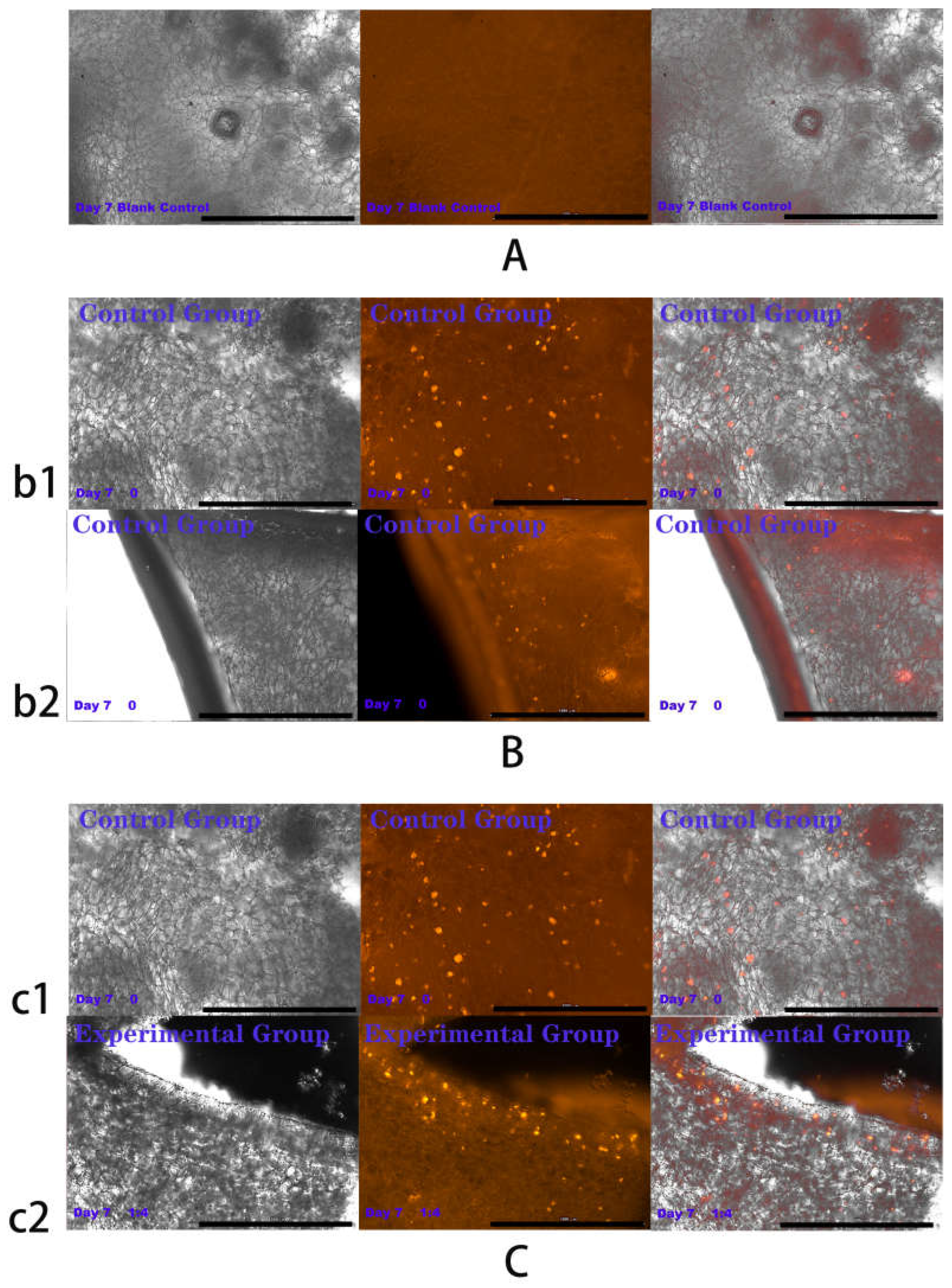
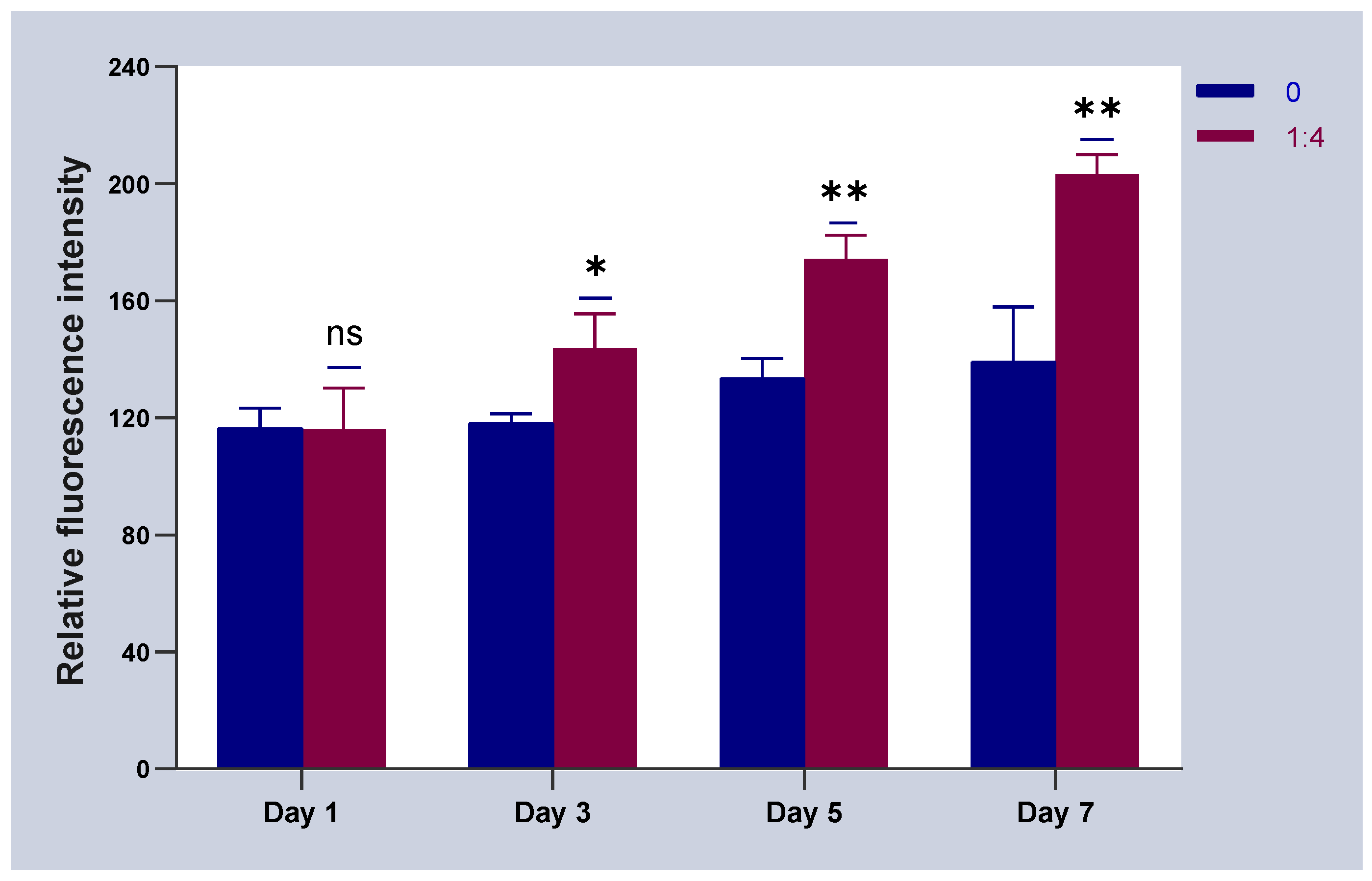
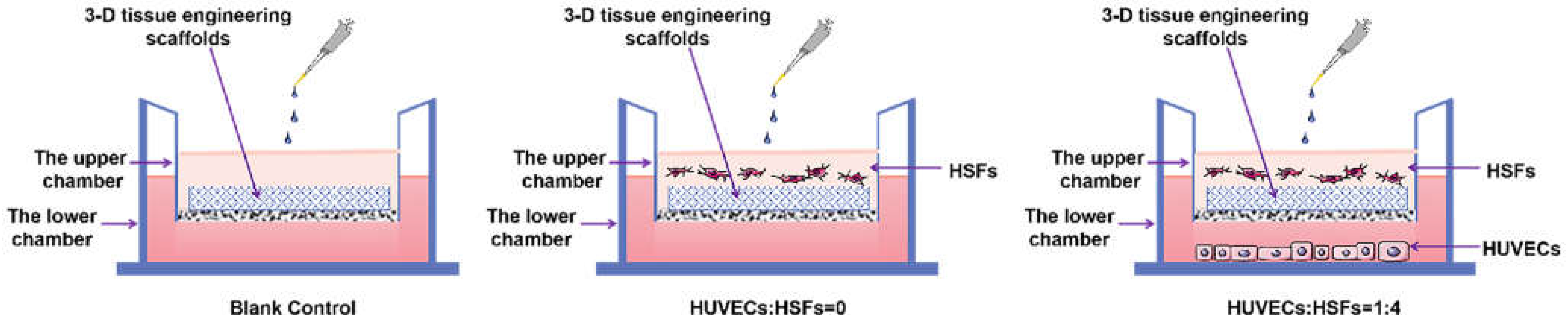
2.4. Results of HSFs Growth on Tissue Engineering Scaffolds
3. Discussion
4. Materials and Methods
4.1. Reagents and Apparatus
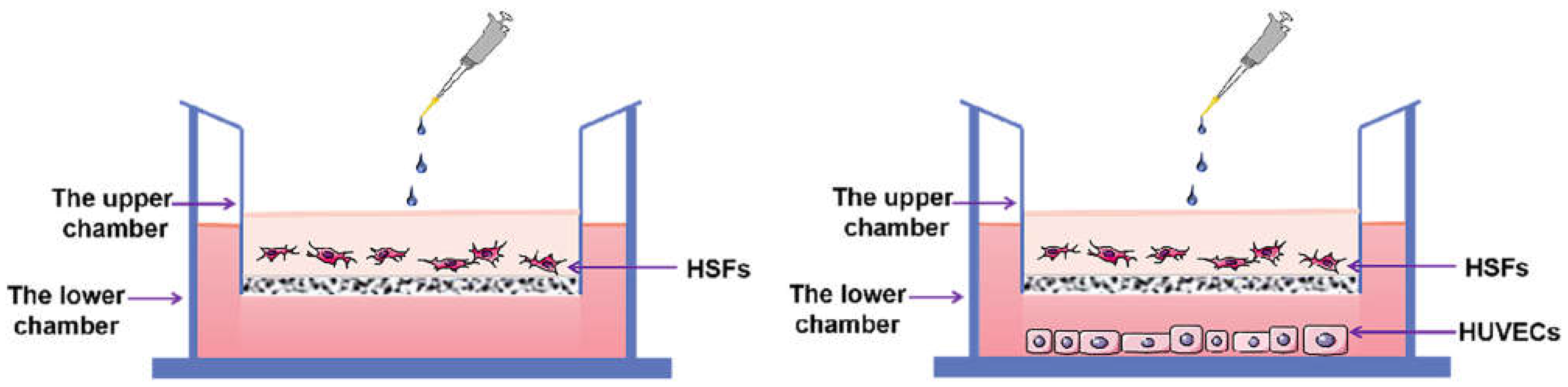
4.2. Establishment of Cell Co-Culture Model
4.3. Detection of HSFs Activity
4.4. Experiment on the Migration Ability of HSFs
4.4.1. Transwell Cell Migration Experiment
4.4.2. Detection of Migration Factor HMGB1 Gene Expression by RT-qPCR
4.4.3. The Expression of Migration Factor HMGB1 Protein was Detected by WB and Immunofluorescence
4.5. Detection of HSFs Vascularization Factor-VEGFA and FGF2
4.5.1. Detection of VEGFA and FGF2 Expression by RT-qPCR
4.5.2. Detection of VEGFA and FGF2 Expression by WB and Immunofluorescence
4.6. Detection of HSFs Growth on Tissue Engineering Scaffolds
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rezaie, F.; Momeni-Moghaddam, M.; Naderi-Meshkin, H. Regeneration and Repair of Skin Wounds: Various Strategies for Treatment. Int. J. Low. Extrem. Wounds 2019, 18, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Towler, M.A.; Rush, E.W.; Richardson, M.K.; Williams, C.L. Randomized, Prospective, Blinded-Enrollment, Head-to-Head Venous Leg Ulcer Healing Trial Comparing Living, Bioengineered Skin Graft Substitute (Apligraf) with Living, Cryopreserved, Human Skin Allograft (TheraSkin). Clin. Podiatr. Med. Surg. 2018, 35, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Morimoto, N.; Inoie, M.; Takahagi, M.; Ogino, S.; Jinno, C.; Suzuki, S. Cultured Human Epidermis Combined with Meshed Skin Autografts Accelerates Epithelialization and Granulation Tissue Formation in a Rat Model. Ann. Plast. Surg. 2017, 78, 651–658. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Keck, M.; Gugerell, A.; Kober, J. Engineering a Multilayered Skin Substitute with Keratinocytes, Fibroblasts, Adipose-Derived Stem Cells, and Adipocytes. In Skin Tissue Engineering; Humana: New York, NY, USA, 2019; pp. 149–157. [Google Scholar]
- Li, B.; Hu, W.; Ma, K.; Zhang, C.; Fu, X. Are hair follicle stem cells promising candidates for wound healing? Expert Opin. Biol. Ther. 2019, 19, 119–128. [Google Scholar] [CrossRef]
- Ud-Din, S.; Bayat, A. Non-invasive objective devices for monitoring the inflammatory, proliferative and remodelling phases of cutaneous wound healing and skin scarring. Exp. Dermatol. 2016, 25, 579–585. [Google Scholar] [CrossRef]
- Goodarzi, P.; Falahzadeh, K.; Nematizadeh, M.; Farazandeh, P.; Payab, M.; Larijani, B.; Beik, A.T.; Arjmand, B. Tissue-Engineered Skin Substitutes. Plast. Reconstr. Surg. 2018, 1107, 143–188. [Google Scholar]
- Rogovaya, O.S.; Zupnik, A.O.; Izmailova, L.S.; Vorotelyak, E. Morphofunctional Characteristics of Fibroblasts of the Papillary and Reticular Layers of Human Skin Dermis. Mosc. Univ. Biol. Sci. Bull. 2021, 76, 225–231. [Google Scholar] [CrossRef]
- Li, G.; Liu, Y.; Yao, J.; Song, K.; Wang, G.; Zhou, L.; Chen, G.; Liu, L. Biophysical model for high-throughput tumor and epithelial cell co-culture in complex biochemical microenvironments. Chin. Phys. B 2021, 31, 028703. [Google Scholar] [CrossRef]
- Yu, L.; Liang, X.H.; Ferrara, N. Comparing protein VEGF inhibitors: In Vitro biological studies. Biochem. Biophys. Res. Commun. 2011, 408, 276–281. [Google Scholar] [CrossRef]
- Wu, Y.X.; Ma, H.; Wang, J.L.; Qu, W. Production of chitosan scaffolds by lyophilization or electrospinning: Which is better for peripheral nerve regeneration? Neural Regen. Res. 2021, 16, 1093–1098. [Google Scholar]
- Sobhani, S.; Khaboushan, A.S.; Jafarnezhad-Ansariha, F.; Azimzadeh, A.; Danesh Payeh, M.; Kajbafzadeh, A.M. Off-the-shelf acellular fetal skin scaffold as a novel alternative to buccal mucosa graft: The development and characterization of human tissue-engineered fetal matrix in rabbit model of hypospadiasis. Int. Urol. Nephrol. 2022, 54, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Ma, H.; Gao, X.; Sun, H.; Wang, L.; An, M. Study on Long-Term Tracing of Fibroblasts on Three-Dimensional Tissue Engineering Scaffolds Based on Graphene Quantum Dots. Int. J. Mol. Sci. 2022, 23, 11040. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Kwon, Y.W.; Kong, J.S.; Park, G.T.; Gao, G.; Han, W.; Kim, M.-B.; Lee, H.; Cho, D.W. 3D cell printing of in vitro stabilized skin model and in vivo pre-vascularized skin patch using tissue-specific extracellular matrix bioink: A step towards advanced skin tissue engineering. Biomaterials 2018, 168, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Frueh, F.S.; Menger, M.D.; Lindenblatt, N.; Giovanoli, P.; Laschke, M.W. Current and emerging vascularization strategies in skin tissue engineering. Crit. Rev. Biotechnol. 2017, 37, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Gantwerker, E.A.; Hom, D.B. Skin: Histology and Physiology of Wound Healing. Clin. Plast. Surg. 2012, 39, 85–97. [Google Scholar] [CrossRef]
- Pazyar, N.; Yaghoobi, R.; Rafiee, E.; Mehrabian, A.; Feily, A. Skin Wound Healing and Phytomedicine: A Rewiew. Ski. Pharmacol. Physiol. 2014, 27, 303–310. [Google Scholar] [CrossRef]
- Oryan, A.; Alemzadeh, E.; Zarei, M. Basic concepts, current evidence, and future potential for gene therapy in managing cutaneous wounds. Biotechnol. Lett. 2019, 41, 889–898. [Google Scholar] [CrossRef]
- Park, G.S.; An, M.K.; Yoon, J.H.; Park, S.S.; Koh, S.H.; Mauro, T.M.; Cho, E.B.; Park, E.J.; Kim, K.J. Botulinum toxin type A suppresses pro-fibrotic effects via the JNK signaling pathway in hypertrophic scar fibroblasts. Arch. Dermatol. Res. 2019, 311, 807–814. [Google Scholar] [CrossRef]
- Karahan, A.; AAbbasoğlu, A.; Işık, S.A.; Çevik, B.; Saltan, Ç.; Elbaş, N.Ö.; Yalılı, A. Factors Affecting Wound Healing in Individuals with Pressure Ulcers: A Retrospective Study. Ostomy/Wound Manag. 2018, 64, 32–39. [Google Scholar] [CrossRef]
- Ning, X.L.; Li, D.; Sthle, M. Transition from inflammation to proliferation: A critical step during wound healing. Cell. Mol. Life Sci. 2016, 73, 3861–3885. [Google Scholar]
- Detmar, M. The role of VEGF and thrombospondins in skin angiogenesis. J. Dermatol. Sci. 2001, 24 (Suppl. S1), S78–S84. [Google Scholar] [CrossRef]
- Mishra, S.R.; Bharati, J.; Rajesh, G.; Chauhan, V.S.; Sharma, G.T.; Bag, S.; Maurya, V.P.; Singh, G.; Sarkar, M. Fibroblast growth factor 2 (FGF2) and vascular endothelial growth factor A (VEGFA) synergistically promote steroidogenesis and survival of cultured buffalo granulosa cells. Anim. Reprod. Sci. 2017, 179, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Terman, B.I.; Dougher-Vermazen, M.; Carrion, M.E.; Dimitrov, D.; Armellino, D.C.; Gospodarowicz, D.; Böhlen, P. Identification of the KDR tyrosine kinase as a receptor for vascular endothelial cell growth factor. Biochem. Biophys. Res. Commun. 1992, 187, 1579–1586. [Google Scholar] [CrossRef]
- Korntner, S.; Lehner, C.; Gehwolf, R.; Wagner, A.; Grütz, M.; Kunkel, N.; Tempfer, H.; Traweger, A. Limiting angiogenesis to modulate scar formation. Adv. Drug Deliv. Rev. 2019, 146, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Steringer, J.P.; Bleicken, S.; Andreas, H.; Zacherl, S.; Laussmann, M.; Temmerman, K.; Contreras, F.X.; Bharat, T.A.; Lechner, J.; Müller, H.M.; et al. Phosphatidylinositol 4,5-Bisphosphate (PI(4,5)P2)-dependent Oligomerization of Fibroblast Growth Factor 2 (FGF2) Triggers the Formation of a Lipidic Membrane Pore Implicated in Unconventional Secretion. J. Biol. Chem. 2012, 287, 27659–27669. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Hanwate, M.; Deb, K.; Sharma, V.; Totey, S. FGF2 secreting human fibroblast feeder cells: A novel culture system for human embryonic stem cells. Mol. Reprod. Dev. 2008, 75, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Takabayashi, Y.; Kuwabara, M.; Sato, Y.; Ishihara, M.; Takikawa, M.; Nakamura, S.; Fukuda, K.; Hiruma, S.; Kiyosawa, T. FGF-2-containing dalteparin/protamine nanoparticles (FGF-2&D/P NPs) ameliorate UV-induced skin photoaging in hairless mice. J. Plast. Surg. Hand Surg. 2018, 52, 375–381. [Google Scholar]
- Xu, J.; Yin, L.; Cao, S.; Zhan, H.; Zhang, J.; Zhou, Q.; Gong, K. Application of WALANT technique for repairing finger skin defect with a random skin flap. J. Orthop. Surg. Res. 2021, 16, 164. [Google Scholar] [CrossRef]
- Milan, P.B.; Pazouki, A.; Joghataei, M.T.; Mozafari, M.; Amini, N.; Kargozar, S.; Amoupour, M.; Latifi, N.; Samadikuchaksaraei, A. Decellularization and preservation of human skin: A platform for tissue engineering and reconstructive surgery. Methods 2020, 171, 62–67. [Google Scholar] [CrossRef]
- L’Heureux, N.; Germain, L.; Auger, F.A. Tissue engineering. Science 1999, 284, 1621–1622. [Google Scholar] [CrossRef]
- Sasaki, K.; Akagi, T.; Asaoka, T.; Eguchi, H.; Fukuda, Y.; Iwagami, Y.; Yamada, D.; Noda, T.; Wada, H.; Gotoh, K.; et al. Construction of three-dimensional vascularized functional human liver tissue using a layer-by-layer cell coating technique. Biomaterials 2017, 133, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Tait, A.; Proctor, T.; Hamilton, N.J.; Birchall, M.A.; Lowdell, M.W. GMP compliant isolation of mucosal epithelial cells and fibroblasts from biopsy samples for clinical tissue engineering. Sci. Rep. 2021, 11, 12392. [Google Scholar] [CrossRef] [PubMed]
- Merfeld-Clauss, S.; Lupov, I.P.; Lu, H.; March, K.L.; Traktuev, D.O. Adipose Stromal Cell Contact with Endothelial Cells Results in Loss of Complementary Vasculogenic Activity Mediated by Induction of Activin A. Stem Cells 2015, 33, 3039–3051. [Google Scholar] [CrossRef] [PubMed]
- Panina, Y.A.; Yakimov, A.S.; Komleva, Y.K.; Morgun, A.V.; Lopatina, O.L.; Malinovskaya, N.A.; Shuvaev, A.N.; Salmin, V.V.; Taranushenko, T.E.; Salmina, A.B. Plasticity of Adipose Tissue-Derived Stem Cells and Regulation of Angiogenesis. Front. Physiol. 2018, 9, 1656. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.Z. Fibroblasts-derived induced pluripotent stem cells in pulp revascularization: Differentiation into vascular endothelial cells and co-culture with dental pulp stem cells. Chin. J. Tissue Eng. Res. 2018, 22, 3371–3375. [Google Scholar]
- Paschos, N.K.; Brown, W.E.; Eswaramoorthy, R.; Hu, J.C.; Athanasiou, K.A. Advances in tissue engineering through stem cell-based co-culture. J. Tissue Eng. Regen. Med. 2015, 9, 488–503. [Google Scholar] [CrossRef]
- Zhang, H.L.; Sun, H.; Yang, Y.F.; Li, Y.M. Skin substitutes comprised of recombinant human collagen hydrogel promote full-thickness skin defect reconstruction. Burns 2022, 48, 1523–1524. [Google Scholar] [CrossRef]






| Gene Name | Forward Primer (F) Reverse Primer (R) |
|---|---|
| GAPDH | F: CAAGGGCATCCTGGGCTACACT R: CTCTCTCTTCCTCTTGTGCTCTTGC |
| HMGB1 | F: ATGCTTCAGTCAACTTCTCAGA R: CATTTCTCTTTCATAACGGGCC |
| Gene Name | Forward Primer (F) Reverse Primer (R) |
|---|---|
| GAPDH | F: CAAGGGCATCCTGGGCTACACT R: CTCTCTCTTCCTCTTGTGCTCTTGC |
| VEGFA | F: ATCGAGTACATCTTCAAGCCAT R: GTGAGGTTTGATCCGCATAATC |
| FGF-2 | F: CATCAAGCTACAACTTCAA-GCA R: CCGTAACACATTTAGAAGCCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, T.; Du, M.; Gao, X.; An, M. Human Vascular Endothelial Cells Promote the Secretion of Vascularization Factors and Migration of Human Skin Fibroblasts under Co-Culture and Its Preliminary Application. Int. J. Mol. Sci. 2022, 23, 13995. https://doi.org/10.3390/ijms232213995
Hou T, Du M, Gao X, An M. Human Vascular Endothelial Cells Promote the Secretion of Vascularization Factors and Migration of Human Skin Fibroblasts under Co-Culture and Its Preliminary Application. International Journal of Molecular Sciences. 2022; 23(22):13995. https://doi.org/10.3390/ijms232213995
Chicago/Turabian StyleHou, Tian, Miaomiao Du, Xiang Gao, and Meiwen An. 2022. "Human Vascular Endothelial Cells Promote the Secretion of Vascularization Factors and Migration of Human Skin Fibroblasts under Co-Culture and Its Preliminary Application" International Journal of Molecular Sciences 23, no. 22: 13995. https://doi.org/10.3390/ijms232213995
APA StyleHou, T., Du, M., Gao, X., & An, M. (2022). Human Vascular Endothelial Cells Promote the Secretion of Vascularization Factors and Migration of Human Skin Fibroblasts under Co-Culture and Its Preliminary Application. International Journal of Molecular Sciences, 23(22), 13995. https://doi.org/10.3390/ijms232213995






