FOXO1 Is a Key Mediator of Glucocorticoid-Induced Expression of Tristetraprolin in MDA-MB-231 Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Screening of a Natural Compound Library Identified an Enantiomer of Betamethasone 21-Phosphate and Dexamethasone 21-Phosphate as a Potent Inducer of TTP in Cancer Cells
2.2. Betamethasone 21-Phosphate, Dexamethasone 21-Phosphate, and Dexamethasone Induce the Expression of TTP in MDA-MB-231 Cells
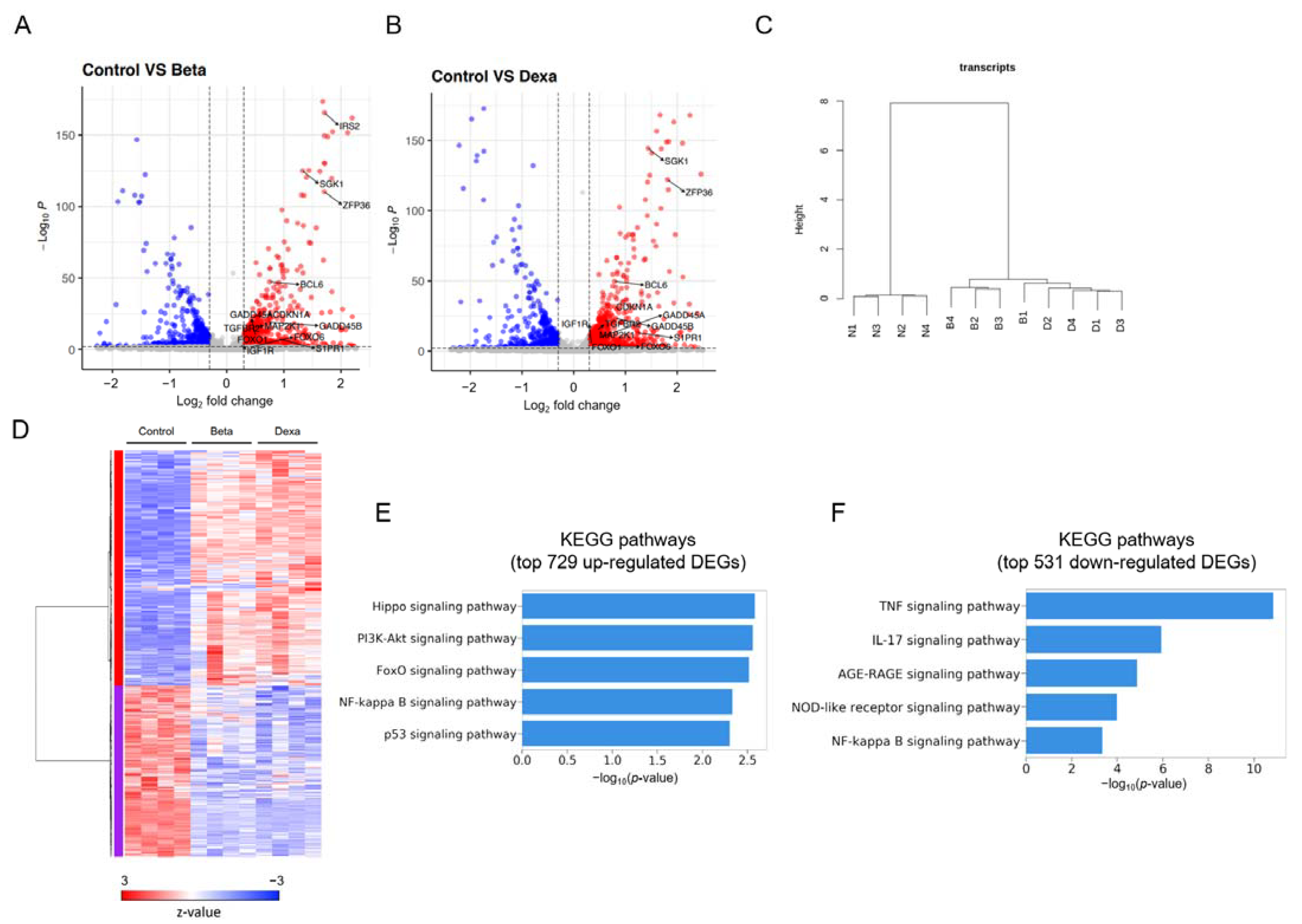
2.3. RNA-Seq Transcriptome Analysis of Betamethasone 21-Phosphate and Dexamethasone-Treated MDA-MB-231 Cells
2.4. FOXO1 Mediates Dexamethasone- and Betamethasone 21-Phosphate-Induced TTP Expression in MDA-MB-231 Cells
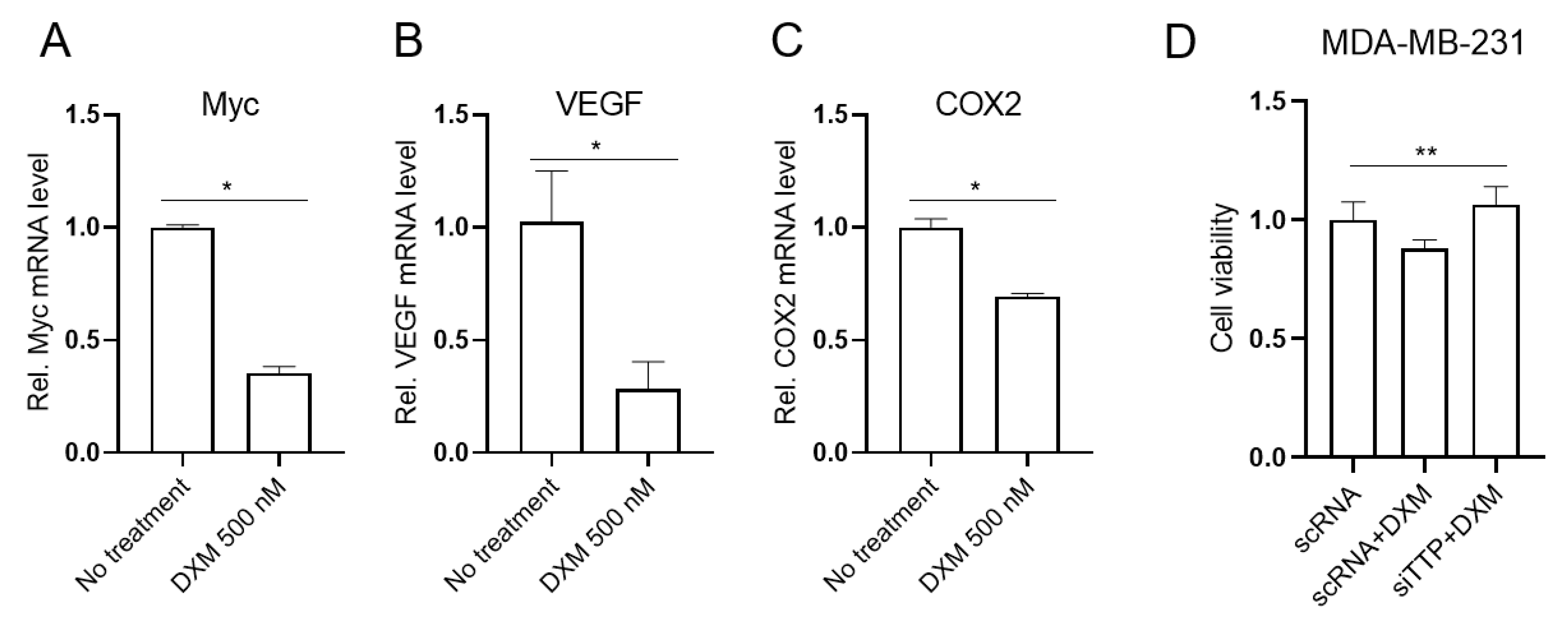
2.5. Dexamethasone-Induced TTP Down-Regulates ARE-Containing Genes in Cancer Cells and Mediates the Anti-Viability Effect of Dexamethasone
3. Discussion
4. Materials and Methods
4.1. Cells and Chemicals
4.2. Cell Viability
4.3. Plasmids, Small Interfering RNAs, Transfections, and Dual-Luciferase Assay
4.4. Screening of the Natural Product Library and Luciferase Assay
4.5. Quantitative Real-Time PCR and Semi-qRT-PCR
4.6. SDS-PAGE and Immunoblotting
4.7. RNA Preparation and RNA-Seq
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Brooks, S.A.; Blackshear, P.J. Tristetraprolin (TTP): Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 2013, 1829, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Carballo, E.; Lai, W.S.; Blackshear, P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 1998, 281, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.; Kamen, R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 1986, 46, 659–667. [Google Scholar] [CrossRef]
- Park, J.M.; Lee, T.H.; Kang, T.H. Roles of Tristetraprolin in Tumorigenesis. Int. J. Mol. Sci. 2018, 19, 3384. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, Z.; Yang, R.; Zhang, S.; Zhang, B.; Tan, Y.; Chen, L.; Li, T.; Tu, J. Tristetraprolin, a Potential Safeguard Against Carcinoma: Role in the Tumor Microenvironment. Front. Oncol. 2021, 11, 632189. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.S.; Stumpo, D.J.; Blackshear, P.J. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 1990, 265, 16556–16563. [Google Scholar] [CrossRef]
- Brennan, S.E.; Kuwano, Y.; Alkharouf, N.; Blackshear, P.J.; Gorospe, M.; Wilson, G.M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009, 69, 5168–5176. [Google Scholar] [CrossRef]
- Marderosian, M.; Sharma, A.; Funk, A.P.; Vartanian, R.; Masri, J.; Jo, O.D.; Gera, J.F. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene 2006, 25, 6277–6290. [Google Scholar] [CrossRef]
- Young, L.E.; Sanduja, S.; Bemis-Standoli, K.; Pena, E.A.; Price, R.L.; Dixon, D.A. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 2009, 136, 1669–1679. [Google Scholar] [CrossRef]
- Lee, H.H.; Son, Y.J.; Lee, W.H.; Park, Y.W.; Chae, S.W.; Cho, W.J.; Kim, Y.M.; Choi, H.J.; Choi, D.H.; Jung, S.W.; et al. Tristetraprolin regulates expression of VEGF and tumorigenesis in human colon cancer. Int. J. Cancer 2010, 126, 1817–1827. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, H.J.; Yoon, N.A.; Lee, W.H.; Min, Y.J.; Ko, B.K.; Lee, B.J.; Lee, A.; Cha, H.J.; Cho, W.J.; et al. Tumor suppressor p53 plays a key role in induction of both tristetraprolin and let-7 in human cancer cells. Nucleic Acids Res. 2013, 41, 5614–5625. [Google Scholar] [CrossRef]
- Rounbehler, R.J.; Fallahi, M.; Yang, C.; Steeves, M.A.; Li, W.; Doherty, J.R.; Schaub, F.X.; Sanduja, S.; Dixon, D.A.; Blackshear, P.J.; et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell 2012, 150, 563–574. [Google Scholar] [CrossRef]
- Soussi, T.; Beroud, C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer 2001, 1, 233–240. [Google Scholar] [CrossRef]
- Cole, M.D. The myc oncogene: Its role in transformation and differentiation. Ann. Rev. Genet. 1986, 20, 361–384. [Google Scholar] [CrossRef] [PubMed]
- Smoak, K.; Cidlowski, J.A. Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Mol. Cell Biol. 2006, 26, 9126–9135. [Google Scholar] [CrossRef]
- Ishmael, F.T.; Fang, X.; Galdiero, M.R.; Atasoy, U.; Rigby, W.F.; Gorospe, M.; Cheadle, C.; Stellato, C. Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation. J. Immunol. 2008, 180, 8342–8353. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Chivers, J.E.; Giembycz, M.A.; Newton, R. Long-acting beta2-adrenoceptor agonists synergistically enhance glucocorticoid-dependent transcription in human airway epithelial and smooth muscle cells. Mol. Pharmacol. 2008, 73, 203–214. [Google Scholar] [CrossRef] [PubMed]
- King, E.M.; Kaur, M.; Gong, W.; Rider, C.F.; Holden, N.S.; Newton, R. Regulation of tristetraprolin expression by interleukin-1 beta and dexamethasone in human pulmonary epithelial cells: Roles for nuclear factor-kappa B and p38 mitogen-activated protein kinase. J. Pharmacol. Exp. Ther. 2009, 330, 575–585. [Google Scholar] [CrossRef]
- Miller, W.L.; Auchus, R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011, 32, 81–151. [Google Scholar] [CrossRef]
- Sapolsky, R.M.; Romero, L.M.; Munck, A.U. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 2000, 21, 55–89. [Google Scholar]
- Rhen, T.; Cidlowski, J.A. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. N. Engl. J. Med. 2005, 353, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Oakley, R.H.; Cidlowski, J.A. The biology of the glucocorticoid receptor: New signaling mechanisms in health and disease. J. Allergy Clin. Immunol. 2013, 132, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Vandewalle, J.; Luypaert, A.; De Bosscher, K.; Libert, C. Therapeutic Mechanisms of Glucocorticoids. Trends Endocrinol. Metab. 2018, 29, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.P.; Zilliacus, J.; McEwan, I.J.; Dahlman-Wright, K.; Almlof, T.; Carlstedt-Duke, J.; Gustafsson, J.A. Structure and function of the glucocorticoid receptor. J. Steroid Biochem. Mol. Biol. 1993, 47, 11–19. [Google Scholar] [CrossRef]
- Kadmiel, M.; Cidlowski, J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013, 34, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Beato, M.; Chalepakis, G.; Schauer, M.; Slater, E.P. DNA regulatory elements for steroid hormones. J. Steroid Biochem. 1989, 32, 737–747. [Google Scholar] [CrossRef]
- Lim, H.W.; Uhlenhaut, N.H.; Rauch, A.; Weiner, J.; Hubner, S.; Hubner, N.; Won, K.J.; Lazar, M.A.; Tuckermann, J.; Steger, D.J. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015, 25, 836–844. [Google Scholar] [CrossRef]
- Diamond, M.I.; Miner, J.N.; Yoshinaga, S.K.; Yamamoto, K.R. Transcription factor interactions: Selectors of positive or negative regulation from a single DNA element. Science 1990, 249, 1266–1272. [Google Scholar] [CrossRef]
- Tuckermann, J.P.; Kleiman, A.; McPherson, K.G.; Reichardt, H.M. Molecular mechanisms of glucocorticoids in the control of inflammation and lymphocyte apoptosis. Crit. Rev. Clin. Lab. Sci. 2005, 42, 71–104. [Google Scholar] [CrossRef]
- Ratman, D.; Vanden Berghe, W.; Dejager, L.; Libert, C.; Tavernier, J.; Beck, I.M.; De Bosscher, K. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol. Cell Endocrinol. 2013, 380, 41–54. [Google Scholar] [CrossRef]
- Song, I.H.; Buttgereit, F. Non-genomic glucocorticoid effects to provide the basis for new drug developments. Mol. Cell Endocrinol. 2006, 246, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Wang, Y.; Wang, F.; Kim, M.S.; Abrahani, A.; Rodrigues, B. The increase in cardiac pyruvate dehydrogenase kinase-4 after short-term dexamethasone is controlled by an Akt-p38-forkhead box other factor-1 signaling axis. Endocrinology 2010, 151, 2306–2318. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, R.C.; Carr, A.J.; Hulley, P.A. Protection against glucocorticoid-induced damage in human tenocytes by modulation of ERK, Akt, and forkhead signaling. Endocrinology 2011, 152, 503–514. [Google Scholar] [CrossRef]
- Kaiser, G.; Gerst, F.; Michael, D.; Berchtold, S.; Friedrich, B.; Strutz-Seebohm, N.; Lang, F.; Haring, H.U.; Ullrich, S. Regulation of forkhead box O1 (FOXO1) by protein kinase B and glucocorticoids: Different mechanisms of induction of beta cell death in vitro. Diabetologia 2013, 56, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Xiao, J.J.; Liu, L.; Jiao, H.C.; Lin, H. Excessive glucocorticoid-induced muscle MuRF1 overexpression is independent of Akt/FoXO1 pathway. Biosci. Rep. 2017, 37, BSR20171056. [Google Scholar] [CrossRef]
- Felice, F.; Cesare, M.M.; Fredianelli, L.; De Leo, M.; Conti, V.; Braca, A.; Di Stefano, R. Effect of Tomato Peel Extract Grown under Drought Stress Condition in a Sarcopenia Model. Molecules 2022, 27, 2563. [Google Scholar] [CrossRef]
- Arden, K.C. Multiple roles of FOXO transcription factors in mammalian cells point to multiple roles in cancer. Exp. Gerontol. 2006, 41, 709–717. [Google Scholar] [CrossRef]
- Fu, Z.; Tindall, D.J. FOXOs, cancer and regulation of apoptosis. Oncogene 2008, 27, 2312–2319. [Google Scholar] [CrossRef]
- Miyamoto, K.; Araki, K.Y.; Naka, K.; Arai, F.; Takubo, K.; Yamazaki, S.; Matsuoka, S.; Miyamoto, T.; Ito, K.; Ohmura, M.; et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem. Cell 2007, 1, 101–112. [Google Scholar] [CrossRef]
- Zhang, X.; Yong, W.; Lv, J.; Zhu, Y.; Zhang, J.; Chen, F.; Zhang, R.; Yang, T.; Sun, Y.; Han, X. Inhibition of forkhead box O1 protects pancreatic beta-cells against dexamethasone-induced dysfunction. Endocrinology 2009, 150, 4065–4073. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Matkar, S.; He, X.; Hua, X. FOXO family in regulating cancer and metabolism. Semin. Cancer Biol. 2018, 50, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Kalfeist, L.; Galland, L.; Ledys, F.; Ghiringhelli, F.; Limagne, E.; Ladoire, S. Impact of Glucocorticoid Use in Oncology in the Immunotherapy Era. Cells 2022, 11, 770. [Google Scholar] [CrossRef]
- Almawi, W.Y.; Saouda, M.S.; Stevens, A.C.; Lipman, M.L.; Barth, C.M.; Strom, T.B. Partial mediation of glucocorticoid antiproliferative effects by lipocortins. J. Immunol. 1996, 157, 5231–5239. [Google Scholar]
- Almawi, W.Y.; Tamim, H. Posttranscriptional mechanisms of glucocorticoid antiproliferative effects: Glucocorticoids inhibit IL-6-induced proliferation of B9 hybridoma cells. Cell Transpl. 2001, 10, 161–164. [Google Scholar] [CrossRef]
- Gruber, A.R.; Fallmann, J.; Kratochvill, F.; Kovarik, P.; Hofacker, I.L. Aresite: A database for the comprehensive investigation of au-rich elements. Nucleic Acids Res. 2011, 39, D66–D69. [Google Scholar] [CrossRef]
- Khabar, K.S.A. Hallmarks of cancer and AU-rich elements. Wiley Interdiscip. Rev. RNA 2017, 8, 1368. [Google Scholar] [CrossRef]
- Bisogno, L.S.; Keene, J.D. RNA regulons in cancer and inflammation. Curr. Opin. Genet. Dev. 2018, 48, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Wagner, E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005, 19, 351–361. [Google Scholar] [CrossRef]
- Jing, Q.; Huang, S.; Guth, S.; Zarubin, T.; Motoyama, A.; Chen, J.; Padova, F.D.; Lin, S.; Gram, H.; Han, J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005, 120, 623–634. [Google Scholar] [CrossRef]
- Surjit, M.; Ganti, K.P.; Mukherji, A.; Ye, T.; Hua, G.; Metzger, D.; Li, M.; Chambon, P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 2011, 145, 224–241. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.; Ganti, K.P.; Chambon, P. Glucocorticoid-induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/NCoR1-HDAC3 repressing complex. Proc. Natl. Acad. Sci. USA 2016, 113, E635–E643. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.L.; Maler, B.A.; Yamamoto, K.R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell 1983, 33, 489–499. [Google Scholar] [CrossRef]
- Schiller, B.J.; Chodankar, R.; Watson, L.C.; Stallcup, M.R.; Yamamoto, K.R. Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol. 2014, 15, 418. [Google Scholar] [CrossRef]
- Obradović, M.M.S.; Hamelin, B.; Manevski, N.; Couto, J.P.; Sethi, A.; Coissieux, M.M.; Münst, S.; Okamoto, R.; Kohler, H.; Schmidt, A.; et al. Glucocorticoids promote breast cancer metastasis. Nature 2019, 567, 540–544. [Google Scholar] [CrossRef]
- Crozier, M.; Tubman, J.; Fifield, B.; Ferraiuolo, R.; Ritchie, J.; Zuccato, K.; Mailloux, E.; Sinha, I.; Hamm, C.; Porter, L.A. Frequently used antiemetic agent dexamethasone enhances the metastatic behaviour of select breast cancer cells. PLoS ONE 2022, 17, e0274675. [Google Scholar] [CrossRef]
- Yoon, N.A.; Jo, H.G.; Lee, U.H.; Park, J.H.; Yoon, J.E.; Ryu, J.; Kang, S.S.; Min, Y.J.; Ju, S.A.; Seo, E.H.; et al. Tristetraprolin suppresses the EMT through the down-regulation of Twist1 and Snail1 in cancer cells. Oncotarget 2016, 7, 8931–8943. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; The International Natural Product Sciences Taskforce; Claudiu, T.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Dis. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]




| Genes | Primer Sequences (5′-3′) | ||
|---|---|---|---|
| Gene expression analysis | β-actin | F | ATCGTGCGTGACATTAAGGAGAAG |
| R | AGGAAGGAAGGCTGCAAG | ||
| BCL6 | F | CATGCAGAGATGTGCCTCCACA | |
| R | TCAGAGAAGCGGCAGTCACACT | ||
| CDKN1A | F | AGGTGGACCTGGAGACTCTCAG | |
| R | TCCTCTTGGAGAAGATCAGCCG | ||
| FOXO1 | F | CTACGAGTGGATGGTCAAGAGC | |
| R | CCAGTTCCTTCATTCTGCACACG | ||
| GADD45A | F | CTGGAGGAAGTGCTCAGCAAAG | |
| R | AGAGCCACATCTCTGTCGTCGT | ||
| GAPDH | F | AATCCCATCACCATCTTCCAG | |
| R | AAATGAGCCCCAGCCTTC | ||
| IRS2 | F | CCTGCCCCCTGCCAACACCT | |
| R | TGTGACATCCTGGTGATAAAGCC | ||
| S1PR1 | F | CCTGTGACATCCTCTTCAGAGC | |
| R | CACTTGCAGCAGGACATGATCC | ||
| SGK1 | F | GCTGAAATAGCCAGTGCCTTGG | |
| R | GTTCTCCTTGCAGAGTCCGAAG | ||
| TTP | F | TCTTCGAGGCGGGAGTTTTT | |
| R | TGCGATTGAAGATGGGGAGTC | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, D.Y.; Jeong, S.Y.; Lee, J.W.; Kim, J.; Kim, J.H.; Chu, H.S.; Jeong, W.J.; Lee, B.J.; Ahn, B.; Kim, J.; et al. FOXO1 Is a Key Mediator of Glucocorticoid-Induced Expression of Tristetraprolin in MDA-MB-231 Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 13673. https://doi.org/10.3390/ijms232213673
Jeon DY, Jeong SY, Lee JW, Kim J, Kim JH, Chu HS, Jeong WJ, Lee BJ, Ahn B, Kim J, et al. FOXO1 Is a Key Mediator of Glucocorticoid-Induced Expression of Tristetraprolin in MDA-MB-231 Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(22):13673. https://doi.org/10.3390/ijms232213673
Chicago/Turabian StyleJeon, Do Yong, So Yeon Jeong, Ju Won Lee, Jeonghwan Kim, Jee Hyun Kim, Hun Su Chu, Won Jin Jeong, Byung Ju Lee, Byungyong Ahn, Junil Kim, and et al. 2022. "FOXO1 Is a Key Mediator of Glucocorticoid-Induced Expression of Tristetraprolin in MDA-MB-231 Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 22: 13673. https://doi.org/10.3390/ijms232213673
APA StyleJeon, D. Y., Jeong, S. Y., Lee, J. W., Kim, J., Kim, J. H., Chu, H. S., Jeong, W. J., Lee, B. J., Ahn, B., Kim, J., Choi, S. H., & Park, J. W. (2022). FOXO1 Is a Key Mediator of Glucocorticoid-Induced Expression of Tristetraprolin in MDA-MB-231 Breast Cancer Cells. International Journal of Molecular Sciences, 23(22), 13673. https://doi.org/10.3390/ijms232213673








