Abstract
The longevity phenomenon is entirely controlled by the insulin signaling pathway (IIS-pathway). Both vertebrates and invertebrates have IIS-pathways that are comparable to one another, though no one has previously described de novo transcriptome assembly of IIS-pathway-associated genes in termites. In this research, we analyzed the transcriptomes of both reproductive (primary kings “PK” and queens “PQ”, secondary worker reproductive kings “SWRK” and queens “SWRQ”) and non-reproductive (male “WM” and female “WF” workers) castes of the subterranean termite Reticulitermes chinensis. The goal was to identify the genes responsible for longevity in the reproductive and non-reproductive castes. Through transcriptome analysis, we annotated 103,589,264 sequence reads and 184,436 (7G) unigenes were assembled, GC performance was measured at 43.02%, and 64,046 sequences were reported as CDs sequences. Of which 35 IIS-pathway-associated genes were identified, among 35 genes, we focused on the phosphoinositide-dependent kinase-1 (Pdk1), protein kinase B2 (akt2-a), tuberous sclerosis-2 (Tsc2), mammalian target of rapamycin (mTOR), eukaryotic translation initiation factor 4E (EIF4E) and ribosomal protein S6 (RPS6) genes. Previously these genes (Pdk1, akt2-a, mTOR, EIF4E, and RPS6) were investigated in various organisms, that regulate physiological effects, growth factors, protein translation, cell survival, proliferation, protein synthesis, cell metabolism and survival, autophagy, fecundity rate, egg size, and follicle number, although the critical reason for longevity is still unclear in the termite castes. However, based on transcriptome profiling, the IIS-pathway-associated genes could prolong the reproductive caste lifespan and health span. Therefore, the transcriptomic shreds of evidence related to IIS-pathway genes provide new insights into the maintenance and relationships between biomolecular homeostasis and remarkable longevity. Finally, we propose a strategy for future research to decrypt the hidden costs associated with termite aging in reproductive and non-reproductive castes.
1. Introduction
The insulin signaling pathway (IIS-pathway) comprises all the proteins and components involved in the action of insulin within the body [1,2,3]. Insulin is the most potent physiological anabolic agent that has ever been discovered (1921) [4,5]. It is responsible for storing and synthesizing lipids, protein, and carbohydrates, and it prevents the breakdown of these macromolecules and their release into circulation [6,7]. The IIS-pathway has been thoroughly investigated in different organisms to regulate their life span [8,9]. The IIS-pathway is responsible for mediating and transducing signals across cell membranes and regulating the physiological aspects of reproduction [10,11,12].
The inhibition of upstream IIS-pathway components (daf-2 and age-1) significantly prolongs the life and health span of Caenorhabditis elegans [13]. On the other hand, IGF-I and IGF-II monitor growth and growth factors (GF) [13]; nutrient sensing, stress replication [14], cell functions, and metabolic regulation is gradually deteriorating due to aging [14,15]. Similarly, IIS-related genes are also responsible for activating hormones and the motility of cells [16,17].
Researchers previously reported that IIS-associated genes that prolong the lifespan of Drosophila melanogaster [18,19], dauer state in C. elegans [20,21], and reproductives diapause in Culex pipiens mosquito [22]. It also indicated that squirrels live for twenty-five years, rats live for three years, and C. elegans, D. longispina, and D. melanogaster live only for a few weeks [23,24]. Long-lived queens of insects, including Ephemera simulans [23], Pogonomyrmex owyheei [25], and Lasius niger can live up to ten years [26]. Polistes canadensis (a social insect) has a lifespan and genetic variation up to the genus level within a single colony [27]. However, there is still a significant gap between what we know and what we do not know regarding the enormous variety of aging rates among different animals and the mechanisms that might be responsible for this diversity [28]. Similarly, the royal caste of termites lives for 18–30 years, ultimately making termites an emerging model for longevity [29] and therefore requires further attention to explore its molecular pathways. However, there are no appropriate mechanisms available for determining the life span of a termite reproductive group [30,31].
We followed the IIS-pathway (PI3K-Akt are the main effector pathways in IIS-signalling) in insects [12,32,33,34] to determine the IIS-pathway in primary and secondary worker reproductives (many subterranean termite colonies consist of former worker termites or immatures that have developed into larvae and eventually become wingless reproductives to supplement the colony), and non-reproductive subterranean termites [29,35,36]. Longevity in Reticulitermes chinensis was determined through next-generation sequencing (NGS) or transcriptomic [36]. A detailed evaluation of transcriptome sequences was examined with developmental processes, biological, and physiological changes in R. chinensis castes [29]. The de novo transcriptome assembly of Reticulitermes chinensis, IIS-pathway genes are enriched, contributing to termite longevity and health. In addition, the reported IIS-pathway and longevity genes in secondary worker reproductive kings (SWRK) and queens (SWRQ), primary kings (PK) and queens (PK), and male (WM) and female (WF) workers were analyzed to determine their life extension mechanism and co-evolutionary process. To better understand the genetic basis for longevity, healthy lifespan, and growth regulation in social insects and other insects, this study presents a comprehensive analysis of the IIS-pathway-associated genes implicated in R. chinensis longevity.
2. Results
2.1. Illumina Data Sequencing and De Novo Transcriptome Assembly
RNA-seq libraries were established from R. chinensis caste (PK, PQ, SWRQ, SWRK, WM, and WF). We annotated a total of 103,589,264 assembled sequence reads with an average length of 561 bp and a minimum of 201 bp (Figure S2) via the Illumina HiSeqTM 4000 platform (Figure S3A,B). The total number of CDs sequences was reported as 64,046 sequences. Through the Trinity system (trinityrnaseq r2012-04-27), 184,436 unigenes were assembled from 201 to 43,214 bp length from precise transcriptome data (7G) (Table S2). The assembly pipeline of Trinity is made up of six castes that come one after the other: PK (PK-1, PK-2, PK-3), PQ (PQ-1, PQ-2, PQ-3), SWRK (SWRK-1, SWRK-2, SWRK-3), SWRQ (SWRQ-1, SWRQ-2, SWRQ-3), WM (WM-1, WM-2, WM-3), WF (WF-1, WF-2, WF-3). The GC content was estimated at 43.02%, suggesting that sequence quality and development are adequate for homogeneous studies [37].
2.2. Functional Annotation of R. chinensis
The Non-redundant (NR) protein database was used for R. chinensis annotation applied for BLASTx (http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 15 March 2021)) program. A total of 184,436 unigenes were annotated with an e-threshold value of 1 × 10−5 (Figure S4). In total, the NR database (34.44%), the Kyoto Genes and Genomes Encyclopedia (KEGG) database (23.64%), Swiss-Prot (17.22%), and the Clusters of Orthologous Groups (COG) database (15%) had unigenes which exhibited influential matches. The Pie chart designated excellent insect genome hits and marked the termite Zootermopsis nevadensis (42%) (Figure S6). In this study, we investigate the metabolic pathways that affect the longevity of the termite castes through transcriptome sequencing. Therefore, 400 termite flagellates have been previously reported to be absorbed and metabolized in cellulose metabolism. Reticulitermes termites rely on intestinal flagellates to digest cellulose and produce the end products (acetate, CO2, and H2) of cellulose fermentation by mixed flagellates (Tritrichomonas species). Additionally, several databases show their relatedness with R. chinensis unigenes like Zootermopsis nevadensis (42%), Tritrichomonas foetus (27%), Trichomonas vaginalis G3 (14%), Coptotermes formosanus (5%), Acanthamoeba glabripennis (2%), and Acanthamoeba castellanli (2%). The genes 120,113 (65.12%) of the sequences were compiled and unable to identify due to the lack of annotations of R. chinensis genome and short gene sequences. These short sequences include R. chinensis genes, unigenes, or short fragments, principally from the untranslated region (5′ and 3′ UTRs) or non-conserved region of protein-coding transcriptomes (Figure S5).
2.3. COG, KEGG, and GO Ontology Classifications
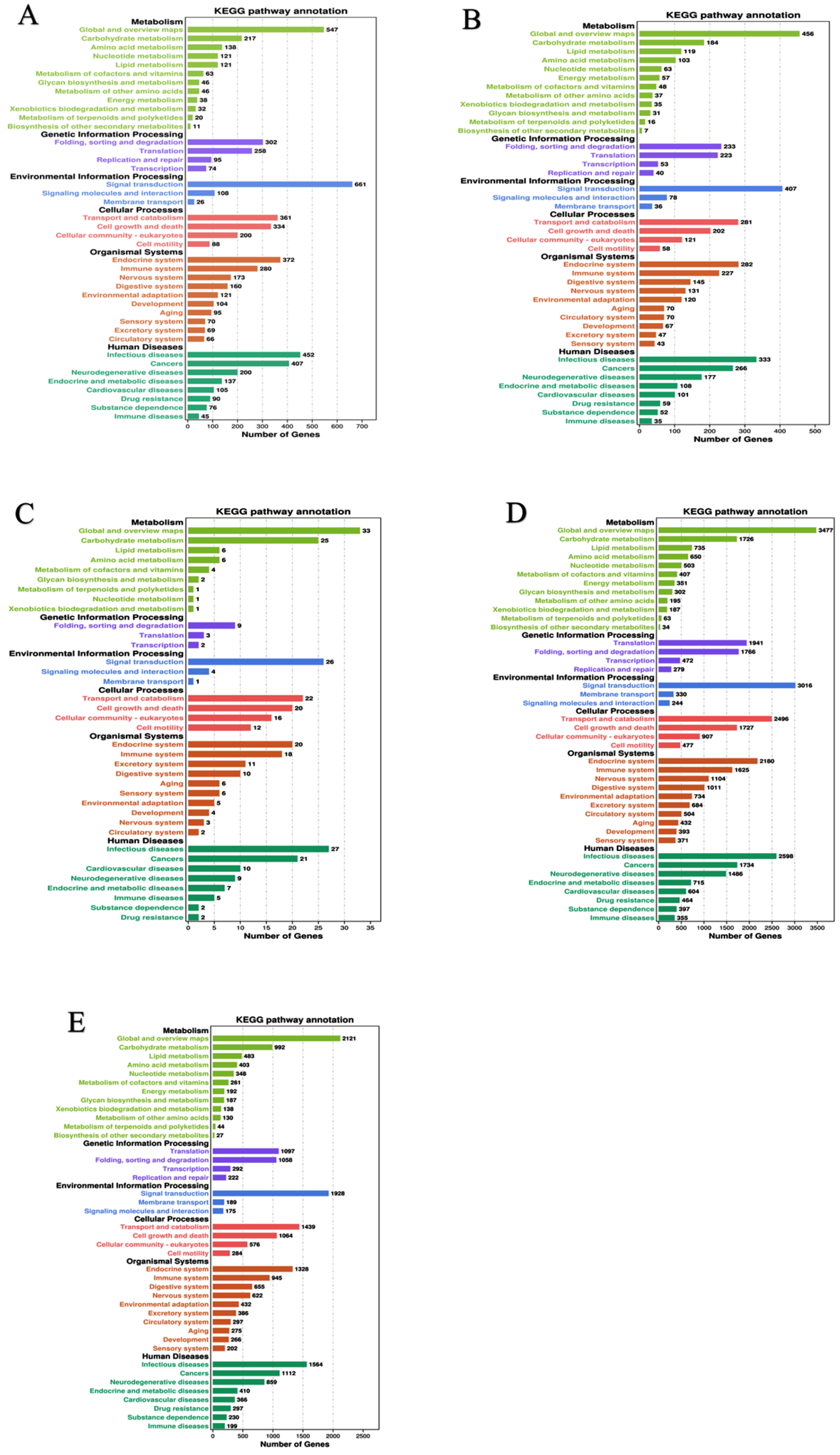
In the functional COG classification with 25 categories, additionally annotated 36,681 unigenes, the general functional prediction had 5707 unigenes (the colossal group), followed by cell motility (97) and nuclear structure (93), the minuscule group (Table 1). We have mapped the unique sequences into KEGG to understand the biological pathways of R. chinensis, including the mitogen-activated protein kinase (MAPK) pathway and the PI3K-Akt pathway, the two pathways that are involved in insulin signaling. However, the PI3K pathway is the most prominent insect insulin signaling pathway. From total castes, 93,078 unigenes were analyzed in IIS-pathway using transcriptome sequencing, and 343 genes were allocated to KEGG pathways (Figure 1A–E).

Table 1.
The table represents clusters of the orthologous group (COG) classifications. In 25 categories of COG classification, a total of 36,681 unigenes were recorded.
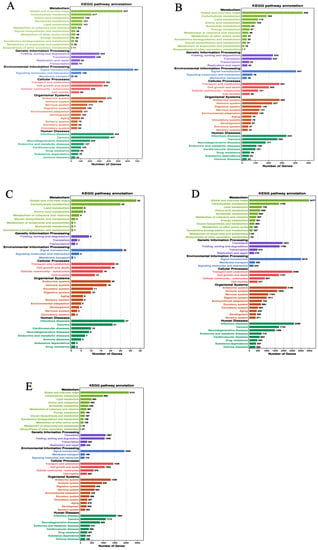
Figure 1.
KEGG classifications of the R. chinensis unigenes. (A) PK-vs-SWRK; (B) PQ-vs-SWRQ; (C) SWRK-vs-SWRQ; (D) WF-vs-SWRQ; (E) WM-vs-SWRK. PKs (primary king), PQs (primary queen), SWRK (secondary worker reproductive king), SWRQ (secondary worker reproductive queen), WMs (workers male), and WFs (female workers), with 93,078 unigenes were assigned to 343 pathways.
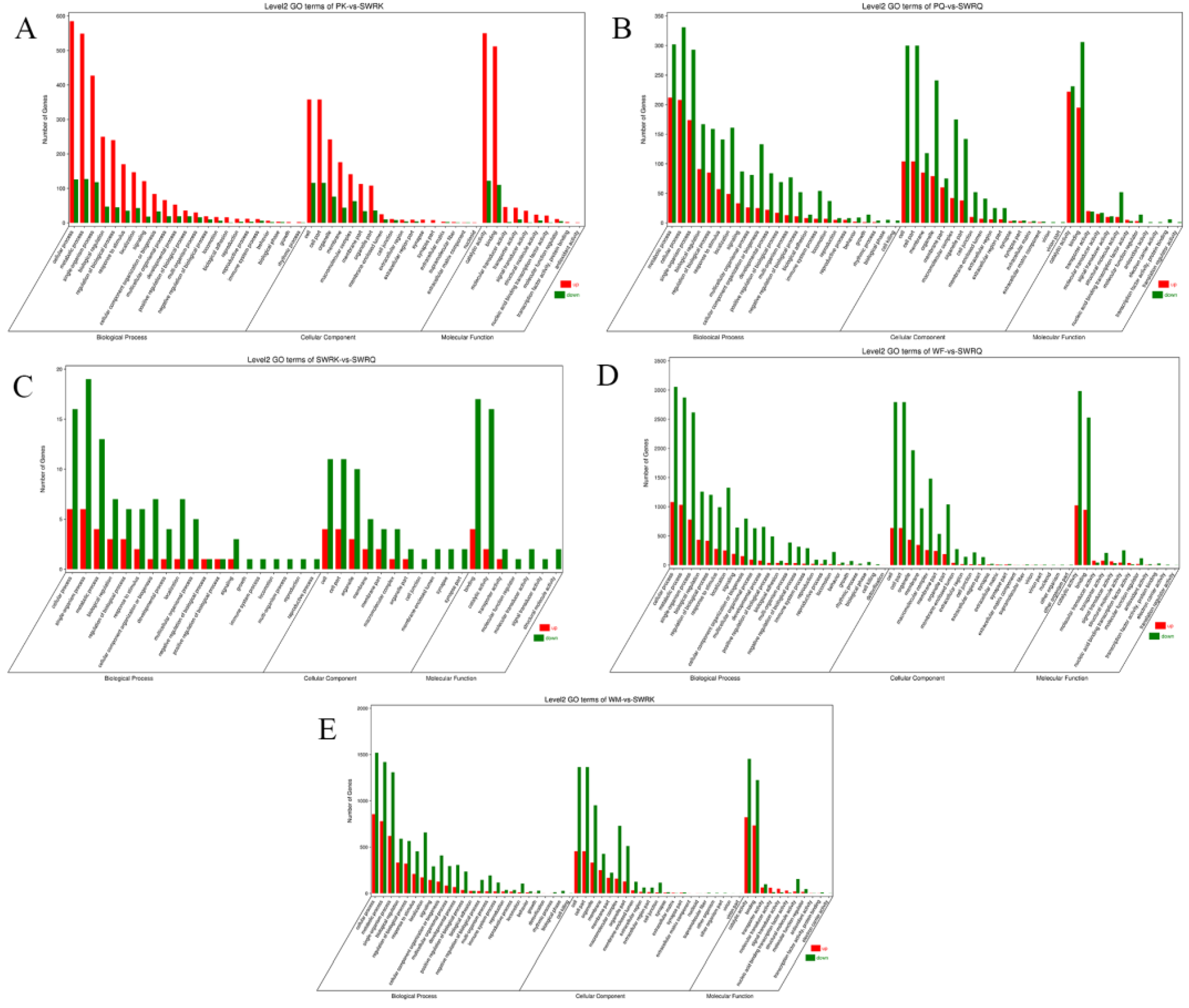
All DEGs in this study were mapped to terms in the GO database and determined the functions of the differentially expressed genes. Among 59 GO terms, the level2 GO enrichment analysis classified genes according to their molecular function (MF), cellular components (CC), and biological process (PB). The results indicated that PK-vs-SWRK castes exhibited significantly up-regulated unigenes. The categorical presentation suggests that a maximum number of up-regulated genes are reported in BP (cellular processes), followed by CC (cell and cell part), and MF (catalytic activity) (Figure 1A). Among PQ-vs-SWRQ castes, the level2 GO terms were reported significantly as down-regulated. The maximum number of down-regulated are cellular processes at BP, followed by cell and cell part in CC and binding at MF (Figure 1B).
Furthermore, level 2 GO terms of SWRK-vs-SWRQ individuals are additionally considerably reported as down-regulated genes shown at BP (Figure 1C). The level2 GO terms for WF-vs-SWRQ and WM-vs-SWRK were significantly down-regulated. The categorical cluster indicates that the BP possessed cellular processes, followed by CC (cell and cell part) and catalytic activity at MF (Figure 1D,E).
2.4. DEGs Analysis, Protein-Coding Region Prediction (CDS) in SWRK, SWRQ, PQ, PK, WF, and WM
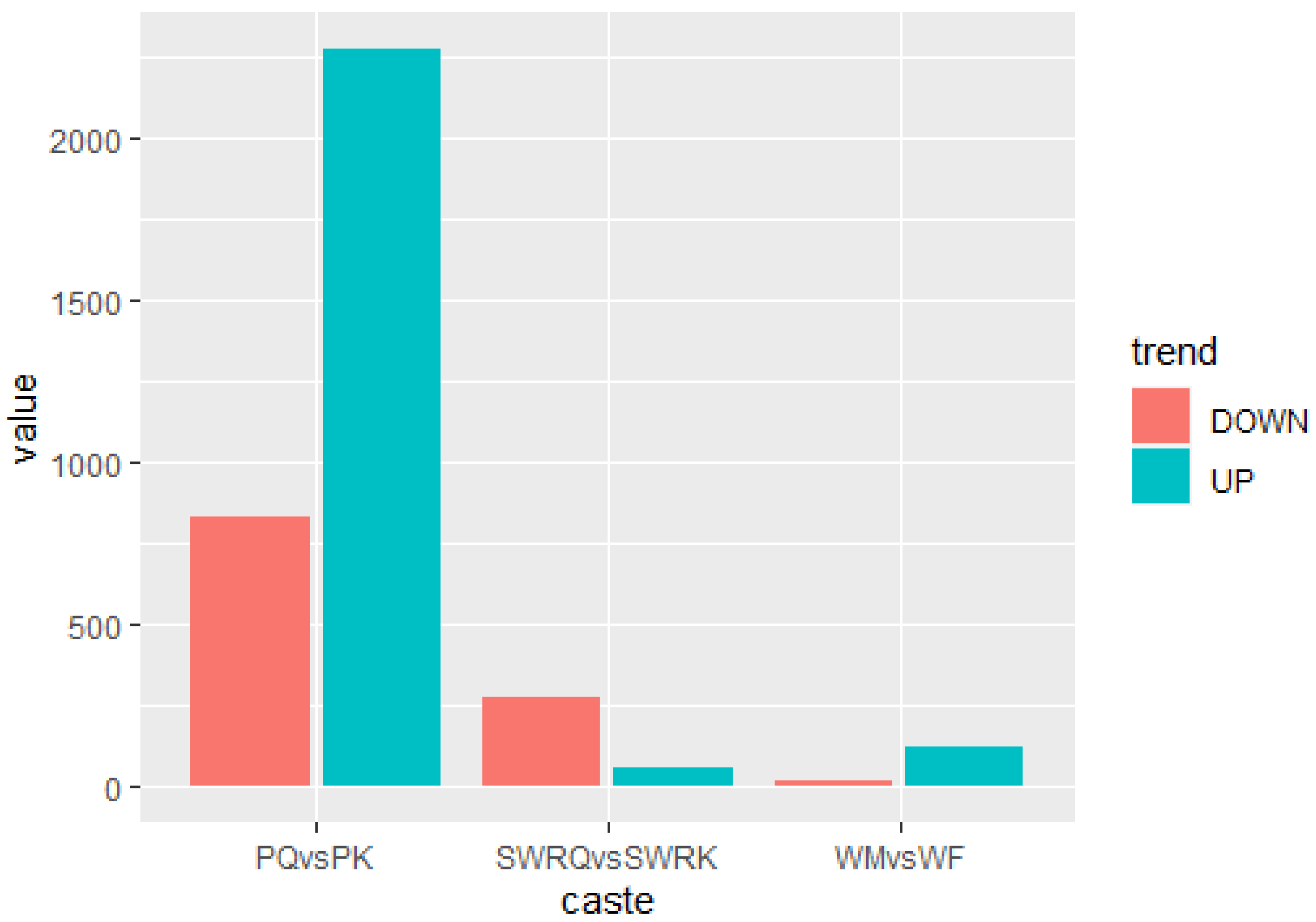
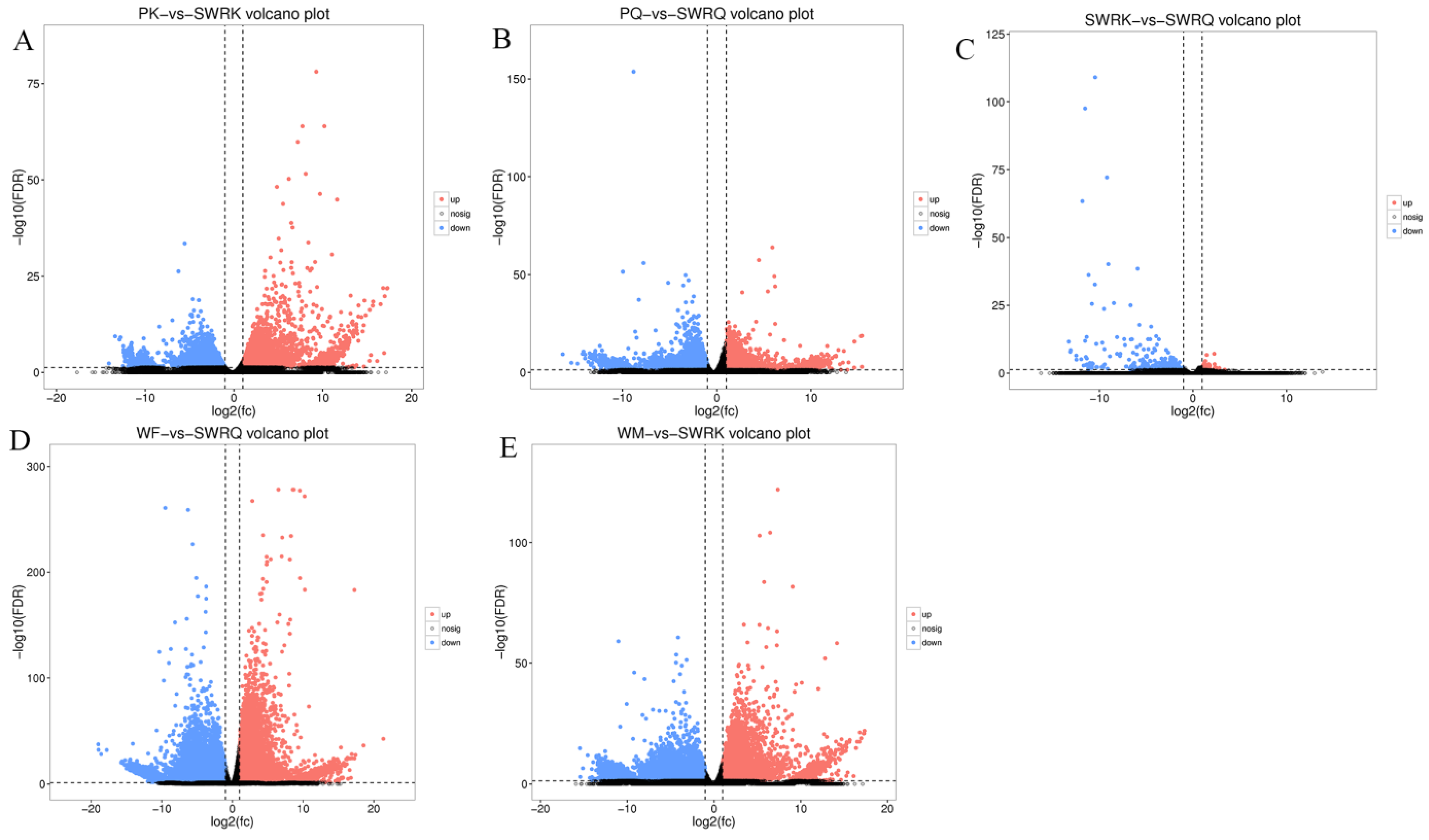
The DEGs for R. chinensis were calculated as up-regulated and downregulated genes through DEseq2. Furthermore, 173,990 DEGs were annotated from SWRK, SWRQ, PK, PQ, WM, and WF. Up-regulated DEGs were 48,354 (27.79%) and 125,636 (72.21%) down-regulated DEGs have been reported. The maximum up-regulated DEGs were identified in WF-vs-SWRQ 20,586 (42.57%) and WM-vs-SWRK 14,165 (29.86%). Similarly, the maximum down-regulated DEGs were identified in WF-vs-SWRQ 71,950 (58.27%) and 32,887 (26.17%) in WM-vs-SWRK castes (Figure 2). The significant DEGs were reported from PQ-vs-PK (2277 up-regulated and 832 downregulated), followed by SWRQ-vs-SWRK (55 up-regulated and 272 downregulated) and WM (13 up-regulated and 124 downregulated) (Figure 3). In DEGs with a p-value < 0.05, the expression levels of GO and KEGG pathways showed a significantly increased. The total number of CDs was 64,046 sequences (Figure S5). The transcriptome sequence analysis and annotations of R. chinensis caste provided valuable evidence for evaluating all unigenes (Figure 4A–E). Transcriptome review showed high expression genes cognate to the IIS-pathway in SWRK, SWRQ, PQ, PK, WM, and WF castes.
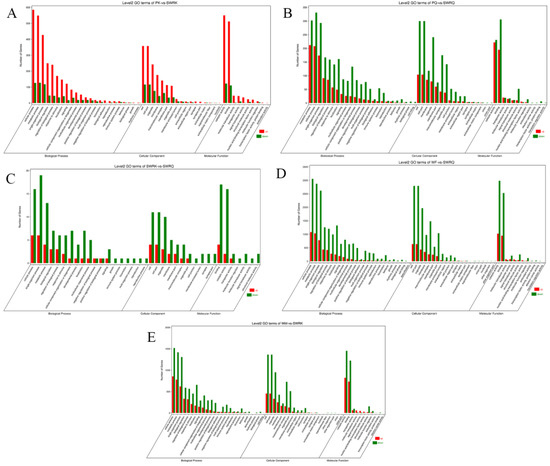
Figure 2.
Histogram presentation of the Gene Ontology (GO) classification in each caste. (A) PK-vs-SWRK; (B) PQ-vs-SWRQ; (C) SWRK-vs-SWRQ; (D) WF-vs-SWRQ; (E) WM-vs-SWRK. PKs (primary king), PQs (primary queen), SWRK (secondary worker reproductive king), SWRQ (secondary worker reproductive queen), WMs (workers male), and WFs (female workers). The figure represents the up (red) and down (green) categorical presentation of biological processes, cellular components, and molecular functions. The x-axis indicates the names of genes in a category, and the y-axis shows the number of genes in the main category.
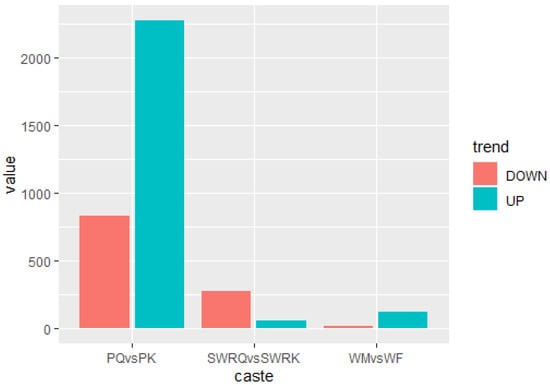
Figure 3.
Differential expression of genes (DEGs) analysis across distinct castes of R. chinensis (SWRQ-vs-SWRK, PQ-vs-PK, and WM-vs-WF). The red column represents DEGs that are down-regulated, whereas the green column indicates DEGs that are up-regulated. To determine the significance of gene expression changes, FDR ≤ 0.001 and log2Ratio ≥ 1 were employed as thresholds.
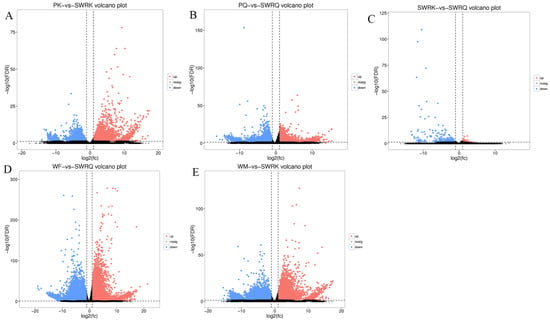
Figure 4.
Analysis of DEGs for enhancement across different castes (A–E). PK (primary king), PQ (primary queen), SWRK (secondary worker reproductive king), SWRQ (secondary worker reproductive queen), WM (male worker), and WF (female worker). The volcano plots indicate enriched DEGs expressing genes in the PK, PQ, SWRK, SWRQ, WM, and WF, with a significantly up-regulated (red) and down-regulated (blue). The threshold level for judging the significance of differences in gene expression, FDR ≤ 0.001 and log2Ratio ≥ 1 parameter were used.
2.5. Caste-Specific Expression-Genes Analysis Related to the IIS-Pathway
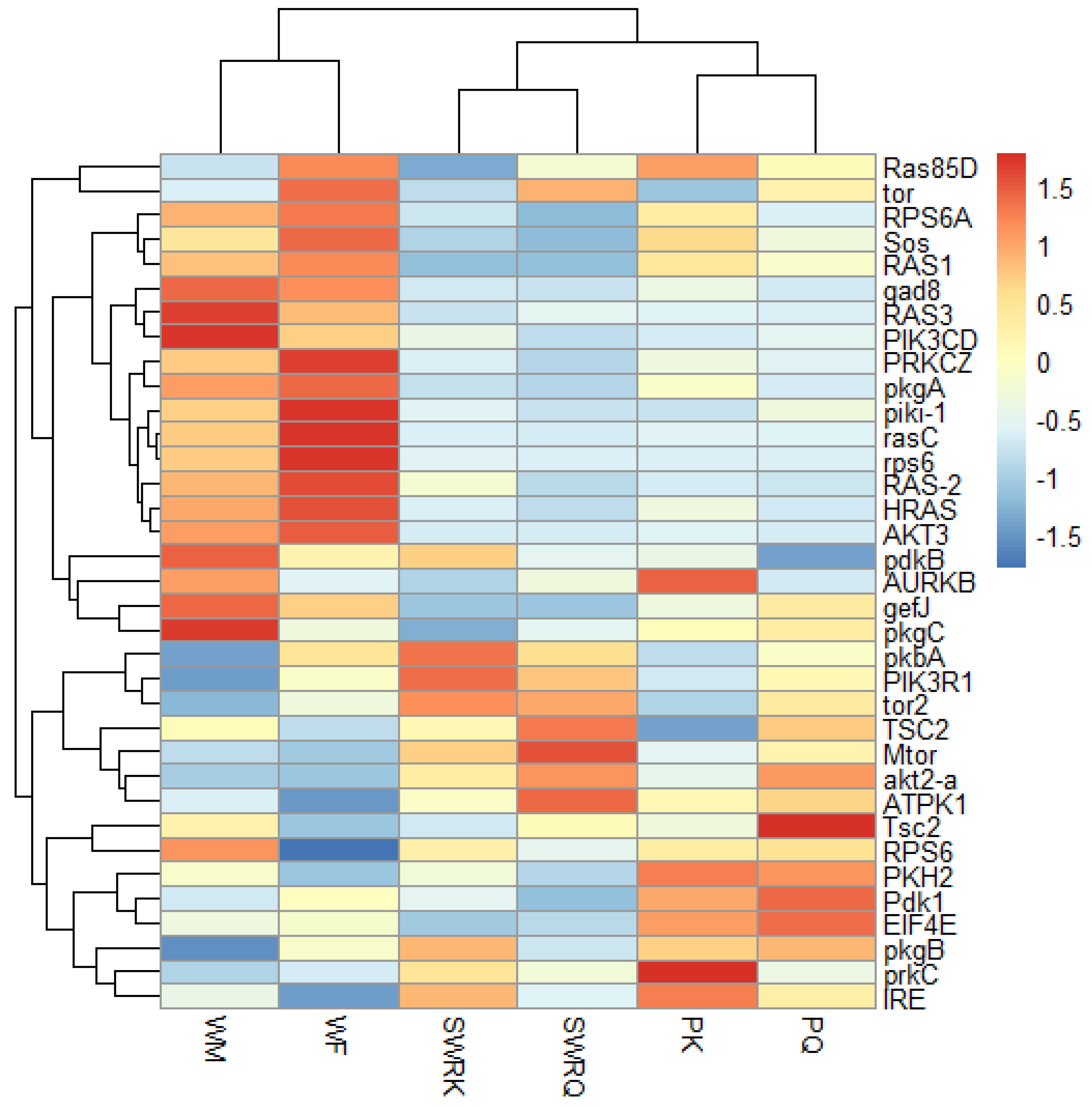
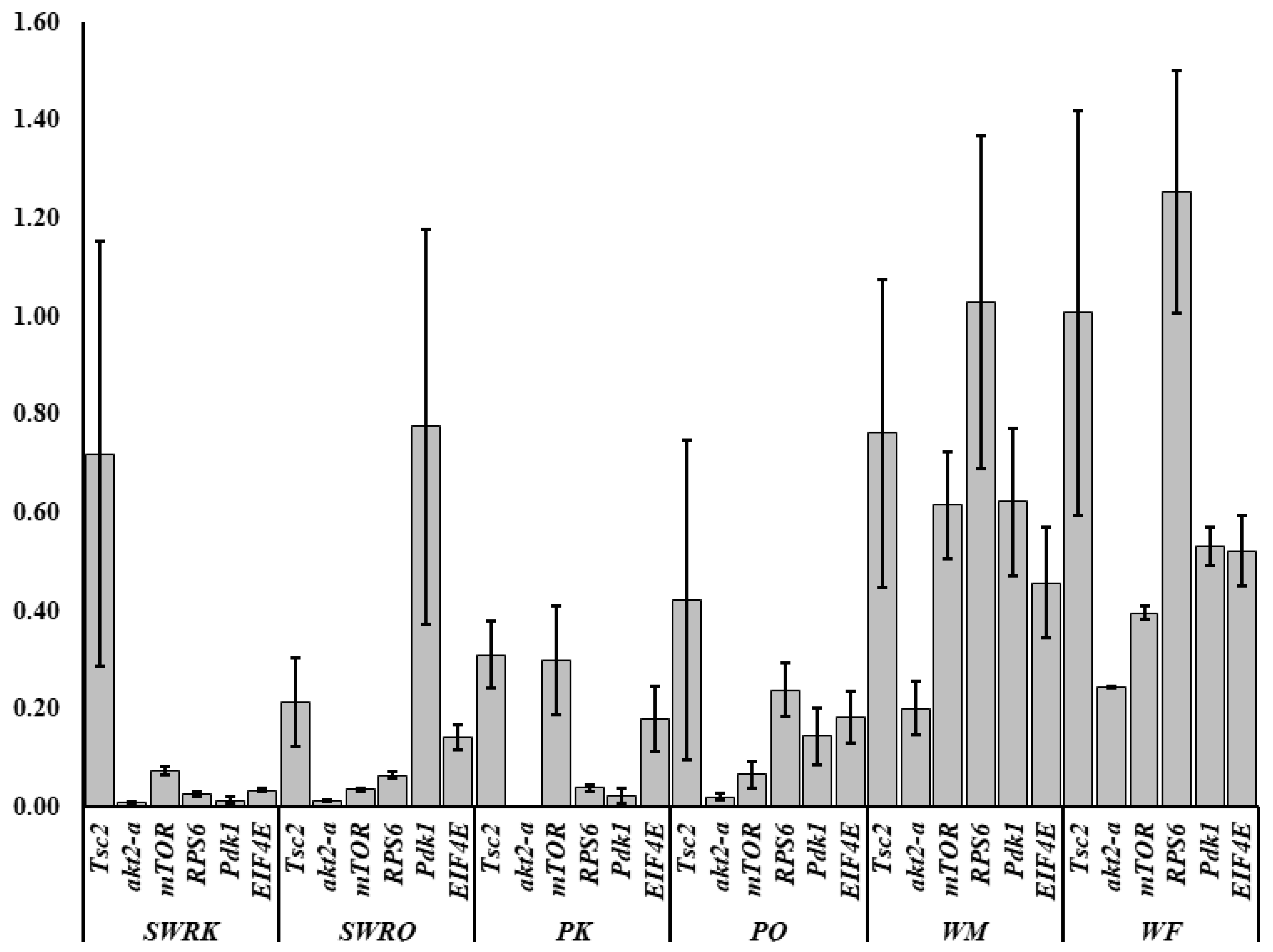
A total of 35 IIS-pathway cognate genes (Figure 5) were examined during the transcriptome analysis. Of them, we selected six genes (Pdk1, akt2-a, Tsc2, mTOR, EIF4E, and RPS6) to validate the fold changes in these genes. A low level of akt2-a in reproductive castes may inhibit tumour invasion and metastasis. The Tuberous Sclerosis Complex (TSC) is a genetic condition that is inherited in an autosomal dominant manner. It is caused by a mutation in either the TSC1 or TSC2 gene and is characterized by the development of tumours or hamartomas in many organs. mTOR, EIF4E, and RPS6 also boost growth factors, physiological processes, cell metabolism and survival, autophagy, fecundity, egg size, and follicle numbers. Therefore, we test the RT-qPCRs analysis in each caste (PK, PQ, SWRQ, SWRK, WM, and WF) of R. chinensis. The average 2−∆∆Ct levels in SWRK are as follows: akt2-a 0.01 (the average of three replicates value calculated by 2−∆∆Ct) (p ˂ 0.00), RPS6 0.03 (p ˂ 0.00), and EIF4E 0.03 (p ˂ 0.00) were reported significantly, followed by Pdk1 0.01 (p ˂ 0.01) and mTOR 0.08 (p ˂ 0.01). In SWRQ, the akt2-a 0.01 (p ˂ 0.00), EIF4E 0.03 (p ˂ 0.00), and mTOR 0.04 (p ˂ 0.01); followed by RPS6 0.07 (p ˂ 0.01). In PK, the average levels of akt2-a, 0.00 (p ˂ 0.00), Pdk1 0.02 (p ˂ 0.01), and RPS6 0.04 (p ˂ 0.01), followed by EIF4E 0.14 (p ≤ 0.03) were reported significantly. In PQ, the akt2-a 0.2 (p ˂ 0.01), mTOR 0.07 (p ≤ 0.03), RPS6 0.24 (p ≥ 0.05), and EIF4E were 0.18 (p ≥ 0.05) reported significantly. In WM, akt2-a 0.20 (p ≥ 0.05), while in WF akt2-a 0.25 (p ˂ 0.00) followed by mTOR 0.40 (p ˂ 0.01) and Pdk1 0.53 (p ≤ 0.04) were reported significantly (Figure 6). The expression of these genes: mTOR (SWRK 0.08; SWRQ 0.04), Pdk1 (SWRK 0.01), akt2-a (SWRK 0.01; SWRQ 0.01), and EIF4E (SWRK 0.03; SWRQ 0.03) (Figure 6), was reported enormously in SWRK and SWRQ than non-reproductive (WM and WF). These results indicate that secondary worker reproductive like the king and queen have a more successful life span than non-reproductive castes. According to these findings, a relatively conserved protein in the insulin signaling system significantly delays or prevents age-related disorders and aging mechanisms, structures, and associated pathways.
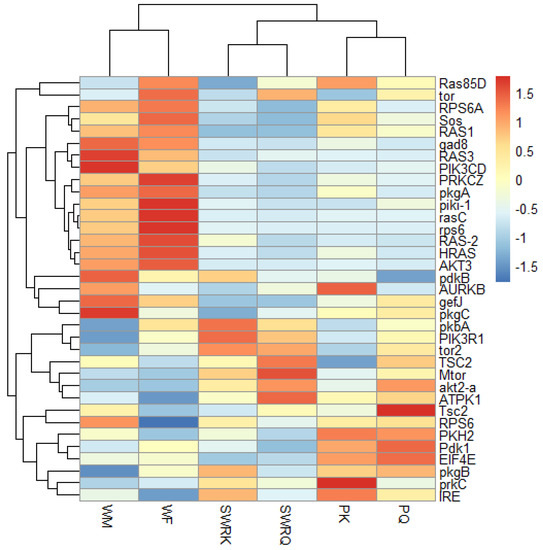
Figure 5.
The heat map indicates the differentially expressed gene (DEGs) involved in the insulin signaling pathway in different castes of R. chinensis. The castes are SWRK, SWRQ, PQ, PK, WM, and WF. A change in color from red to blue indicates the gene expression level.
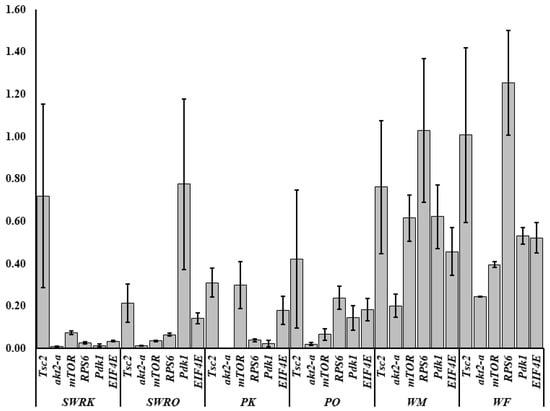
Figure 6.
Log2 fold changes in the insulin signaling pathway in R. chinensis castes SWRK, SWRQ, PK, PQ, WM, and WF. Differential expression of genes in each group is shown as log2 fold changes compared to the reference group. The y-axis represents the log2 fold changes, and the x-axis shows the gene name and individual caste name.
3. Discussion
The transcriptome sequences of R. chinensis castes were compiled to determine the longevity-related genes that contribute to extending the lifespan of SWRK, SWRQ, PK, and PQ caste’s. Transcriptomically, we confirmed that several genes (Pdk1, akt2-a, Tsc2, mTOR, EIF4E, and RPS6) in R. chinensis had extended the lifespan like D. melanogaster [18,19] and C. elegans [20]. The SWRK and SWRQ live longer (18–30 years) than WM and WF (a few weeks to months) [19,33]. It is challenging to sort out and estimate the lifespan of termites. Therefore, we evaluated transcriptomic sequences with next-generation sequencing (NGS) associated with developmental, biological, and physiological changes in cells or tissues [29,38]. The transcriptome research was determined through quantitative RT-qPCR. However, the current study is the second endeavor to longevity-cognate genes of R. chinensis castes [29,38]. According to the transcriptome data, 35 DEGs involved in the longevity-related genes; Pdk1, akt2-a, Tsc2, mTOR, EIF4E, and RPS6 are involved in the longevity of invertebrates and vertebrates. The insulin/insulin-like growth factor signaling pathway is a hormonally mediated cell-signaling pathway, that involves insulin-like peptides, transmembrane receptors of their cognate cell surface, and downstream effectors [19,33], which shreds of evidence annexing genetic and biochemical changes [39]. The PI3K-Akt signaling pathway plays a principal role in insects’ longevity [12,33,34].
Downstream Akt recruited and activated phosphorylation at S473 and T308 at the plasma membrane [40]. Permitted Akt controls cell survival, proliferation, and protein synthesis [41], and activates mTORC2 and Pdk1 [42,43]. In many organisms like yeast, C. elegans, and D. melanogaster, Pdk1 is important in cell survival and evolution [44,45]. Additionally, for murine embryonic development, Pdk1 is essential; mice without a Pdk1 gene died at 9.5 days of the embryo and showed abnormalities in different tissues [46]. Pdk1 hypomorphic mice have smaller cardiovascular organ volumes, and Pdk1 conditional abstraction in muscle cells leads to heart failure and minimizes lifespan [47]. Pdk1 has direct effectors on Akt, S6K, and RSK, causing embryonic stem cells to activate all three of these kinases [46]. However, Akt/PKB activations are involved in several cellular replications to magnification factor signalings, such as apoptosis bulwark, glucose transporter (GLT), and glycogen synthesis [10], while in D. melanogaster, it regulates lifespan, reproductive status, growth, and metabolism [48,49,50]. Moreover, Pdk1 in neuronal IIS-pathway can influence chemotaxis and learning [50]. PI3K-Akt and FOXO proteins are also essential in Drosophila, Nilaparvata lugens, and Sogatella furcifera flight muscle genes [51,52]. In C. elegans, Pdk1 controls insulin physiological effects, and growth factor (GF) enters the staggering dauer phase and elongates their life span when Pdk1 becomes dormant [53]. The expression of the Pdk1 gene through transcriptome and RT-PCRs analysis was significantly higher in SWRK than in any other castes. As a consequence of these findings, our results indicated that Pdk1 serves the same active function in the life span regulation of R. chinensis termites as it does in C. elegans, S. furcifera, Drosophila, and N. lugens.
Tuberous sclerosis (TSC) and its domains (Tsc1 and Tsc2) are responsible for autosomal-dominating disorders, including epilepsy, skin, retina, heart, kidney, and the central nervous system [54,55]. A germ-line mutation in the rat homologous human Tsc2 gene makes the Eker rat prone to many tumors and death in the intermediate stage. Tetracycline-dependent conditions and overexpression of Tsc2 inhibited the proliferation of an Eker rat-derived kidney tumor cell line [55]. Tsc1 in the Drosophila homolog is associated with the cell mutation study (mosaic screens) and polyploidy intended to trigger a cell switch [56]. Current RT-qPCR findings in this study showed that Tsc2 is abundantly conserved among all the castes.
Protein kinase B and its three paralogs, akt1, akt2, and akt3, exhibit intensively deliberated cancer and metabolism [41], and also regulate the plasma membrane [57,58] and cell size mutations in Drosophila [56]. Akt activates considerable tissue growth/size and is essential for the PTEN facility to act as a tumor suppressor in Drosophila [10,59]. The expression of akt2-a may also help longevity and remove tumor cell incursion and metastasis in reproductive castes of R. chinensis. mTOR was first identified and cloned within the budding yeast Saccharomyces cerevisiae [60,61]. Extracellular signaling activates the mTORC1 and mTORC2 network in innate immune cells [11,62,63] and regulates translation, protein synthesis, mRNA translation, anabolic cell growth, and metabolism [64]. Knock-down of mTOR regulates low fertility in N. lugens and activates vitellogenin gene (Vg) expression in A. aegypti [65,66], also essential for transduction alimentation during mosquito egg development [67,68,69]. mTOR also controls ovary size in Drosophila [70], which promotes the number of follicles and owns diphenic development in Apis mellifera [71]. The previously reported results indicate that mTOR increases life span, fecundity, and GF in reproductive termite castes. The same strategy (RT-PCR analysis) is used for mTOR in WM, WF, PK, PQ, SWRK, and SWRQ, where it shows a possibility of prolonging the lifespan of termite castes.
The RPS6 deficiency cells did not significantly reduce global protein translation or 5′-TOP mRNAs [9]. However, functional analysis and mutations in eIF4E-1 result in embryonic defects and lethality [72,73]. Our results suggest that IIS-related pathway genes increase reproductive castes’ life expectancy and control a metabolic pathway in D. melanogaster, R. chinensis, C. elegans, and fly. In addition, their nutritional reactions and sensory compensation for IIS-pathways and insulin peptides are similar [13,74,75].
In the current investigation, the hypothesized findings (RT-qPCR results) were obtained from the reproductive castes (PK, PQ, SWRK, and SWRQ), with much higher expression than in the non-reproductive castes (WM and WF). Our findings ultimately shed new insight into the maintenance and significance of biomolecular homeostasis in reproductive and nonproductive R. chinensis castes. As previously described by a number of researchers, IIS pathway-related genes desperately promote growth factors, physiological processes, cell metabolism and survival, autophagy, fecundity rate, egg size, and follicle number. These findings imply that a relatively conserved protein in the insulin signaling system significantly prolongs or avoids age-related diseases, processes, structures, and associated pathways. Further research is required to determine the biological function of genes connected to the insulin signaling pathway using genetic tools such as RNA interference and transgenic structures in order to determine how these genes contribute to the continued existence of termites. In the end, the findings we were able to obtain new shreds of insight into the processes involved in preserving biomolecule homeostasis and its connection to remarkable longevity.
4. Materials and Methods
4.1. Collection and Rearing of Reticulitermes Chinensis
Termite Reticulitermes chinensis colonies were dug from Chengdu in April–May 2014 (Table 2) and transferred to the laboratory via plastic boxes (25 cm × 18 cm × 15 cm). Isolated colonies were reared for 6 years under generous (munificent) and crowded conditions (the lab was 24 h open) at room temperature (25–28 ± 1 °C) at Northwest University (http://english.nwu.edu.cn/ (accessed on 7 June 2021), Xian, Shaanxi, China. A total of 250 monohybrid colonies were established; each colony corresponds to male × female alates 78% (195/250) and female × female alates 22% (55/250) using pine sawdust (50–60% humidity) in specially designed plastic boxes (80 mm × 65 mm × 40 mm) [76]. At an early stage, active and mature colonies were selected for the experiment, from where we accumulated R. chinensis reproductive (PQ and PK; SWRQ and SWRK) and non-reproductive (WF and WM) castes [29]. These collections were done randomly from different colonies (not specific to the quantities and targeted colonies), where we found the primary and secondary reproductives and non-reproductives castes. Anyhow, many factors influence the division of labour in an insect community, including the size of the colony, castes and instar demography, food availability, and competition [77]. For example, the size of the Rhytidoponera metallica (Smith) ant and Coptotermes formosanus [78] colony influences the existence of age polyethism [79]. Colonies of R. chinensis grow from a mated pair (30–40 days start egg laying) to an immature colony (7–25 months), a juvenile colony (3–6 years), and a sexually mature colony (after 6 years). In a single colony, each caste performs its own role (e.g., food foraging and colony carrying). Due to the fact that, during the rearing of the colonies, a variety of colonies died due to fungus attacks (paralyzing the termites till death), while some colonies laid eggs very late (an average period for eggs laying was observed from 30–40 days). According to these fundamental reasons, the termites were collected randomly.

Table 2.
The information of R. chinensis wild colonies and the collected number of different castes during 2014.
4.2. Experimental Samples
During the study, R. chinensis colonies were subjected to synchronization procedures immediately after six years of rearing of these colonies because we were looking for selected castes, i.e., primary king (PK) and queen (PQ), secondary worker reproductive king (SWRK) and queen (SWRQ), worker male (WM) and female (WF). Total RNA was extracted from the whole body (for RT-PCR, we used heads of individuals free from protozoans) of PQ, PK, SWRQ, SWRK, WF, and WM for RNA Illumina sequencing [29]. Three technical (e.g., PK-1, PK-2, and PK-3) and five biological replicates (e.g., PK-1 had five individuals) (randomly collected from 250 colonies) of each caste (six in number “PQ, PK, SWRQ, SWRK, WF, and WM”) were used.
4.3. Total RNA Extraction, cDNA Synthesis, and Illumina Sequencing
The whole body of R. chinensis castes was micro-dissected (into the head, legs, thorax, and wing (of an adult) to extract DNA and RNA), and it was promptly stored in liquid nitrogen.
We used TRIzol reagent and an Agilent 2100 Bioanalyzer in order to collect sufficient RNA for Illumina sequencing (Agilent Technologies, Palo Alto, CA, USA). The Lysate RZ (TRIzol reagent) and RNAsimple Total RNA-Kit (Tiangen Biotech “Beijing” Co., Ltd., Beijing, China) were applied to the tissue to homogenize and prevent the tissue from degrading [76]. Tissue homogenates were heated to temperatures ranging from 15–30 °C to separate the nucleic acid-protein complex. Afterward, the tissue homogenate was transferred to RNase-free centrifuge tubes (for centrifuge spinning) to separate the aqueous phase, precipitation, deproteinized, and abstracted residual liquid. An adequate quantity (for Illumina sequencing and RT-PCR experiments) of total RNA was extracted and stored at −80 °C for further experiments. Finally, with the help of a spectrophotometer (NanoReady “Model: F-1100”, Shanghai, China), the total amount of RNA (protein: 260/280 and salt: 260/230) was checked to quantify and verify the entire amount of RNA integrity [29].
After that, the NEB-Next Prep-Kit for Illumina (NEB) sequences [80] was used to revert cDNA in accordance with the manufacturer protocol. Furthermore, we used the QiaQuick PCR extraction kit for cDNA fragments emasculated. The poly(A) end-paired were tailed and combined with the sequence of the Illumina adapter. We obtained sequence readings of an average length (561 bp) and a minimum length (201 bp) by using the Illumina HiSeqTM 4000 platform (Figure S1). The ligation product was selected by size from the Gene Denovo Biotechnology Co (Guangzhou, China). In addition, the Trinity system (trinityrnaseq r2012-04-27) was used for each test simultaneously in order to obtain an adequate number of clear unigenes [81,82].
4.4. Transcriptome Assembly and Reads Mapping
Raw data evaluation affects the quality and screening; therefore, we used fastp (version 0.18.0) before and after filtering and using the following parameters (˃10% of unknown and ˂50% inference of reads) for readings that contain adapters of low-quality q-value (≤20 bases) [83]. Quality clean reads of the transcriptome de novo was assembled with the short-read assembly through the trinityrnaseq r2012-04-27 assembly program [81]. Further data on the development of transcriptomic assemblies using a de Bruijn graph algorithm from short-read sequences have been provided in supplementary data [84].
4.5. Read Alignments, Normalization, and the Gene Expression Level
Short sequences and readings have been mapped into a reference SOAPaligner/soap2 tool [85,86]. The edgeR (Bioconductor package version 2.4.0) was used to generate read counts for genomic features and summarize short reads (edgeR is a method that is based on the weighted mean of log ratios). This method normalizes data in the beginning by calculating size and normalization factors from raw reads of the IlluminaTM sequence. The program implements precisely established statistical approaches for multigroup experiments [87]. These genes were tenacious by the quantifications allocated exclusively to the exon region per million mapped reads (RPKM) [76]. The R-package (http://www.r-project.org (accessed on 15 March 2021)) was used to calculate all statistical data expression and visualization [37].
4.6. Differentially Expressed Genes (DEGs), Identification, Validation, and Functional Enrichment Analyses
After reading alignments and normalization of genes, finally, we obtained DEGs, using DEseq2 (because each caste has three replicates); the relative expression level of each gene was calculated [88]. These statistical methods distinguish between digital gene expression data (electronic data) of a negative binomial distribution model, fold changes (FC > 2), and the false discovery rate (FDR < 0.01) to justify the threshold level (p-value). The value of log2ratio ≥ 1 is an absolute value in calculations of significant DEGs between pooling samples. Annotations from the Gene Ontology (GO), the Kyoto Genes and Genomes Encyclopedia (KEGG) (http://www.genome.jp/kegg (accessed on 15 March 2021), the Clusters of Orthologous Groups (COG) (https://www.ncbi.nlm.nih.gov/COG/ (accessed on 15 March 2021) and the Non-redundant (NR) (http://www.ncbi.nlm.nih.gov (accessed on 15 March 2021) database were all relegated [86]. Blast2GO software assigned the GO annotation (obtained from the Nr database profile) [89]. In addition, WEGO software was used to classify unigenes functions [90].
4.7. Quantitative Real-Time PCR (RT-qPCR) Assay
The RT-qPCR assay was performed in a 20 μL reaction volume SuperReal PreMix Plus (Tiangen, Sichuan, China) with CFX 96 (Bio-Rad system) following the manufacturer’s instructions. The Beta-actin gene averted fluctuations in quality and quantity for Reticulitermes termites [35,83] and was selected as a housekeeping gene [37,91]. We designed primer pairs for each IIS pathway-cognate gene using Primer3 v1.1.4 (Table S1) [29]. For RT-qPCR, the total RNA was extracted from the head (free from protozoans) of each caste (PK, PQ, SWRQ, SWRK, WM, and WF), using an RNAsimple Total RNA kit (Tiangen). Three technical (eg., PK-1, PK-2, and PK-3) and five biological replicates (randomly collected from different colonies) were used (e.g., six castes, three replicates, and each replicate consisted of five individuals). We used Fast-King RT-Kit (Tiangen) for cDNA library construction (cDNA kept for a further experiment at −20 °C). The cDNA (4 μL), SYBR Premix-Ex Taq-TMII (10 μL), and ddH2O (4.8 μL) were performed in a quantitative reaction (20 μL) on life technologies/Vii7, 0.6 μL of the reverse and forward primers. RT-qPCR was amplified at 95 °C (15 min), 95 °C (10 s), 60 °C (20 s), and 72 °C (32 s). Following amplification, the melt curve (at CFX 96 Bio-Rad system) analysis detected a gene-specific and nonspecific amplification.
4.8. Statistical Data Analysis
A standard method 2−∆∆Ct (formulae are given below) was used to calculate the relative gene expression of the mRNA genes used with each replicate [92,93]. The RT-qPCR experiment quantifying and counting results were statistically analyzed using IBM Corp. Released 2019 (IBM SPSS Statistics for Windows, Version 26.0. Armonk, NY, USA: IBM Corp.) to determine the relationship between groups (reproductive and non-reproductives) using a t-test [29]. All values were expressed as mean ± standard deviation (p-values < 0.05) and statistical figures constructed with Microsoft Excel (0365) and OriginPro (2018).
whereas and
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232113660/s1.
Author Contributions
H. and Y.-X.L.: design, analysis, data collection, manuscript writing. C.-X.Y. and J.S.: data collection, analysis. G.N. and X.-H.S.: critical review and editing. L.-X.X.: conception, funding, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31870389), Xi’an Science and Technology Planning Project (21NYYF0015) and Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education (ZSK2017002).
Data Availability Statement
The transcriptome data will be available under the accession number (BioProject: PRJNA592596) at the NCBI database. The corresponding author will answer any question upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cheatham, B.; Kahn, C.R. Insulin action and the insulin signaling network. Endocr. Rev. 1995, 16, 117–142. [Google Scholar] [PubMed]
- Puig, O.; Michael, T.M.; Ruhf, M.L.; Robert, T. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2006, 17, 2006–2020. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, L.; Horman, S.; Beauloye, C.; Vanoverschelde, J.L. Insulin signalling in the heart. Cardiovasc. Res. 2008, 79, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Gerald, I.S. Mechanisms of insulin action and insulin resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef]
- Jarosinski, M.A.; Dhayalan, B.; Chen, Y.S.; Chatterjee, D.; Varas, N.; Weiss, M.A. Structural principles of insulin formulation and analog design: A century of innovation. Mol. Metab. 2021, 52, 101325. [Google Scholar] [CrossRef] [PubMed]
- Dallman, M.F.; Strack, A.M.; Akana, S.F.; Bradbury, M.J.; Hanson, E.S.; Scribner, K.A.; Smith, M. Feast and famine: Critical role of glucocorticoids with insulin in daily energy flow. Front. Neuroendocrinol. 1993, 14, 303–347. [Google Scholar] [CrossRef]
- Peppas, N.A.; Kavimandan, N.J. Nanoscale analysis of protein and peptide absorption: Insulin absorption using complexation and pH-sensitive hydrogels as delivery vehicles. Eur. J. Pharm. Sci. 2006, 29, 183–197. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Chiocchetti, A.; Zhou, J.; Zhu, H.; Karl, T.; Haubenreisser, O.; Rinnerthaler, M.; Heeren, G.; Oender, K.; Bauer, J.; Hintner, H.; et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp. Gerontol. 2007, 42, 275–286. [Google Scholar] [CrossRef]
- Shioi, T.; Mcmullen, J.R.; Kang, P.M.; Douglas, P.S.; Obata, T.; Franke, T.F.; Cantley, L.C.; Izumo, S. Akt/Protein kinase B promotes organ growth in transgenic mice. Mol. Cell. Biol. 2002, 22, 2799–2809. [Google Scholar] [CrossRef]
- Lehman, J.A.; Calvo, V.; Gomez-Cambronero, J. Mechanism of ribosomal p70s6 kinase activation by granulocyte macrophage colony-stimulating factor in neutrophils. Cooperation of a MEK-related, Thr421/Ser424 kinase and a rapamycin-sensitive, mTOR-related Thr389 kinase. J. Biol. Chem. 2003, 278, 28130–28138. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, M.; Coffer, P.J. The role of the PI3K-PKB signaling module in regulation of hematopoiesis. Cell Cycle 2009, 8, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.D.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. daf-2, An insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 1997, 277, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Mills, K.; Le Cessie, S.; Noordam, R.; Van Heemst, D. Ageing, age-related diseases and oxidative stress: What to do next? Ageing Res. Rev. 2020, 57, 100982. [Google Scholar] [CrossRef]
- Ros, M.; Carrascosa, J.M. Current nutritional and pharmacological anti-aging interventions. Biochim. Biophys. Acta-Mol. Basis Dis. 2020, 1866, 165612. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Paciga, J.E.; Feldman, R.I.; Yuan, Z.-Q.; Coppola, D.; Lu, Y.Y.; Shelley, S.A.; Nicosia, S.V.; Cheng, J.Q. Phosphatidylinositol-3-OH kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor α (ERα) via interaction between ERα and PI3K. Cancer Res. 2001, 61, 5985–5991. [Google Scholar]
- Yoeli-Lerner, M.; Toker, A.J.C.C. Akt/PKB signaling in cancer: A function in cell motility and invasion. Cell Cycle 2006, 5, 603–605. [Google Scholar] [CrossRef]
- Sim, C.; Denlinger, D.L. Insulin signaling and the regulation of insect diapause. Front. Physiol. 2013, 4, 189. [Google Scholar] [CrossRef]
- Altintas, O.; Park, S.; Lee, S.J.V. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep. 2016, 49, 81–92. [Google Scholar] [CrossRef]
- Wang, J.; Kim, S.K. Global analysis of dauer gene expression in Caenorhabditis elegans. Development 2003, 130, 1621–1634. [Google Scholar] [CrossRef]
- Tissenbaum, H.A. Genetics, life span, health span, and the aging process in Caenorhabditis elegans. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2012, 67A, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.S.; Denlinger, D.L.; Sim, C. Suppression of allatotropin simulates reproductive diapause in the mosquito Culex pipiens. J. Insect Physiol. 2014, 64, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Carey, J.R. Longevity minimalists: Life table studies of two species of northern Michigan adult mayflies. Exp. Gerontol. 2002, 37, 567–570. [Google Scholar] [CrossRef]
- Jemielity, S.; Chapuisat, M.; Parker, J.D.; Keller, L. Long live the queen: Studying aging in social insects. Age 2005, 27, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Porter, S.D.; Jorgensen, C.D. Foragers of the harvester ant, Pogonomyrmex owyheei: A disposable caste? Behav. Ecol. Sociobiol. 1981, 9, 247–256. [Google Scholar] [CrossRef]
- Kramer, B.H.; Schaible, R.; Scheuerlein, A. Worker lifespan is an adaptive trait during colony establishment in the long-lived ant Lasius niger. Exp. Gerontol. 2016, 85, 18–23. [Google Scholar] [CrossRef]
- Southon, R.J.; Bell, E.F.; Graystock, P.; Sumner, S. Long live the wasp: Adult longevity in captive colonies of the eusocial paper wasp Polistes canadensis (L.). PeerJ 2015, 3, e848. [Google Scholar] [CrossRef]
- Korb, J.; Karen, M.; Denise, A.; Abel, B.; Daniel, E.; Barbara, F.; Susanne, F. Comparative transcriptomic analysis of the mechanisms underpinning ageing and fecundity in social insects. Philos. Trans. R. Soc. B 2021, 376, 20190728. [Google Scholar] [CrossRef]
- Haroon; Ma, X.M.; Li, Y.X.; Zhang, H.X.; Liu, Q.; Su, X.H.; Xing, L.X. Transcriptomic evidence that insulin signalling pathway regulates the ageing of subterranean termite castes. Sci. Rep. 2020, 10, 8187. [Google Scholar] [CrossRef]
- Carey, J.R. Demographic mechanisms for the evolution of long life in social insects. Exp. Gerontol. 2001, 36, 713–722. [Google Scholar] [CrossRef]
- Vargo, E.L.; Husseneder, C. Biology of subterranean termites: Insights from molecular studies of Reticulitermes and Coptotermes. Annu. Rev. Entomol. 2009, 54, 379–403. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Badisco, L.; Van Wielendaele, P.V.; Broeck, J.V. Eat to reproduce: A key role for the insulin signaling pathway in adult insects. Front. Physiol. 2013, 4, 202. [Google Scholar] [CrossRef]
- Kakanj, P.; Moussian, B.; Grönke, S.; Bustos, V.; Eming, S.A.; Partridge, L.; Leptin, M. Insulin and TOR signal in parallel through FOXO and S6K to promote epithelial wound healing. Nat. Commun. 2016, 7, 12972. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Haroon; Li, Y.X.; Ye, C.X.; Ma, X.Q.; Su, J.; Su, X.H.; Xing, L.X. Genome-wide profiling and identification of insulin signaling pathway genes of subterranean termite castes. Entomol. Res. 2021, 51, 462–476. [Google Scholar] [CrossRef]
- Ye, C.; Rasheed, H.; Ran, Y.; Yang, X.; Xing, L.; Su, X. Transcriptome changes reveal the genetic mechanisms of the reproductive plasticity of workers in lower termites. BMC Genom. 2019, 20, 702. [Google Scholar] [CrossRef]
- Steijger, T.; Abril, J.F.; Engström, P.G.; Kokocinski, F.; Akerman, M.; Alioto, T.; Ambrosini, G.; Antonarakis, S.E.; Behr, J.; Bertone, P.; et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [Google Scholar] [CrossRef]
- Pan, H.; Finkel, T. Key proteins and pathways that regulate lifespan. J. Biol. Chem. 2017, 292, 6452–6460. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB signaling: Navigating the network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Moon, Z.; Wang, Y.; Aryan, N.; Mousseau, D.D.; Scheid, M.P. Serine 396 of PDK1 is required for maximal PKB activation. Cell. Signal. 2008, 20, 2038–2049. [Google Scholar] [CrossRef]
- Hu, T.; Li, C.; Wang, L.; Zhang, Y.; Peng, L.; Cheng, H.; Chu, Y.; Wang, W.; Ema, H.; Gao, Y.; et al. PDK1 plays a vital role on hematopoietic stem cell function. Sci. Rep. 2017, 7, 4943. [Google Scholar] [CrossRef] [PubMed]
- Paradis, S.; Ailion, M.; Toker, A.; Thomas, J.H.; Ruvkun, G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999, 13, 1438–1452. [Google Scholar] [CrossRef]
- Cho, K.S.; Lee, J.H.; Kim, S.; Kim, D.; Koh, H.; Lee, J.; Kim, C.; Kim, J.; Chung, J. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 6144–6149. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, M.A.; Mora, A.; Ashby, P.R.; Williams, M.R.; Murray-Tait, V.; Malone, L.; Prescott, A.R.; Lucocq, J.M.; Alessi, D.R. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002, 21, 3728–3738. [Google Scholar] [CrossRef]
- Mora, A.; Davies, A.M.; Bertrand, L.; Sharif, I.; Budas, G.R.; Jovanović, S.; Mouton, V.; Kahn, C.R.; Lucocq, J.M.; Gray, G.A.; et al. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. EMBO J. 2003, 22, 4666–4676. [Google Scholar] [CrossRef]
- Orgogozo, V.; Broman, K.W.; Stern, D.L. High-resolution quantitative trait locus mapping reveals sign epistasis controlling ovariole number between two Drosophila species. Genetics 2006, 173, 197–205. [Google Scholar] [CrossRef][Green Version]
- Tomioka, M.; Adachi, T.; Suzuki, H.; Kunitomo, H.; Schafer, W.R.; Iino, Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 2006, 51, 613–625. [Google Scholar] [CrossRef]
- Wang, Y.; Amdam, G.V.; Rueppell, O.; Wallrichs, M.A.; Fondrk, M.K.; Kaftanoglu, O.; Page, R.E. PDK1 and HR46 gene homologs tie social behavior to ovary signals. PLoS ONE 2009, 4, e4899. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Bao, Y.Y.; Li, B.L.; Cheng, Y.B.; Peng, Z.Y.; Liu, H.; Xu, H.J.; Zhu, Z.R.; Lou, Y.G.; Cheng, J.A.; et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS ONE 2010, 5, e14233. [Google Scholar] [CrossRef]
- Yang, X.; Liu, X.; Xu, X.; Li, Z.; Li, Y.; Song, D.; Yu, T.; Zhu, F.; Zhang, Q.; Zhou, X. Gene expression profiling in winged and wingless cotton aphids, Aphis gossypii (Hemiptera: Aphididae). Int. J. Biol. Sci. 2014, 10, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Rintelen, F.; Stocker, H.; Thomas, G.; Hafen, E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 15020–15025. [Google Scholar] [CrossRef] [PubMed]
- Miloloza, A.; Rosner, M.; Nellist, M.; Halley, D.; Bernaschek, G.; Hengstschläger, M. The TSC1 gene product, hamartin, negatively regulates cell proliferation. Hum. Mol. Genet. 2000, 9, 1721–1727. [Google Scholar] [CrossRef][Green Version]
- Hengstschläger, M.; Rodman, D.M.; Miloloza, A.; Hengstschläger-Ottnad, E.; Rosner, M.; Kubista, M. Tuberous sclerosis gene products in proliferation control. Mutat. Res.-Rev. Mutat. Res. 2001, 488, 233–239. [Google Scholar] [CrossRef]
- Potter, C.J.; Pedraza, L.G.; Huang, H.; Xu, T. The tuberous sclerosis complex (TSC) pathway and mechanism of size control. Biochem. Soc. Trans. 2003, 31, 584–586. [Google Scholar] [CrossRef]
- James, P.; Halladay, J.; Craig, E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1996, 144, 1425–1436. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K pathway in human disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Verdu, J.; Buratovicht, M.A.; Wilder, E.L.; Birnbaum, M.J. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1999, 1, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Albers, M.W.; Bum Shin, T.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef]
- Chiu, M.I.; Katz, H.; Berlin, V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 1994, 91, 12574–12578. [Google Scholar] [CrossRef]
- Haidinger, M.; Poglitsch, M.; Geyeregger, R.; Kasturi, S.; Zeyda, M.; Zlabinger, G.J.; Pulendran, B.; Hörl, W.H.; Säemann, M.D.; Weichhart, T. A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. J. Immunol. 2010, 185, 3919–3931. [Google Scholar] [CrossRef] [PubMed]
- Sathaliyawala, T.; O’gorman, W.E.; Greter, M.; Bogunovic, M.; Konjufca, V.; Hou, Z.E.; Nolan, G.P.; Miller, M.J.; Merad, M.; Reizis, B. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity 2010, 33, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T.; Hengstschläger, M.; Linke, M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015, 15, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Guan, K.L. Expanding mTOR signaling. Cell Res. 2007, 17, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Sun, Z.; Zhang, J.; Kang, K.; Chen, J.; Zhang, W. Activation of the TOR signalling pathway by glutamine regulates insect fecundity. Sci. Rep. 2015, 5, 10694. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Park, J.H.; Peng, Q.; Raikhel, A.S. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc. Natl. Acad. Sci. USA 2004, 101, 10626–10631. [Google Scholar] [CrossRef]
- Attardo, G.M.; Hansen, I.A.; Raikhel, A.S. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem. Mol. Biol. 2005, 35, 661–675. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Roy, S.G.; Raikhel, A.S. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J. Biol. Chem. 2005, 280, 20565–20572. [Google Scholar] [CrossRef]
- Zhang, Y.; Billington, C.J.; Pan, D.; Neufeld, T.P. Drosophila target of rapamycin kinase functions as a multimer. Genetics 2006, 172, 355–362. [Google Scholar] [CrossRef]
- Harwood, G.P.; Ihle, K.E.; Salmela, H.; Amdam, G.V. Regulation of Honeybee Worker (Apis mellifera) Life Histories by Vitellogenin; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Lachance, P.E.D.; Miron, M.; Raught, B.; Sonenberg, N.; Lasko, P. Phosphorylation of Eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 2002, 22, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Lafever, L.; Feoktistov, A.; Hsu, H.J.; Drummond-Barbosa, D. Specific roles of Target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development 2010, 137, 2117–2126. [Google Scholar] [CrossRef] [PubMed]
- Brogiolo, W.; Stocker, H.; Ikeya, T.; Rintelen, F.; Fernandez, R.; Hafen, E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001, 11, 213–221. [Google Scholar] [CrossRef]
- Garofalo, R.S. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 2002, 13, 156–162. [Google Scholar] [CrossRef]
- Su, X.; Liu, H.; Yang, X.; Chen, J.; Zhang, H.; Xing, L.; Zhang, X. Characterization of the transcriptomes and cuticular protein gene expression of alate adult, brachypterous neotenic and adultoid reproductives of Reticulitermes labralis. Sci. Rep. 2016, 6, 34183. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.E. Regulation of division of labor in insect societies. Annu. Rev. Entomol. 1992, 37, 637–665. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Chouvenc, T.; Su, N.Y. Development of age polyethism with colony maturity in Coptotermes formosanus (Isoptera: Rhinotermitidae). Environ. Entomol. 2017, 46, 311–318. [Google Scholar]
- Thomas, M.L.; Elgar, M.A. Colony size affects division of labourin the ponerine ant Rhytidoponera metallica. Naturwissenschaften 2003, 90, 88–92. [Google Scholar] [CrossRef]
- Jia, Z.Q.; Liu, D.; Peng, Y.C.; Han, Z.J.; Zhao, C.Q.; Tang, T. Identification of transcriptome and fluralaner responsive genes in the common cutworm Spodoptera litura Fabricius, based on RNA-seq. BMC Genom. 2020, 21, 120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, K.; Maekawa, K. Ovarian development of female-female pairs in the termite, Reticulitermes speratus. J. Insect Sci. 2010, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Mitaka, Y.; Kobayashi, K.; Matsuura, K. Caste-, sex-, and age-dependent expression of immune-related genes in a Japanese subterranean termite, Reticulitermes speratus. PLoS ONE 2017, 12, e0175417. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, K.; Zeng, S.; Teixeira Da Silva, J.A.; Zhao, X.; Tian, C.E.; Xia, H.; Duan, J. Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral development. BMC Genom. 2013, 14, 279. [Google Scholar] [CrossRef]
- Robinson, M.; McCarthy, D.; Chen, Y.; Smyth, G.K. edgeR: Differential expression analysis of digital gene expression data User’s Guide. R Man 2013, 1–76. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).