Hypo-Osmoregulatory Roles of Vasotocinergic and Isotocinergic Systems in the Intestines of Two European Sea Bass Lineages
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Analysis
2.2. Amino Acid Analysis in Regions Involved in Ligand Binding
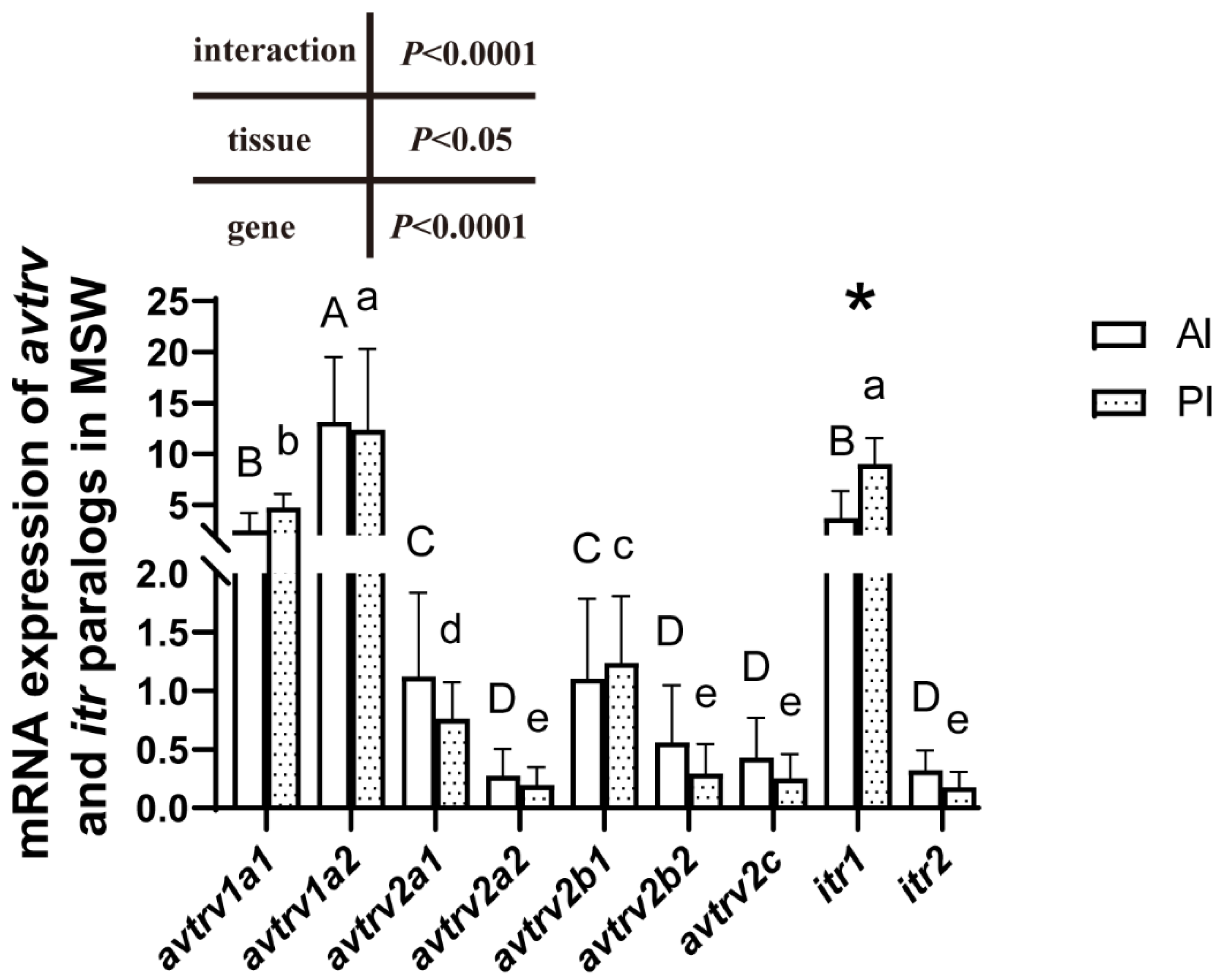
2.3. mRNA Expression of avtr and itr Paralogs
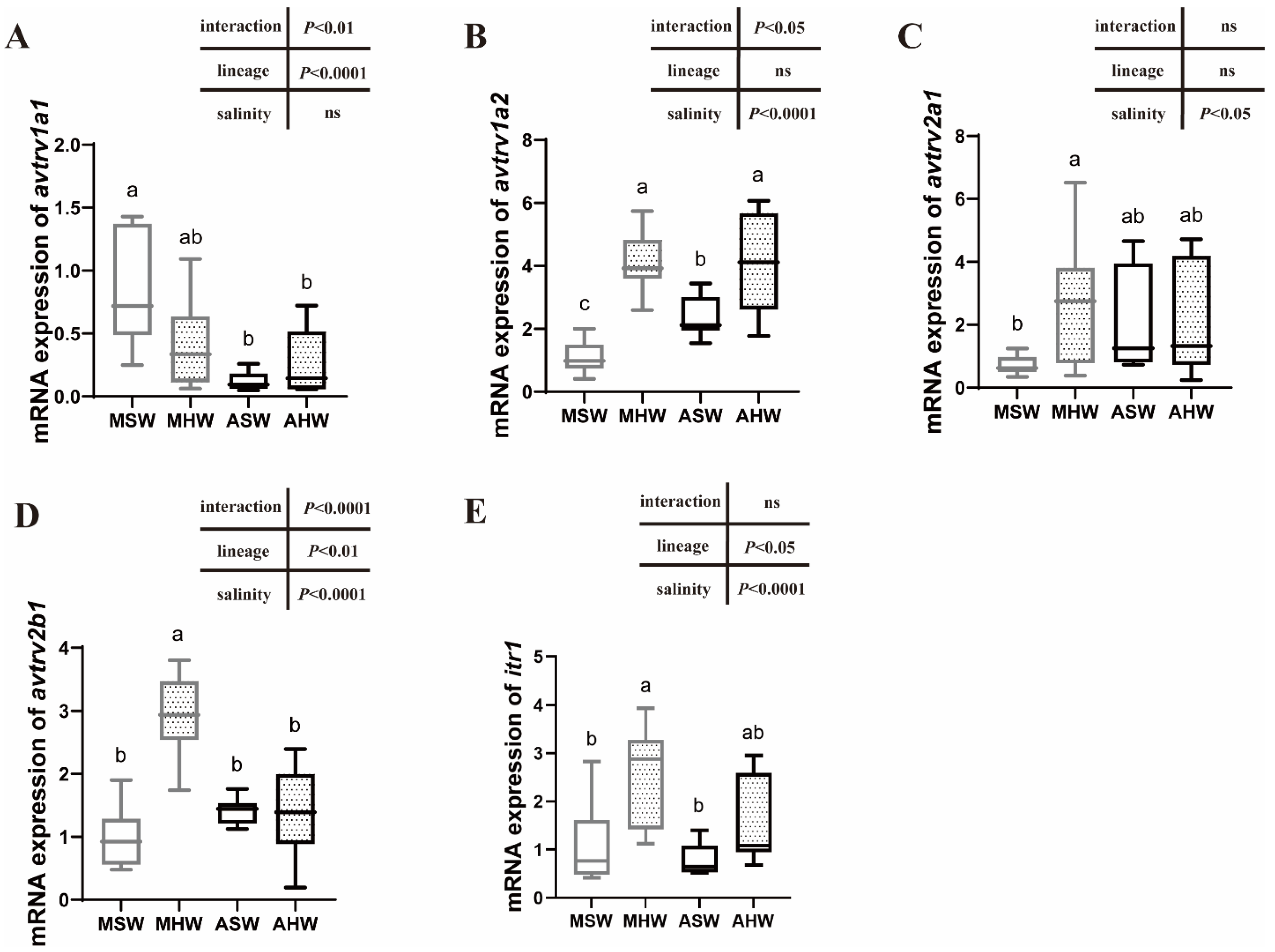
2.4. avtr and itr mRNA Expression in the Anterior Intestine
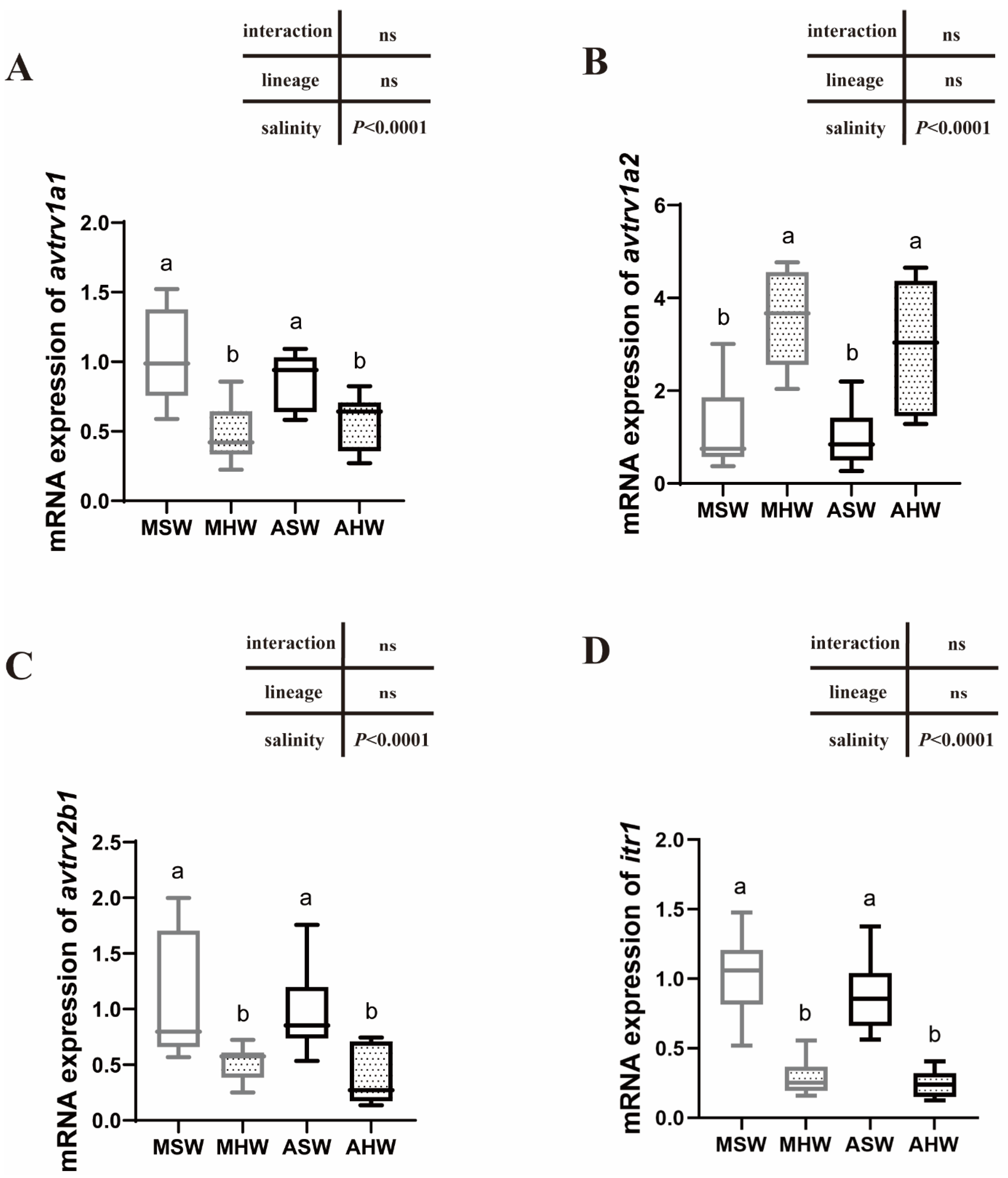
2.5. avtr and itr mRNA Expression in the Posterior Intestine
2.6. Principal Component Analysis (PCA)
3. Discussion
3.1. Diversity of European Sea Bass AVTR and ITR Receptors
3.2. V1A-Type Receptor and ITR1 Are Highly Expressed in the Intestine
3.3. Expression of AVTR Following Hypersalinity Transfer
3.4. Functional Link between AVTR/ITR and Solute-Couped Water Uptake
3.5. Intraspecific Differences in the Expression Patterns of Arginine Vasotocin and Isotocin Receptors
4. Materials and Methods
4.1. Phylogenetic Analysis of AVT and IT Receptors and Amino Acid Analysis
4.2. Experimental Design
4.3. RNA Extraction and Complementary cDNA Synthesis
4.4. Quantitative Real-Time PCR (qPCR)
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamashita, J.; Kawabata, Y.; Okubo, K. Expression of isotocin is male-specifically up-regulated by gonadal androgen in the medaka brain. J. Neuroendocrinol. 2017, 29, e12545. [Google Scholar] [CrossRef] [PubMed]
- Sokołowska, E.; Gozdowska, M.; Kulczykowska, E. Nonapeptides Arginine Vasotocin and Isotocin in Fishes: Advantage of Bioactive Molecules Measurement. Front. Mar. Sci. 2020, 7, 610. [Google Scholar] [CrossRef]
- Lyu, L.K.; Li, J.S.; Wang, X.J.; Yao, Y.J.; Li, J.F.; Li, Y.; Wen, H.S.; Qi, X. Arg-Vasotocin Directly Activates Isotocin Receptors and Induces COX2 Expression in Ovoviviparous Guppies. Front. Endocrinol. 2021, 12, 617580. [Google Scholar] [CrossRef] [PubMed]
- Gesto, M.; Soengas, J.L.; Rodriguez-Illamola, A.; Miguez, J.M. Arginine vasotocin treatment induces a stress response and exerts a potent anorexigenic effect in rainbow trout, Oncorhynchus mykiss. J. Neuroendocrinol. 2014, 26, 89–99. [Google Scholar] [CrossRef]
- Yan, J.J.; Hwang, P.P. Novel discoveries in acid-base regulation and osmoregulation: A review of selected hormonal actions in zebrafish and medaka. Gen. Comp. Endocrinol. 2019, 277, 20–29. [Google Scholar] [CrossRef]
- Aruna, A.; Lin, C.J.; Nagarajan, G.; Chang, C.F. Neurohypophysial Hormones Associated with Osmotic Challenges in the Brain and Pituitary of the Euryhaline Black Porgy, Acanthopagrus schlegelii. Cells 2021, 10, 3086. [Google Scholar] [CrossRef]
- Skrzynska, A.K.; Maiorano, E.; Bastaroli, M.; Naderi, F.; Miguez, J.M.; Martinez-Rodriguez, G.; Mancera, J.M.; Martos-Sitcha, J.A. Impact of Air Exposure on Vasotocinergic and Isotocinergic Systems in Gilthead Sea Bream (Sparus aurata): New Insights on Fish Stress Response. Front. Physiol. 2018, 9, 96. [Google Scholar] [CrossRef]
- Skrzynska, A.K.; Gozdowska, M.; Kulczykowska, E.; Martinez-Rodriguez, G.; Mancera, J.M.; Martos-Sitcha, J.A. The effect of starvation and re-feeding on vasotocinergic and isotocinergic pathways in immature gilthead sea bream (Sparus aurata). J Comp. Physiol. B 2017, 187, 945–958. [Google Scholar] [CrossRef]
- Kleszczynska, A.; Vargas-Chacoff, L.; Gozdowska, M.; Kalamarz, H.; Martinez-Rodriguez, G.; Mancera, J.M.; Kulczykowska, E. Arginine vasotocin, isotocin and melatonin responses following acclimation of gilthead sea bream (Sparus aurata) to different environmental salinities. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006, 145, 268–273. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Wunderink, Y.S.; Gozdowska, M.; Kulczykowska, E.; Mancera, J.M.; Martinez-Rodriguez, G. Vasotocinergic and isotocinergic systems in the gilthead sea bream (Sparus aurata): An osmoregulatory story. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2013, 166, 571–581. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Fuentes, J.; Mancera, J.M.; Martinez-Rodriguez, G. Variations in the expression of vasotocin and isotocin receptor genes in the gilthead sea bream Sparus aurata during different osmotic challenges. Gen. Comp. Endocrinol. 2014, 197, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Bond, H.; Winter, M.J.; Warne, J.M.; McCrohan, C.R.; Balment, R.J. Plasma concentrations of arginine vasotocin and urotensin II are reduced following transfer of the euryhaline flounder (Platichthys flesus) from seawater to fresh water. Gen. Comp. Endocrinol. 2002, 125, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Balment, R.J.; Lu, W.; Weybourne, E.; Warne, J.M. Arginine vasotocin a key hormone in fish physiology and behaviour: A review with insights from mammalian models. Gen. Comp. Endocrinol. 2006, 147, 9–16. [Google Scholar] [CrossRef]
- Chou, M.Y.; Hung, J.C.; Wu, L.C.; Hwang, S.P.; Hwang, P.P. Isotocin controls ion regulation through regulating ionocyte progenitor differentiation and proliferation. Cell. Mol. Life Sci. 2011, 68, 2797–2809. [Google Scholar] [CrossRef] [PubMed]
- Martos-Sitcha, J.A.; Gregorio, S.F.; Carvalho, E.S.; Canario, A.V.; Power, D.M.; Mancera, J.M.; Martinez-Rodriguez, G.; Fuentes, J. AVT is involved in the regulation of ion transport in the intestine of the sea bream (Sparus aurata). Gen. Comp. Endocrinol. 2013, 193, 221–228. [Google Scholar] [CrossRef]
- Lema, S.C.; Washburn, E.H.; Crowley, M.E.; Carvalho, P.G.; Egelston, J.N.; McCormick, S.D. Evidence for a role of arginine vasotocin receptors in the gill during salinity acclimation by a euryhaline teleost fish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 316, R735–R750. [Google Scholar] [CrossRef]
- Amer, S.; Brown, J.A. Glomerular actions of arginine vasotocin in the in situ perfused trout kidney. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1995, 269, R775–R780. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; MartinezRodriguez, G.; Mancera, J.M.; Fuentes, J. AVT and IT regulate ion transport across the opercular epithelium of killifish (Fundulus heteroclitus) and gilthead sea bream (Sparus aurata). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 182, 93–101. [Google Scholar] [CrossRef]
- Hasunuma, I.; Sakai, T.; Nakada, T.; Toyoda, F.; Namiki, H.; Kikuyama, S. Molecular cloning of three types of arginine vasotocin receptor in the newt, Cynops pyrrhogaster. Gen. Comp. Endocrinol. 2007, 151, 252–258. [Google Scholar] [CrossRef]
- Ocampo Daza, D.; Lewicka, M.; Larhammar, D. The oxytocin/vasopressin receptor family has at least five members in the gnathostome lineage, inclucing two distinct V2 subtypes. Gen. Comp. Endocr. 2012, 175, 135–143. [Google Scholar] [CrossRef]
- Tanoue, A.; Ito, S.; Honda, K.; Oshikawa, S.; Kitagawa, Y.; Koshimizu, T.A.; Mori, T.; Tsujimoto, G. The vasopressin V1b receptor critically regulates hypothalamic-pituitary-adrenal axis activity under both stress and resting conditions. J. Clin. Investig. 2004, 113, 302–309. [Google Scholar] [CrossRef]
- Hayashi, M.; Sasaki, S.; Tsuganezawa, H.; Monkawa, T.; Kitajima, W.; Konishi, K.; Fushimi, K.; Marumo, F.; Saruta, T. Expression and distribution of aquaporin of collecting duct are regulated by vasopressin V2 receptor in rat kidney. J. Clin. Investig. 1994, 94, 1778–1783. [Google Scholar] [CrossRef]
- Lema, S.C. Identification of multiple vasotocin receptor cDNAs in teleost fish: Sequences, phylogenetic analysis, sites of expression, and regulation in the hypothalamus and gill in response to hyperosmotic challenge. Mol. Cell. Endocrinol. 2010, 321, 215–230. [Google Scholar] [CrossRef]
- Lema, S.C.; Slane, M.A.; Salvesen, K.E.; Godwin, J. Variation in gene transcript profiles of two V1a-type arginine vasotocin receptors among sexual phases of bluehead wrasse (Thalassoma bifasciatum). Gen. Comp. Endocrinol. 2012, 179, 451–464. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Kaiya, H.; Konno, N.; Iwata, E.; Miyazato, M.; Uchiyama, M.; Hyodo, S. The fifth neurohypophysial hormone receptor is structurally related to the V2-type receptor but functionally similar to V1-type receptors. Gen. Comp. Endocrin. 2012, 178, 519–528. [Google Scholar] [CrossRef]
- Lagman, D.; Daza, D.O.; Widmark, J.; Abalo, X.M.; Sundström, G.; Larhammar, D. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was estab-lished by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol. Biol. 2013, 13, 238. [Google Scholar] [CrossRef]
- Cao, Q.; Liang, F.; Wang, D.; Zhang, X.; Lorin-Nebel, C.; Gu, J.; Yin, S. Dynamic expression of vasotocin and isotocin receptor genes in the marbled eel (Anguilla marmorata) following osmotic challenges. Gene 2018, 677, 49–56. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Campinho, M.A.; Mancera, J.M.; Martinez-Rodriguez, G.; Fuentes, J. Vasotocin and isotocin regulate aquaporin 1 function in the sea bream. J. Exp. Biol. 2015, 218, 684–693. [Google Scholar] [CrossRef]
- Mancera, J.M.; Martinez-Rodriguez, G.; Skrzynska, A.K.; Martos-Sitcha, J.A. Osmoregulatory role of vasotocinergic and isotocinergic systems in the gilthead sea bream (Sparus aurata L.). Gen. Comp. Endocrinol. 2018, 257, 177–183. [Google Scholar] [CrossRef]
- Krahmann, G.; Schott, F. Longterm increases in western Mediterranean salinities and temperatures: Anthropogenic and climatic sources. Geophys. Res. Lett. 1998, 25, 4209–4212. [Google Scholar] [CrossRef]
- Aurelle, D.; Thomas, S.; Albert, C.; Bally, M.; Bondeau, A.; Boudouresque, C.F.; Cahill, A.E.; Carlotti, F.; Chenuil, A.; Cramer, W.; et al. Biodiversity, climate change, and adaptation in the Mediterranean. Ecosphere 2022, 13, e3915. [Google Scholar] [CrossRef]
- Borghini, M.B.H.S. The Mediterranean is becoming saltier. Ocean Sci. 2014, 10, 693–700. [Google Scholar] [CrossRef]
- Margirier, F.; Testor, P.; Heslop, E.; Mallil, K.; Bosse, A.; Houpert, L.; Mortier, L.; Bouin, M.N.; Coppola, L.; D’Ortenzio, F.; et al. Abrupt warming and salinification of intermediate waters interplays with decline of deep convection in the Northwestern Mediterranean Sea. Sci. Rep. 2020, 10, 20923. [Google Scholar] [CrossRef]
- Kulczykowska, E.; Gozdowska, M.; Kalamarz, H.; Kleszczyńska, A.; Nietrzeba, M.; Martínez-Rodríguez, G.; Mancera, J.M. Hypothalamic arginine vasotocin and isotocin are involved in stress response in fish. Comp. Bichem. Physiol. A Mol. Integr. Physiol. 2009, 154, S23–S27. [Google Scholar] [CrossRef]
- Gonzalez, R.J. The physiology of hyper-salinity tolerance in teleost fish: A review. J. Comp. Physiol. B 2012, 182, 321–329. [Google Scholar] [CrossRef]
- Cao, Q.; Blondeau-Bidet, E.; Lorin-Nebel, C. Intestinal osmoregulatory mechanisms differ in Mediterranean and Atlantic European sea bass: A focus on hypersalinity. Sci. Total Environ. 2022, 804, 150208. [Google Scholar] [CrossRef]
- Cao, Q.; Giffard-Mena, I.; Blondeau-Bidet, E.; Hermet, S.; Hu, Y.; Lee, T.; Lorin-Nebel, C. Mechanisms of acclimation to hypersalinity in two European sea bass lineages: A focus on the kidney function. Aquaculture 2021, 534, 736305. [Google Scholar] [CrossRef]
- Koehbach, J.; Stockner, T.; Bergmayr, C.; Muttenthaler, M.; Gruber, C.W. Insights into the molecular evolution of oxytocin receptor ligand binding. Biochem. Soc. Trans. 2013, 41, 197–204. [Google Scholar] [CrossRef]
- Chini, B.; Fanelli, F. Molecular basis of ligand binding and receptor activation in the oxytocin and vasopressin receptor family. Exp. Physiol. 2000, 85, 59S–66S. [Google Scholar] [CrossRef]
- Acharjee, S.; Do-Rego, J.L.; Oh, D.Y.; Ahn, R.S.; Choe, H.; Vaudry, H.; Kim, K.; Seong, J.Y.; Kwon, H.B. Identification of amino acid residues that direct differential ligand selectivity of mammalian and nonmammalian V1a type receptors for arginine vasopressin and vasotocin. Insights into molecular coevolution of V1a type receptors and their ligands. J. Biol. Chem. 2004, 279, 54445–54453. [Google Scholar] [CrossRef]
- Tine, M.; Kuhl, H.; Gagnaire, P.A.; Louro, B.; Desmarais, E.; Martins, R.S.; Reinhardt, R. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef]
- Ocampo Daza, D.; Bergqvist, C.A.; Larhammar, D. The Evolution of Oxytocin and Vasotocin Receptor Genes in Jawed Vertebrates: A Clear Case for Gene Duplications Through Ancestral Whole-Genome Duplications. Front. Endocrinol. 2022, 12, 792644. [Google Scholar] [CrossRef]
- Lema, S.C.; Sanders, K.E.; Walti, K.A. Arginine vasotocin, isotocin and nonapeptide receptor gene expression link to social status and aggression in sex-dependent patterns. J. Neuroendocrinol. 2015, 27, 142–157. [Google Scholar] [CrossRef]
- Mayasich, S.A.; Clarke, B.L. Vasotocin and the origins of the vasopressin/oxytocin receptor gene family. Vitam. Horm. 2020, 113, 1–27. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, N.; Shi, B.; Zhang, S.; Zhang, L.; Zhang, W. Isotocin regulates growth hormone but not prolactin release from the pituitary of ricefield eels. Front. Endocrinol. 2018, 9, 166. [Google Scholar] [CrossRef]
- Cadiz, L.; Roman-Padilla, J.; Gozdowska, M.; Kulczykowska, E.; Martinez-Rodriguez, G.; Mancera, J.M.; Martos-Sitcha, J.A. Cortisol modulates vasotocinergic and isotocinergic pathways in the gilthead sea bream. J. Exp. Biol. 2015, 218, 316–325. [Google Scholar] [CrossRef]
- Abimbola, J.K.M.; Edwards, J.M.; Lema, S.C. Expression profiles of genes encoding arginine vasotocin and isotocin receptors and the leucyl-cystinyl aminopeptidase (LNPEP) nonapeptide degradation enzyme in blue tilapia (Oreochromis aureus) during high salinity acclimation. Mar. Freshw. Behav. Physiol. 2020, 53, 163–191. [Google Scholar] [CrossRef]
- Westin, L.; Nissling, A. Effects of salinity on spermatozoa motility, percentage of fertilized eggs and egg development of Baltic cod (Gadus morhua), and implications for cod stock fluctuations in the Baltic. Mar. Biol. 1991, 108, 5–9. [Google Scholar] [CrossRef]
- Larsen, P.F.; Nielsen, E.E.; Williams, T.D.; Loeschcke, V. Intraspecific variation in expression of candidate genes for osmoregulation, heme biosynthesis and stress resistance suggests local adaptation in European flounder (Platichthys flesus). Heredity (Edinb.) 2008, 101, 247–259. [Google Scholar] [CrossRef]
- Gaggiotti, O.E.; Bekkevold, D.; Jørgensen, H.B.; Foll, M.; Carvalho, G.R.; Andre, C.; Ruzzante, D.E. Disentangling the effects of evolutionary, demographic, and environmental factors influencing genetic structure of natural populations: Atlantic herring as a case study. Evolution 2009, 63, 2939–2951. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y.; Huang, X.; Li, S.; Zhan, A. Local environment-driven adaptive evolution in a marine invasive ascidian (Molgula manhattensis). Ecol. Evol. 2021, 11, 4252–4266. [Google Scholar] [CrossRef]
- Ayala, M.D.; López-Albors, O.; Gil, F.; García-Alcázar, A.; Abellán, E.; Alarcón, J.A.; Álvarez, M.C.; Ramírez-Zarzosa, G.; Moreno, F. Temperature effects on muscle growth in two populations (Atlantic and Mediterranean) of sea bass, Dicentrarchus labrax L. Aquaculture 2001, 202, 359–370. [Google Scholar] [CrossRef]
- Person-Le Ruyet, J.; Mahé, K.; Le Bayon, N.; Le Delliou, H. Effects of temperature on growth and metabolism in a Mediterranean population of European sea bass, Dicentrarchus labrax. Aquaculture 2004, 237, 269–280. [Google Scholar] [CrossRef]
- Doan, Q.K.; Vandeputte, M.; Chatain, B.; Haffray, P.; Vergnet, A.; Breuil, G.; Allal, F. Genetic variation of resistance to Viral Nervous Necrosis and genetic correlations with production traits in wild populations of the European sea bass (Dicentrarchus labrax). Aquaculture 2017, 478, 1–8. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Keane, T.M.; Creevey, C.J.; Pentony, M.M.; Naughton, T.J.; Mclnerney, J.O. Assessment of methods of amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006, 6, 1–17. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 34. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]


| Sequences ID | Target Gene | Primer Name | Sequence (from 5′ to 3′) | Amplicon Size | Efficiency |
|---|---|---|---|---|---|
| DLAgn_00209950 | avtrv1a1 | avtrv1a1-F avtrv1a1-R | CGCGCCAAATTACGCACG TAGCGGGCGGCAGCACG | 259 | 1.858 |
| DLAgn_00170900 | avtrv1a2 | avtrv1a2-F avtrv1a2-R | CAGTCTCTGGTTTTTAGTCCCAC AAGGTTGTGTTGTACATTGCCA | 194 | 2.025 |
| DLAgn_00207720 | avtrv2c | avtrv2c-F avtrv2c-R | AGAGCTGGGATTTTGGGTTTG CAAGTATTTTACGGACCGGC | 209 | 2.062 |
| DLAgn_00125320 | avtrv2a2 | avtrv2a2-F | CAACAACCCTCTGCACCAG | 234 | 2.22 |
| avtrv2a2-R | TGCGCCAGCAGAGGGCAG | ||||
| DLAgn_00092800 | avtrv2b1 | avtrv2b1-F | TCAGTAAAGACTCTGTGAGCAGC | 174 | 2.001 |
| avtrv2b1-R | CTGTCTGTAAAGGCAAAAGTGCT | ||||
| DLAgn_00089180 | avtrv2b2 | avtrv2b2-F | TGCGTCTCTCTCATCTTCGG | 165 | 2.083 |
| avtrv2b2-R | CACCGTGAGAATGGGCAGG | ||||
| DLAgn_00131190 | avtrv2a1 | avtrv2a1-F | GCACGCCTAATGTACAAAGCAG | 213 | 2.03 |
| avtrv2a1-R | CTCCTGCAGCAGTAACATCCAT | ||||
| DLAgn_00093990 | itr2 | itr2-F | CTGCCGACAGTACCTACCCT | 145 | 2.188 |
| itr2-R | TCCTGGCCCTCCATTGCT | ||||
| DLAgn_00093190 | itr1 | itr1-F | AACCTCCAAAGGCAACACGC | 214 | 1.869 |
| itr1-R | CATGGAGATGATGAAGGGCA | ||||
| FM004681 | Fau | fau-F fau-R | GACACCCAAGGTTGACAAGCAG GGCATTGAAGCACTTAGGAGTTG | 150 | 1.851 |
| AJ866727 | ef1α | ef1α-F ef1α-R | GGCTGGTATCTCTAAGAACG CCTCCAGCATGTTGTCTCC | 239 | 1.853 |
| DLAgn_00023060 | l13 | l13-F l13-R | TCTGGAGGACTGTCAGGGGCATGC AGACGCACAATCTTGAGAGCAG | 148 | 1.873 |
| Interaction | Salinity | Lineage | |
|---|---|---|---|
| mRNA expression | |||
| AI-avtrv1a1 | ** | ns | **** |
| AI-avtrv1a2 | * | **** | ns |
| AI-avtrv2a1 | ns | * | ns |
| AI-avtrv2a2 | ns | ns | ns |
| AI-avtrv2b1 | **** | **** | ** |
| AI-avtrv2b2 | ns | ns | ns |
| AI-avtrv2c | ns | ns | ns |
| AI-itr2 | ns | ns | ns |
| AI-itr1 | ns | **** | * |
| PI-avtrv1a1 | ns | **** | ns |
| PI-avtrv1a2 | ns | **** | ns |
| PI-avtrv2a1 | ns | ns | * |
| PI-avtrv2a2 | ns | ns | ns |
| PI-avtrv2b1 | ns | **** | ns |
| PI-avtrv2b2 | ns | ns | ns |
| PI-avtrv2c | ns | ns | ns |
| PI-itr2 | ns | ns | ns |
| PI-itr1 | ns | **** | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Q.; Blondeau-Bidet, E.; Lorin-Nebel, C. Hypo-Osmoregulatory Roles of Vasotocinergic and Isotocinergic Systems in the Intestines of Two European Sea Bass Lineages. Int. J. Mol. Sci. 2022, 23, 13636. https://doi.org/10.3390/ijms232113636
Cao Q, Blondeau-Bidet E, Lorin-Nebel C. Hypo-Osmoregulatory Roles of Vasotocinergic and Isotocinergic Systems in the Intestines of Two European Sea Bass Lineages. International Journal of Molecular Sciences. 2022; 23(21):13636. https://doi.org/10.3390/ijms232113636
Chicago/Turabian StyleCao, Quanquan, Eva Blondeau-Bidet, and Catherine Lorin-Nebel. 2022. "Hypo-Osmoregulatory Roles of Vasotocinergic and Isotocinergic Systems in the Intestines of Two European Sea Bass Lineages" International Journal of Molecular Sciences 23, no. 21: 13636. https://doi.org/10.3390/ijms232113636
APA StyleCao, Q., Blondeau-Bidet, E., & Lorin-Nebel, C. (2022). Hypo-Osmoregulatory Roles of Vasotocinergic and Isotocinergic Systems in the Intestines of Two European Sea Bass Lineages. International Journal of Molecular Sciences, 23(21), 13636. https://doi.org/10.3390/ijms232113636







