Proteomic Analysis of Roots Response to Potassium Deficiency and the Effect of TaHAK1-4A on K+ Uptake in Wheat
Abstract
1. Introduction
2. Results
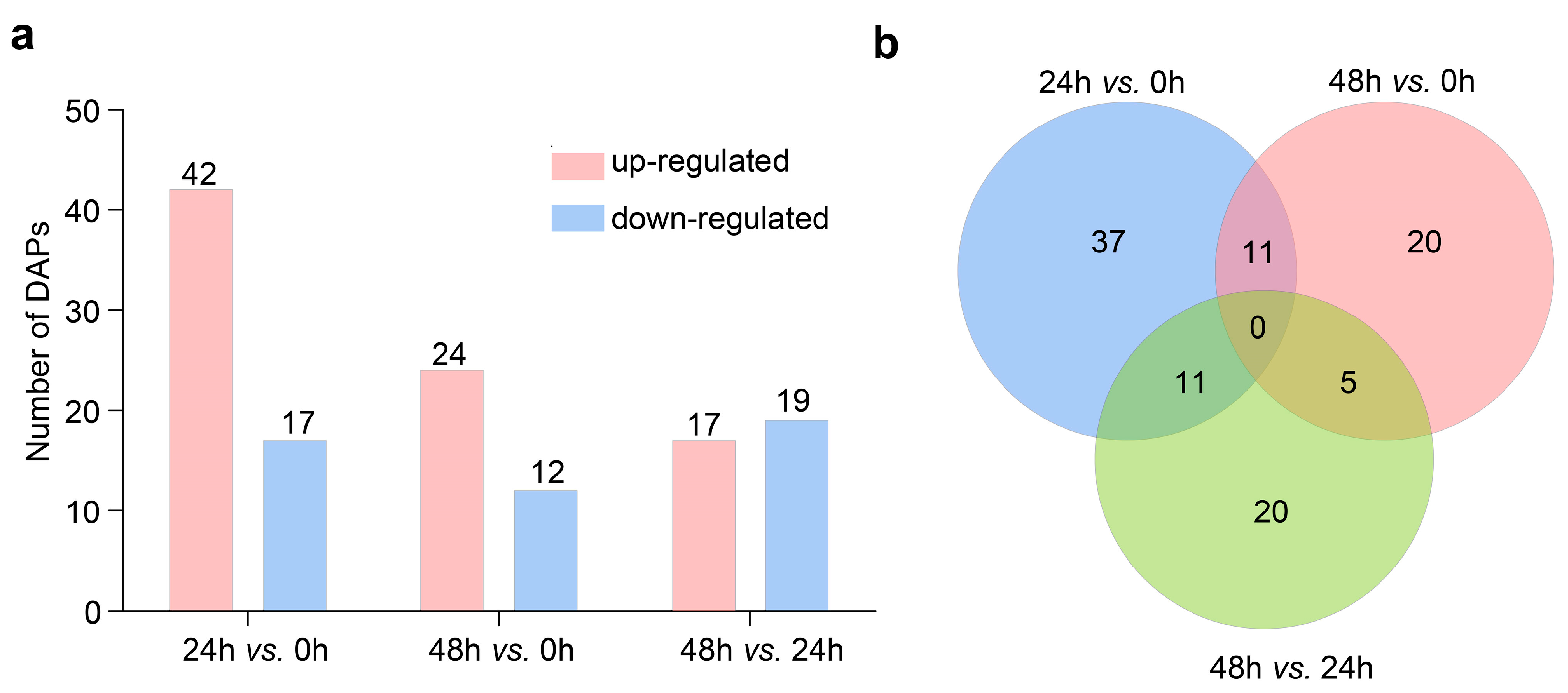
2.1. Wheat Root Proteomic Profiles under LK Conditions
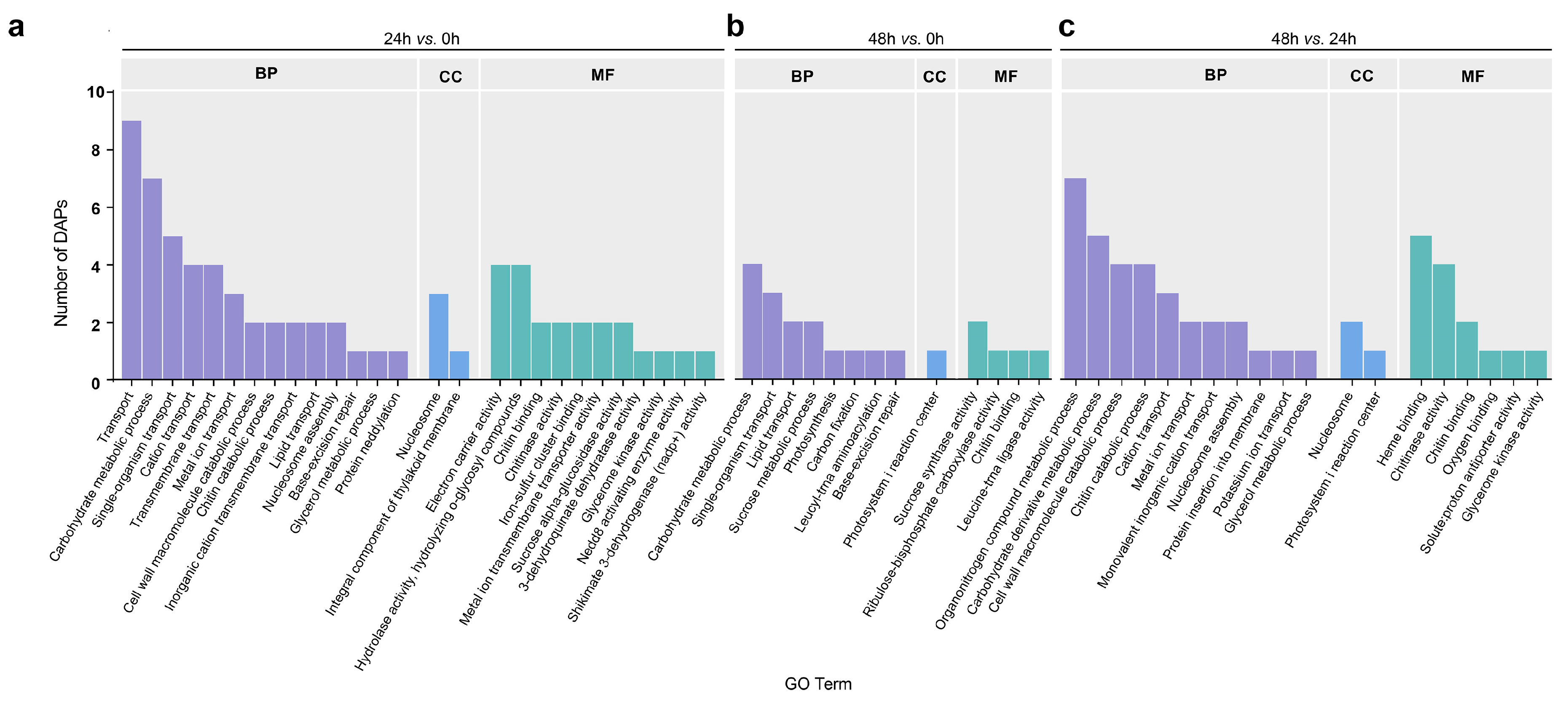
2.2. Functional Analysis of DAPs Related to LK Stress Responses
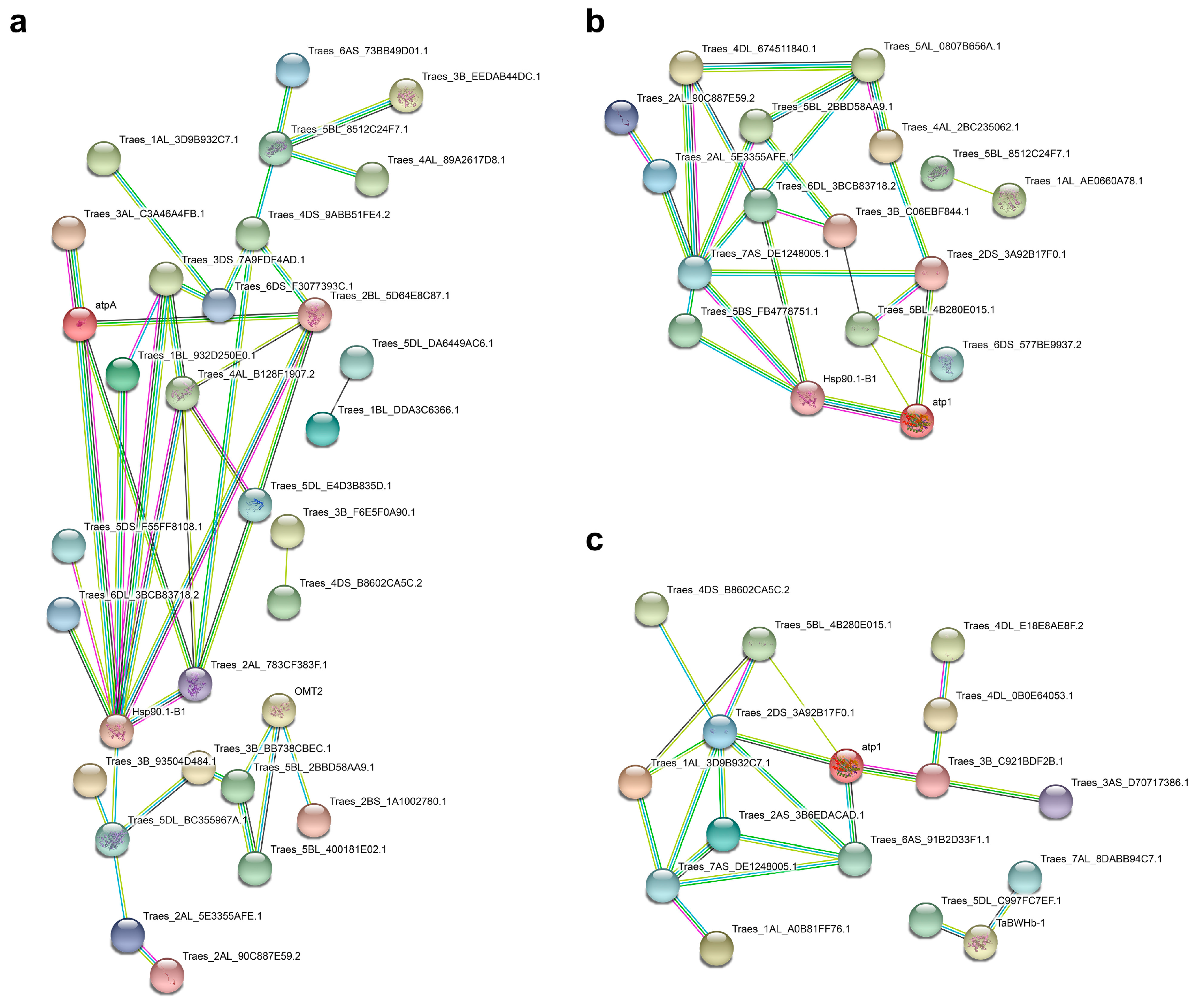
2.3. Protein–Protein Interaction (PPI) Network Analysis of the DAPs Related to LK Stress Responses
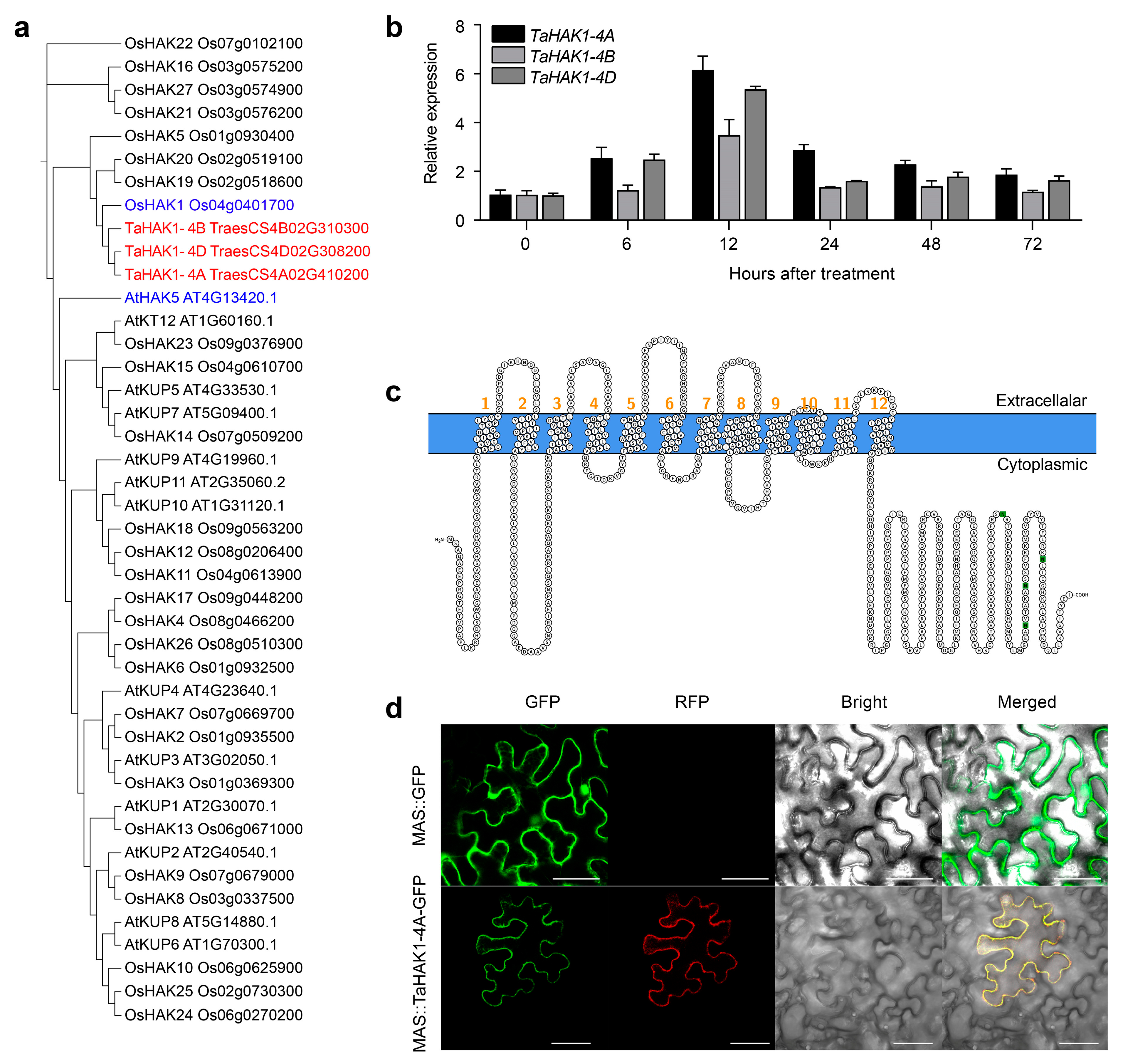
2.4. Expression Analysis and Molecular Characterization of TaHAK1-4A
2.5. Subcellular Localization of TaHAK1-4A
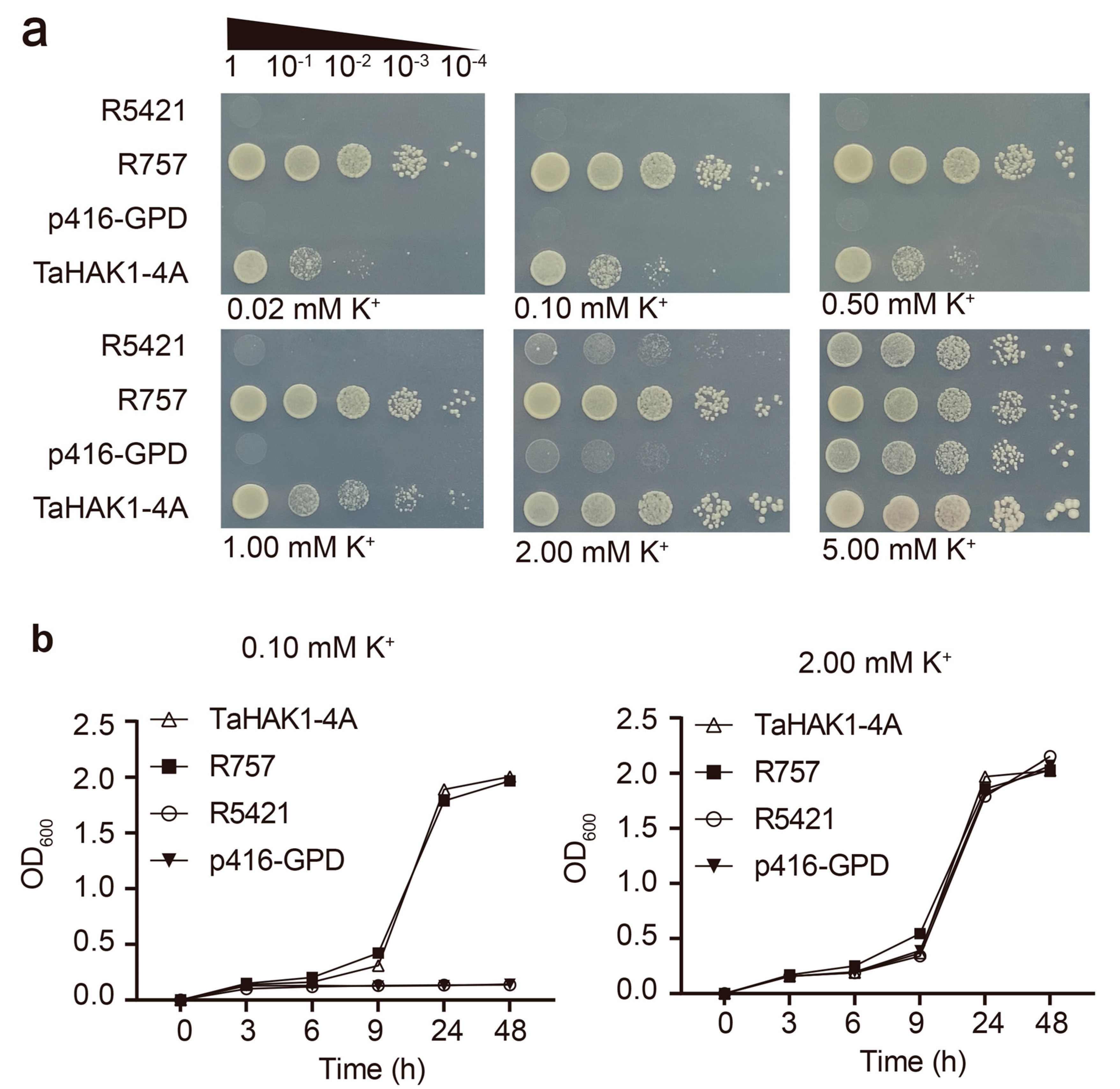
2.6. TaHAK1-4A Mediates K+ Uptake in Yeast
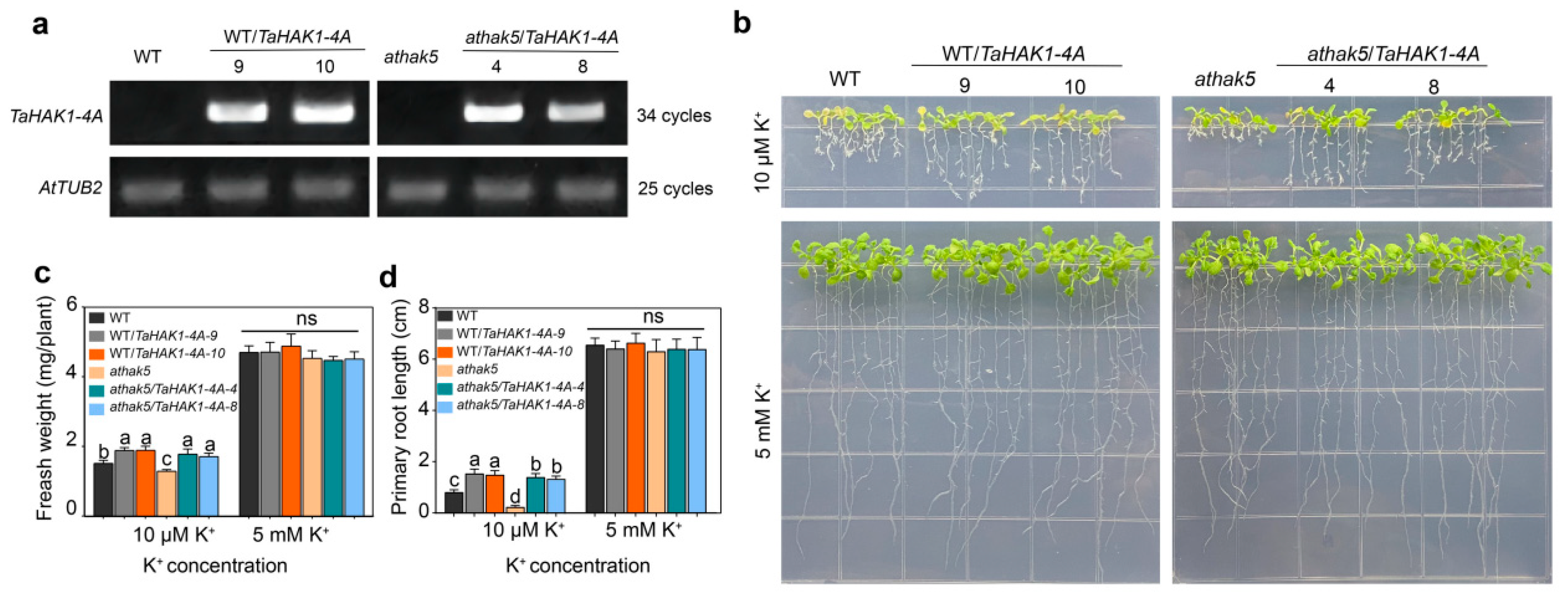
2.7. TaHAK1-4A-Overexpressing Arabidopsis Lines Are Tolerant to LK Stress
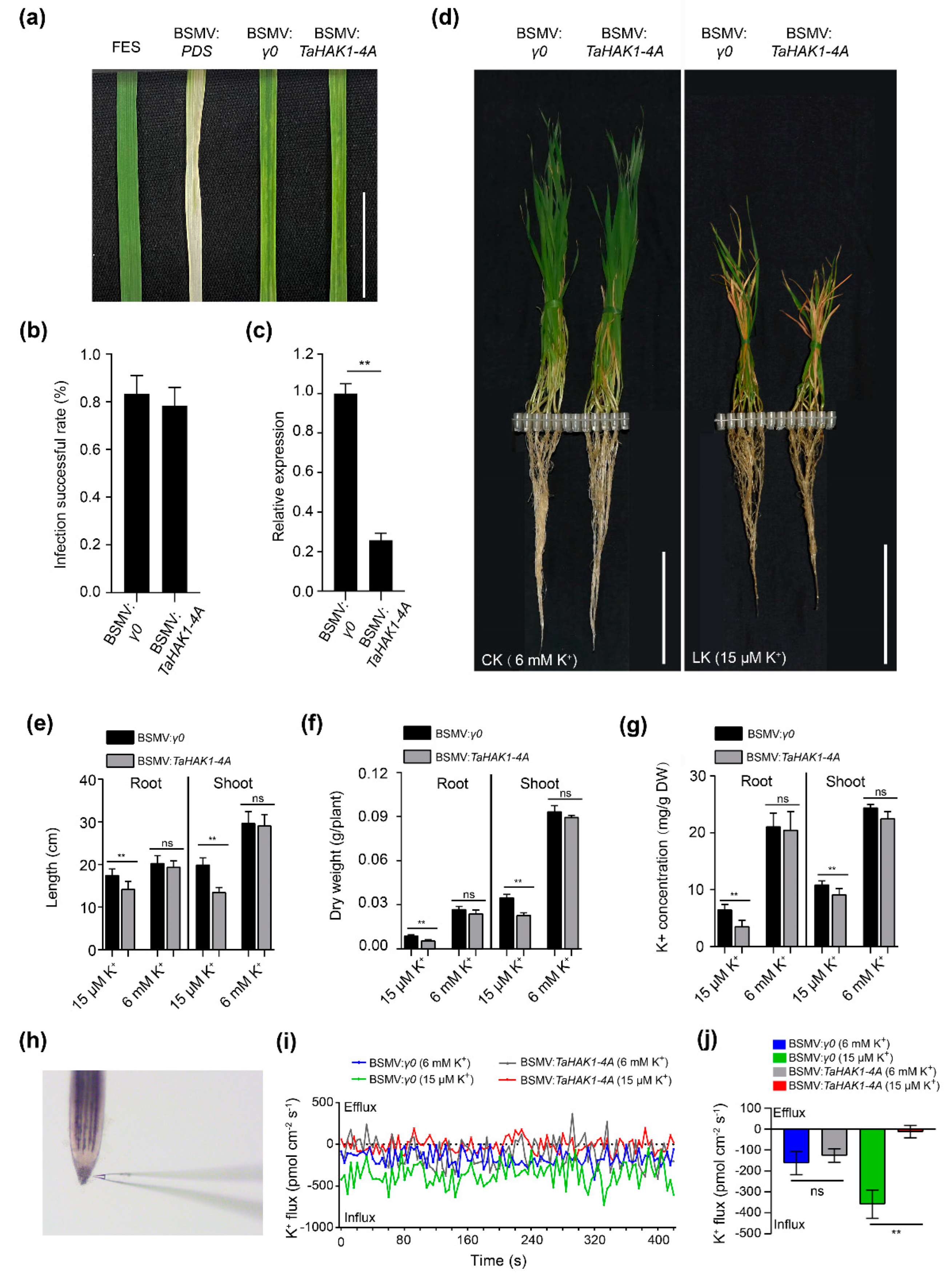
2.8. TaHAK1-4A Promotes K+ Uptake in Wheat
3. Discussion
3.1. Proteins Related to Carbohydrate and Energy Metabolism in Wheat Roots under LK Conditions
3.2. Proteins Related to Stress Responses and Defense in Wheat Roots under LK Conditions
3.3. Proteins Related to Post-Translational Modifications in Wheat Roots under LK Conditions
3.4. Transporters and Channel Proteins in Wheat Roots under LK Conditions
3.5. The K+ Transporter TaHAK1-4A Contributes to the K+ Uptake by Wheat Roots under LK Conditions
4. Materials and Methods
4.1. Wheat Seedling Culture and LK Stress
4.2. Protein Extraction
4.3. Digestion and Tandem Mass Tag (TMT) Labeling of Peptides
4.4. High-pH Reversed-Phase HPLC Fractionation and LC-MS/MS Analysis
4.5. Bioinformatics Analysis of Identified Proteins
4.6. Identification of HAK Gene Family and Expression Profiles of HAKs under the LK Stress in Wheat
4.7. RNA Extraction and Transcript Analysis
4.8. Functional Complementation of TaHAK1-4A in Yeast
4.9. Subcellular Localization of TaHAK1-4A
4.10. Generation of Arabidopsis Transgenic Plants and LK Stress Response Analysis
4.11. Silencing of TaHAK1-4A and LK Stress Response Assay in Wheat
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zorb, C.; Senbayram, M.; Peiter, E. Potassium in agriculture—Status and perspectives. J. Plant Physiol. 2014, 171, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Çolpan, E.; Zengin, M.; Özbahçe, A. The effects of potassium on the yield and fruit quality components of stick tomato. Hortic. Environ. Biote 2013, 54, 20–28. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [PubMed]
- Dinu, M.; Whittaker, A.; Pagliai, G.; Benedettelli, S.; Sofi, F. Ancient wheat species and human health: Biochemical and clinical implications. J. Nutr. Biochem. 2018, 52, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sparks, D. Potassium dynamics in soils. Adv. Soil Sci. 1987, 6, 1–63. [Google Scholar] [CrossRef]
- Ghadam Khani, A.; Enayatizamir, N.; Norouzi Masir, M. Impact of plant growth promoting rhizobacteria on different forms of soil potassium under wheat cultivation. Lett. Appl. Microbiol. 2019, 68, 514–521. [Google Scholar] [CrossRef]
- Wu, X.; Wang, D.; Riaz, M.; Zhang, L.; Jiang, C. Investigating the effect of biochar on the potential of increasing cotton yield, potassium efficiency and soil environment. Ecotoxicol. Environ. Saf. 2019, 182, 109451. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, R.; Liu, H.; Liu, X.; Xu, K.; Xiao, K.; Zhang, S.; Yang, X.; Xue, C. Multi-omics analyses reveal the molecular mechanisms underlying the adaptation of wheat (Triticum aestivum L.) to potassium deprivation. Front. Plant Sci. 2020, 11, 588994. [Google Scholar] [CrossRef]
- Zhang, Z.; Chao, M.; Wang, S.; Bu, J.; Tang, J.; Li, F.; Wang, Q.; Zhang, B. Proteome quantification of cotton xylem sap suggests the mechanisms of potassium-deficiency-induced changes in plant resistance to environmental stresses. Sci. Rep. 2016, 6, 21060. [Google Scholar] [CrossRef]
- Li, L.Q.; Liu, L.; Zhuo, W.; Chen, Q.; Hu, S.; Peng, S.; Wang, X.Y.; Lu, Y.F.; Lu, L.M. Physiological and quantitative proteomic analyses unraveling potassium deficiency stress response in alligator weed (Alternanthera philoxeroides L.) root. Plant Mol. Biol. 2018, 97, 265–278. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Agarwal, R.M. Potassium up-regulates antioxidant metabolism and alleviates growth inhibition under water and osmotic stress in wheat (Triticum aestivum L). Protoplasma 2017, 254, 1471–1486. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Liu, X.; Mao, W.; Zhang, X.; Chen, S.; Zhan, K.; Bi, H.; Xu, H. Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2018, 19, 3969. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Qiu, X.; Wang, L.; Xie, W.; Zhang, C.; Xiong, L.; Lian, X.; Zhang, Q. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genom. 2008, 280, 437–452. [Google Scholar] [CrossRef]
- Song, Z.; Yang, Y.; Ma, R.; Xu, J.; Yu, M. Transcription of potassium transporter genes of KT/HAK/KUP family in peach seedlings and responses to abiotic stresses. Biol. Plant. 2014, 59, 65–73. [Google Scholar] [CrossRef]
- Cai, K.; Zeng, F.; Wang, J.; Zhang, G. Identification and characterization of HAK/KUP/KT potassium transporter gene family in barley and their expression under abiotic stress. BMC Genom. 2021, 22, 317. [Google Scholar] [CrossRef] [PubMed]
- Banuelos, M.A.; Garciadeblas, B.; Cubero, B.; Rodriguez-Navarro, A. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 2002, 130, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Wei, J. Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 2012, 39, 8465–8473. [Google Scholar] [CrossRef]
- Rubio, F.; Nieves-Cordones, M.; Aleman, F.; Martinez, V. Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant 2008, 134, 598–608. [Google Scholar] [CrossRef]
- Chen, G.; Hu, Q.; Luo, L.; Yang, T.; Zhang, S.; Hu, Y.; Yu, L.; Xu, G. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef]
- Feng, H.; Tang, Q.; Cai, J.; Xu, B.; Xu, G.; Yu, L. Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta 2019, 250, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Fulgenzi, F.R.; Peralta, M.L.; Mangano, S.; Danna, C.H.; Vallejo, A.J.; Puigdomenech, P.; Santa-Maria, G.E. The ionic environment controls the contribution of the barley HvHAK1 transporter to potassium acquisition. Plant Physiol. 2008, 147, 252–262. [Google Scholar] [CrossRef]
- Qin, Y.-J.; Wu, W.-H.; Wang, Y. ZmHAK5 and ZmHAK1 function in K uptake and distribution in maize under low K conditions. J. Integr. Plant Biol. 2019, 61, 691–705. [Google Scholar] [CrossRef]
- Rubio, F.; Aleman, F.; Nieves-Cordones, M.; Martinez, V. Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol. Plant 2010, 139, 220–228. [Google Scholar] [CrossRef]
- Gierth, M.; Maser, P.; Schroeder, J.I. The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, M.L.; Ma, T.L.; Wang, Y. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 2016, 28, 3005–3019. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gonzalez, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, 6403. [Google Scholar] [CrossRef]
- Gaber, R.F.; Styles, C.A.; Fink, G.R. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell Biol. 1988, 8, 2848–2859. [Google Scholar] [CrossRef]
- Ruan, L.; Zhang, J.; Xin, X.; Zhang, C.; Ma, D.; Chen, L.; Zhao, B. Comparative analysis of potassium deficiency-responsive transcriptomes in low potassium susceptible and tolerant wheat (Triticum aestivum L.). Sci. Rep. 2015, 5, 10090. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhao, Y.; Gu, J.; Zhou, M.; Gao, L.; Sun, R.; Wang, W.; Zhang, S.; Yang, X. Proteomic analysis reveals the molecular mechanism underlying the cold acclimation and freezing tolerance of wheat (Triticum aestivum L.). Plant Sci. 2022, 318, 111242. [Google Scholar] [CrossRef]
- Liu, Y.; Ji, D.; Turgeon, R.; Chen, J.; Lin, T.; Huang, J.; Luo, J.; Zhu, Y.; Zhang, C.; Lv, Z. Physiological and proteomic responses of mulberry Trees (Morus alba. L.) to combined salt and drought stress. Int. J. Mol. Sci. 2019, 20, 2486. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Y.; Jin, Y.; Kan, L.; Shen, C.; Malladi, A.; Nambeesan, S.; Xu, Y.; Dong, C. Transcriptome analysis of Pyrus betulaefolia seedling root responses to short-term potassium deficiency. Int. J. Mol. Sci. 2020, 21, 8857. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lyu, C.; Li, J.; Tong, Z.; Lu, Y.; Wang, X.; Ni, S.; Yang, S.; Zeng, F.; Lu, L. Physiological analysis and proteome quantification of alligator weed stems in response to potassium deficiency stress. Int. J. Mol. Sci. 2019, 20, 221. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, P.; Sulpice, R.; Miller, A.J.; Stitt, M.; Amtmann, A.; Gibon, Y. Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 2009, 150, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Nemati, F.; Ghanati, F.; Gavlighi, H.A.; Sharifi, M. Fructan dynamics and antioxidant capacity of 4-day-old seedlings of wheat (Triticum aestivum) cultivars during drought stress and recovery. Funct. Plant Biol. 2018, 45, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Livingston, D.P., 3rd; Hincha, D.K.; Heyer, A.G. Fructan and its relationship to abiotic stress tolerance in plants. Cell Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jiao, T.; Zhang, Z.; Yao, Z.; Li, Z.; Wang, S.; Xin, H.; Li, Y.; Wang, A.; Zhu, J. Ectopic Expression of the Allium cepa 1-SST gene in cotton improves drought tolerance and yield under drought stress in the field. Front. Plant Sci. 2021, 12, 783134. [Google Scholar] [CrossRef]
- Kawakami, A.; Sato, Y.; Yoshida, M. Genetic engineering of rice capable of synthesizing fructans and enhancing chilling tolerance. J. Exp. Bot. 2008, 59, 793–802. [Google Scholar] [CrossRef]
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Naz, R.; Bano, A.; Wilson, N.L.; Guest, D.; Roberts, T.H. Pathogenesis-related protein expression in the apoplast of wheat leaves protected against leaf rust following application of plant extracts. Phytopathology 2014, 104, 933–944. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Kasprzewska, A. Plant chitinases--regulation and function. Cell Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 2009, 229, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- He, Y.; Yao, Y.; Li, L.; Li, Y.; Gao, J.; Fan, M. A heat-shock 20 protein isolated from watermelon (ClHSP22.8) negatively regulates the response of Arabidopsis to salt stress via multiple signaling pathways. PeerJ 2021, 9, e10524. [Google Scholar] [CrossRef]
- Bourgine, B.; Guihur, A. Heat shock signaling in land plants: From plasma membrane sensing to the transcription of small heat shock proteins. Front. Plant Sci. 2021, 12, 710801. [Google Scholar] [CrossRef]
- He, Y.; Li, R.; Lin, F.; Xiong, Y.; Wang, L.; Wang, B.; Guo, J.; Hu, C. Transcriptome changes induced by different potassium levels in banana roots. Plants 2019, 9, 11. [Google Scholar] [CrossRef]
- Li, L.; Lyu, C.; Huang, L.; Chen, Q.; Zhuo, W.; Wang, X.; Lu, Y.; Zeng, F.; Lu, L. Physiology and proteomic analysis reveals root, stem and leaf responses to potassium deficiency stress in alligator weed. Sci. Rep. 2019, 9, 17366. [Google Scholar] [CrossRef]
- Filipcik, P.; Curry, J.R.; Mace, P.D. When worlds collide-mechanisms at the interface between phosphorylation and ubiquitination. J. Mol. Biol. 2017, 429, 1097–1113. [Google Scholar] [CrossRef]
- Ragel, P.; Rodenas, R.; Garcia-Martin, E.; Andres, Z.; Villalta, I.; Nieves-Cordones, M.; Rivero, R.M.; Martinez, V.; Pardo, J.M.; Quintero, F.J.; et al. The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 2015, 169, 2863–2873. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Long, Y.; Qi, G.N.; Li, J.; Xu, Z.J.; Wu, W.H.; Wang, Y. The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 2014, 26, 3387–3402. [Google Scholar] [CrossRef] [PubMed]
- Zelazny, E.; Barberon, M.; Curie, C.; Vert, G. Ubiquitination of transporters at the forefront of plant nutrition. Plant Signal. Behav. 2011, 6, 1597–1599. [Google Scholar] [CrossRef]
- Zhou, H.; Zhao, J.; Yang, Y.; Chen, C.; Liu, Y.; Jin, X.; Chen, L.; Li, X.; Deng, X.W.; Schumaker, K.S.; et al. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell 2012, 24, 5106–5122. [Google Scholar] [CrossRef] [PubMed]
- Pozo, J.C.; Timpte, C.; Tan, S.; Callis, J.; Estelle, M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 1998, 280, 1760–1763. [Google Scholar] [CrossRef] [PubMed]
- Mostofa, M.G.; Rahman, M.M.; Ghosh, T.K.; Kabir, A.H.; Abdelrahman, M.; Rahman Khan, M.A.; Mochida, K.; Tran, L.P. Potassium in plant physiological adaptation to abiotic stresses. Plant Physiol. Biochem. 2022, 186, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S. The elements of plant micronutrients. Plant Physiol. 2010, 154, 512–515. [Google Scholar] [CrossRef]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta 2000, 1465, 190–198. [Google Scholar] [CrossRef]
- Arguello, J.M.; Eren, E.; Gonzalez-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 2007, 20, 233–248. [Google Scholar] [CrossRef]
- Subbarayan, S.; Dhanasekaran, K. Effects of different sources and levels of manganese on dry matter production and nutrient uptake by cotton (Gossypium hirsutum L.) in salt affected soil. Life Sci. Arch. 2015, 1, 293–297. [Google Scholar]
- Siddiqui, M.H.; Al-Whaibi, M.H.; Sakran, A.M.; Basalah, M.O.; Ali, H.M. Effect of calcium and potassium on antioxidant system of Vicia faba L. Under cadmium stress. Int. J. Mol. Sci. 2012, 13, 6604–6619. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.N.; Moon, S.; Jung, K.H. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 2014, 171, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Ezaki, B.; Matsumoto, H. A gene encoding multidrug resistance (MDR)-like protein is induced by aluminum and inhibitors of calcium flux in wheat. Plant Cell Physiol. 2002, 43, 177–185. [Google Scholar] [CrossRef]
- Liu, C.; Liao, W. Potassium signaling in plant abiotic responses: Crosstalk with calcium and reactive oxygen species/reactive nitrogen species. Plant Physiol. Biochem. 2022, 173, 110–121. [Google Scholar] [CrossRef]
- Wang, X.; Hao, L.; Zhu, B.; Jiang, Z. Plant calcium signaling in response to potassium deficiency. Int. J. Mol. Sci. 2018, 19, 3456. [Google Scholar] [CrossRef]
- Roche, J.V.; Tornroth-Horsefield, S. Aquaporin Protein-protein interactions. Int. J. Mol. Sci. 2017, 18, 2255. [Google Scholar] [CrossRef]
- Sade, N.; Vinocur, B.J.; Diber, A.; Shatil, A.; Ronen, G.; Nissan, H.; Wallach, R.; Karchi, H.; Moshelion, M. Improving plant stress tolerance and yield production: Is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol. 2009, 181, 651–661. [Google Scholar] [CrossRef]
- Xu, C.; Wang, M.; Zhou, L.; Quan, T.; Xia, G. Heterologous expression of the wheat aquaporin gene TaTIP2;2 compromises the abiotic stress tolerance of Arabidopsis thaliana. PLoS ONE 2013, 8, e79618. [Google Scholar] [CrossRef]
- Bienert, G.P.; Moller, A.L.; Kristiansen, K.A.; Schulz, A.; Moller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef]
- Santa María, G.; Rubio, F.; Dubcovsky, J.; Rodriguez-Navarro, A. The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 1998, 9, 2281–2289. [Google Scholar] [CrossRef]
- Zhang, H.; Xiao, W.; Yu, W.; Jiang, Y.; Li, R. Halophytic Hordeum brevisubulatum HbHAK1 facilitates potassium retention and contributes to salt tolerance. Int. J. Mol. Sci. 2020, 21, 5292. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhao, Y.; Zhao, S.; Liu, H.; Wang, W.; Zhang, S.; Yang, X. Genome-wide identification and low tmperature responsive pattern of Actin Depolymerizing Factor (ADF) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 618984. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, K.; Liu, G.; Li, S.; Zhao, S.; Liu, X.; Yang, X.; Xiao, K. Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 15812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, H.; Zhang, Y.; Wang, Y.; Li, M.; Zhang, J.; Duan, L.; Zhang, M.; Li, Z. Increased abscisic acid levels in transgenic maize overexpressing AtLOS5 mediated root ion fluxes and leaf water status under salt stress. J. Exp. Bot. 2016, 67, 1339–1355. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Zhao, Y.; Yu, Y.; Sun, R.; Wang, W.; Zhang, S.; Yang, X. Proteomic Analysis of Roots Response to Potassium Deficiency and the Effect of TaHAK1-4A on K+ Uptake in Wheat. Int. J. Mol. Sci. 2022, 23, 13504. https://doi.org/10.3390/ijms232113504
Xu K, Zhao Y, Yu Y, Sun R, Wang W, Zhang S, Yang X. Proteomic Analysis of Roots Response to Potassium Deficiency and the Effect of TaHAK1-4A on K+ Uptake in Wheat. International Journal of Molecular Sciences. 2022; 23(21):13504. https://doi.org/10.3390/ijms232113504
Chicago/Turabian StyleXu, Ke, Yong Zhao, Yaxin Yu, Ruoxi Sun, Weiwei Wang, Shuhua Zhang, and Xueju Yang. 2022. "Proteomic Analysis of Roots Response to Potassium Deficiency and the Effect of TaHAK1-4A on K+ Uptake in Wheat" International Journal of Molecular Sciences 23, no. 21: 13504. https://doi.org/10.3390/ijms232113504
APA StyleXu, K., Zhao, Y., Yu, Y., Sun, R., Wang, W., Zhang, S., & Yang, X. (2022). Proteomic Analysis of Roots Response to Potassium Deficiency and the Effect of TaHAK1-4A on K+ Uptake in Wheat. International Journal of Molecular Sciences, 23(21), 13504. https://doi.org/10.3390/ijms232113504





