Molecular Mechanisms of Inhibition of Protein Amyloid Fibril Formation: Evidence and Perspectives Based on Kinetic Models
Abstract
1. Introduction
2. Kinetics and Thermodynamics of Amyloid Fibril Formation
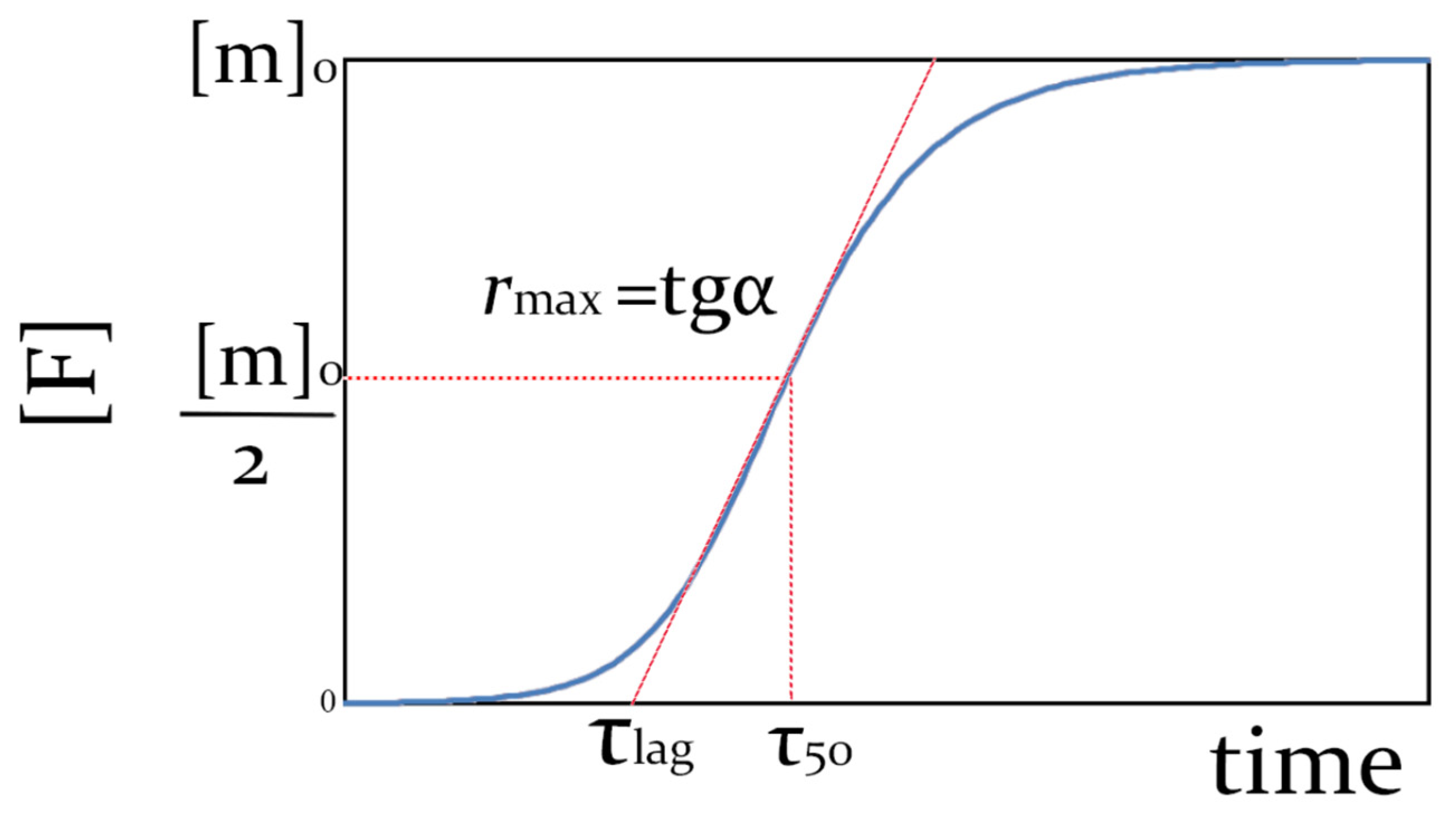
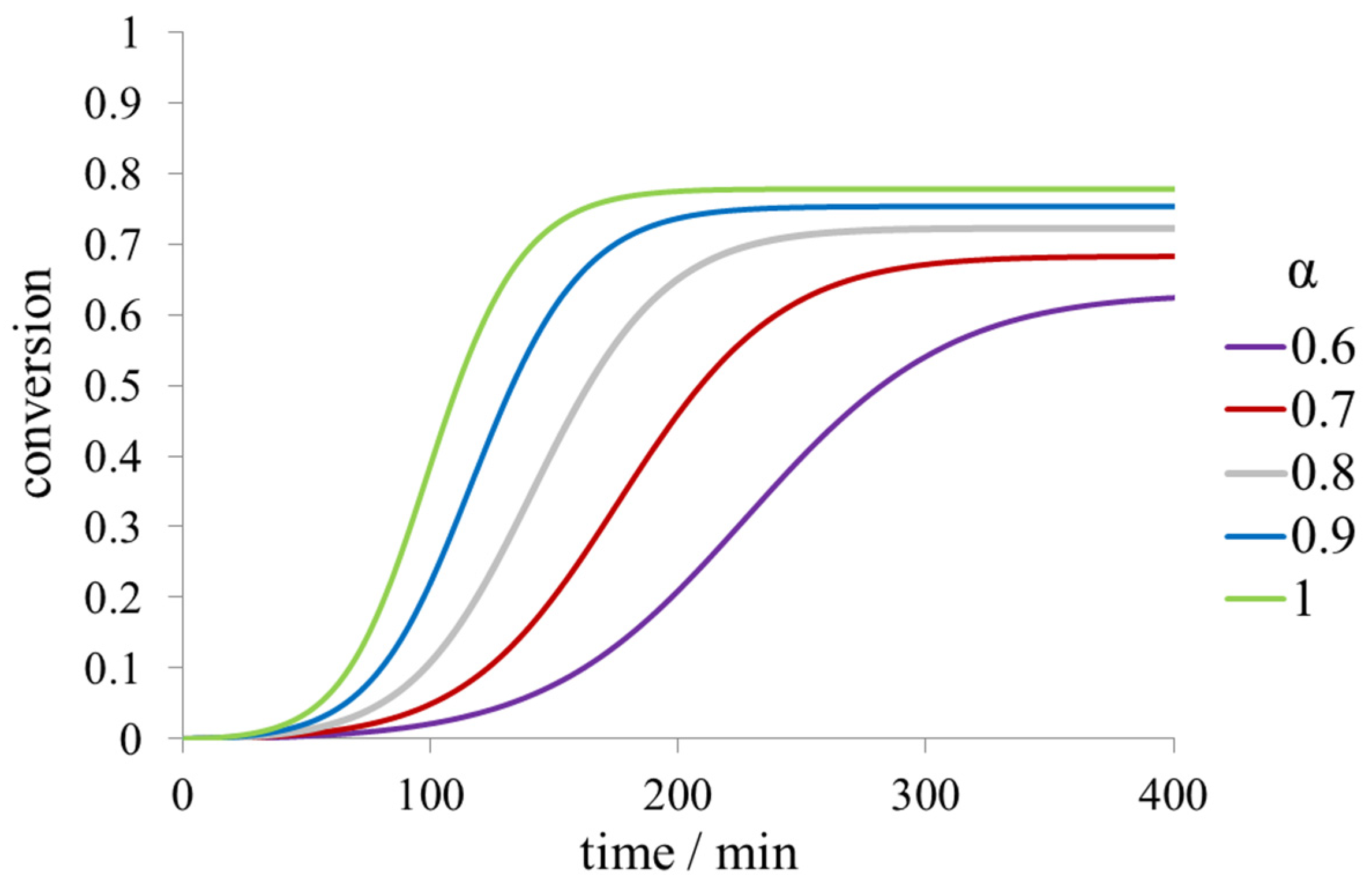
2.1. Kinetic Models of Fibril Formation
2.2. Fibril Formation from the Particular Form of Protein
2.3. Thermodynamic Equilibrium of Fibril Formation
2.4. Fibril Formation without a Lag Period
3. Effect of Inhibitor Binding on Amyloid Fibril Formation
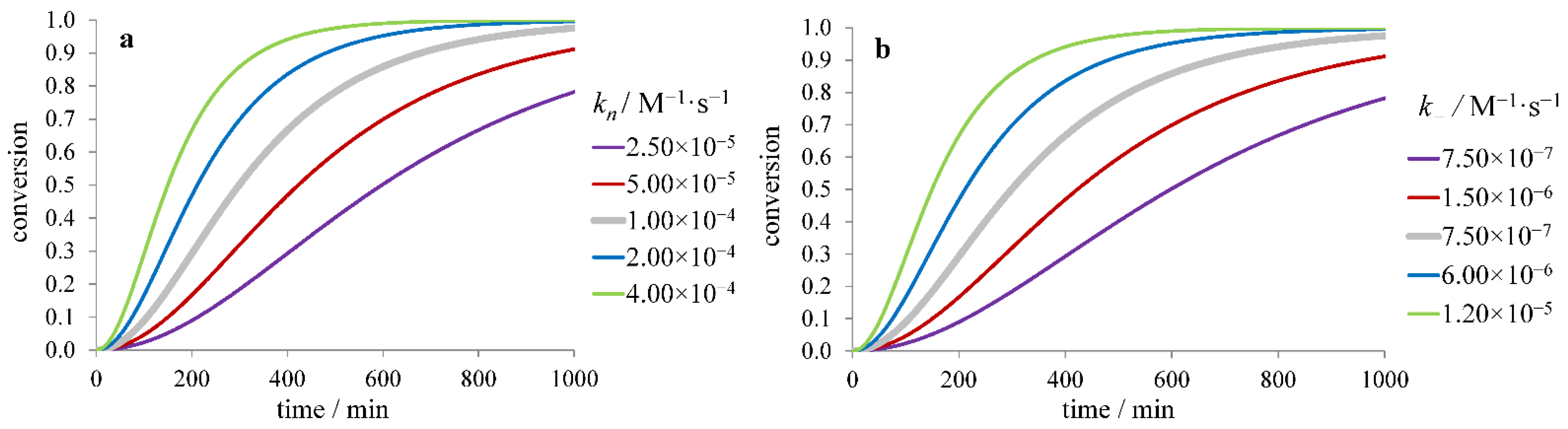
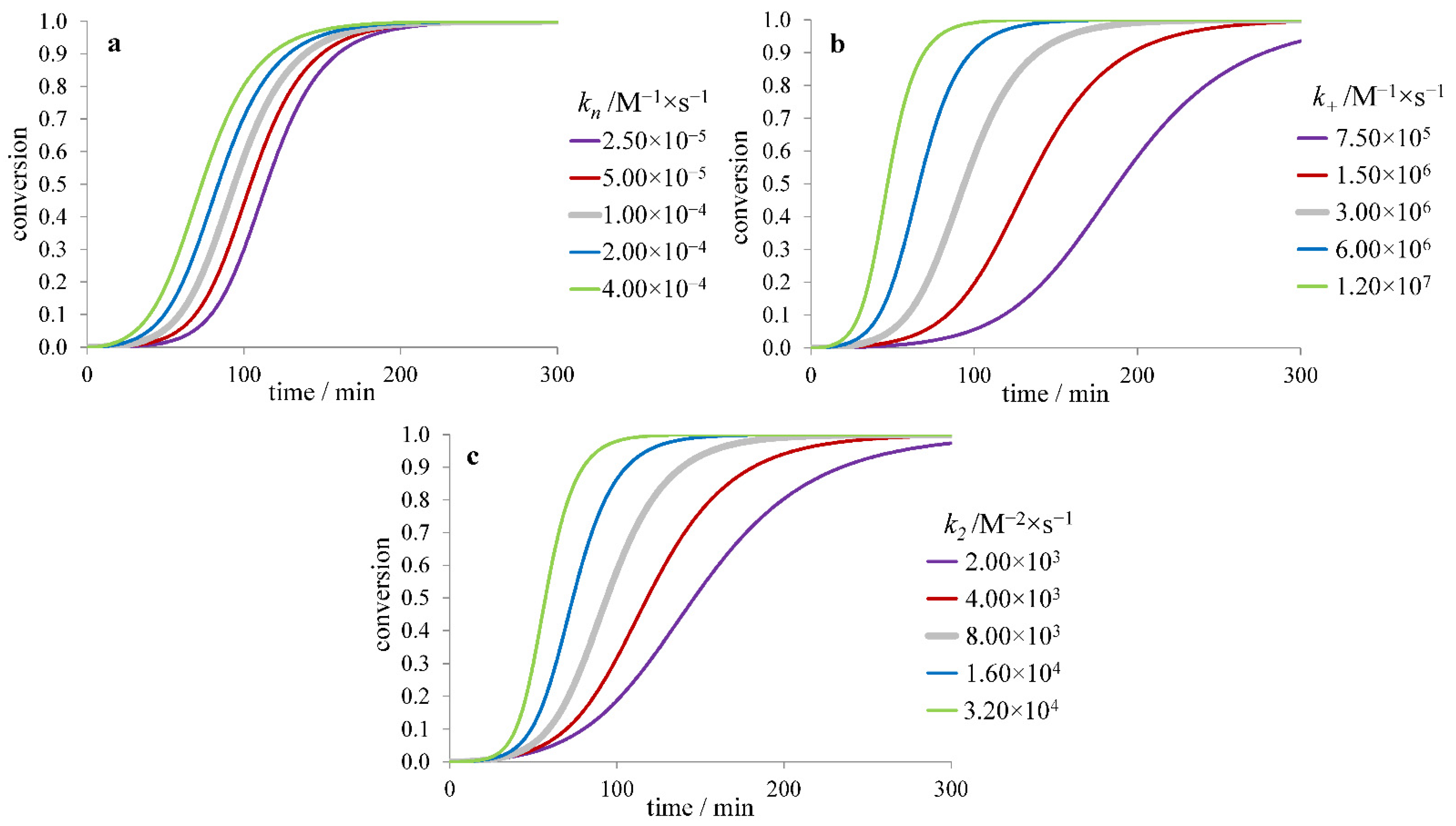
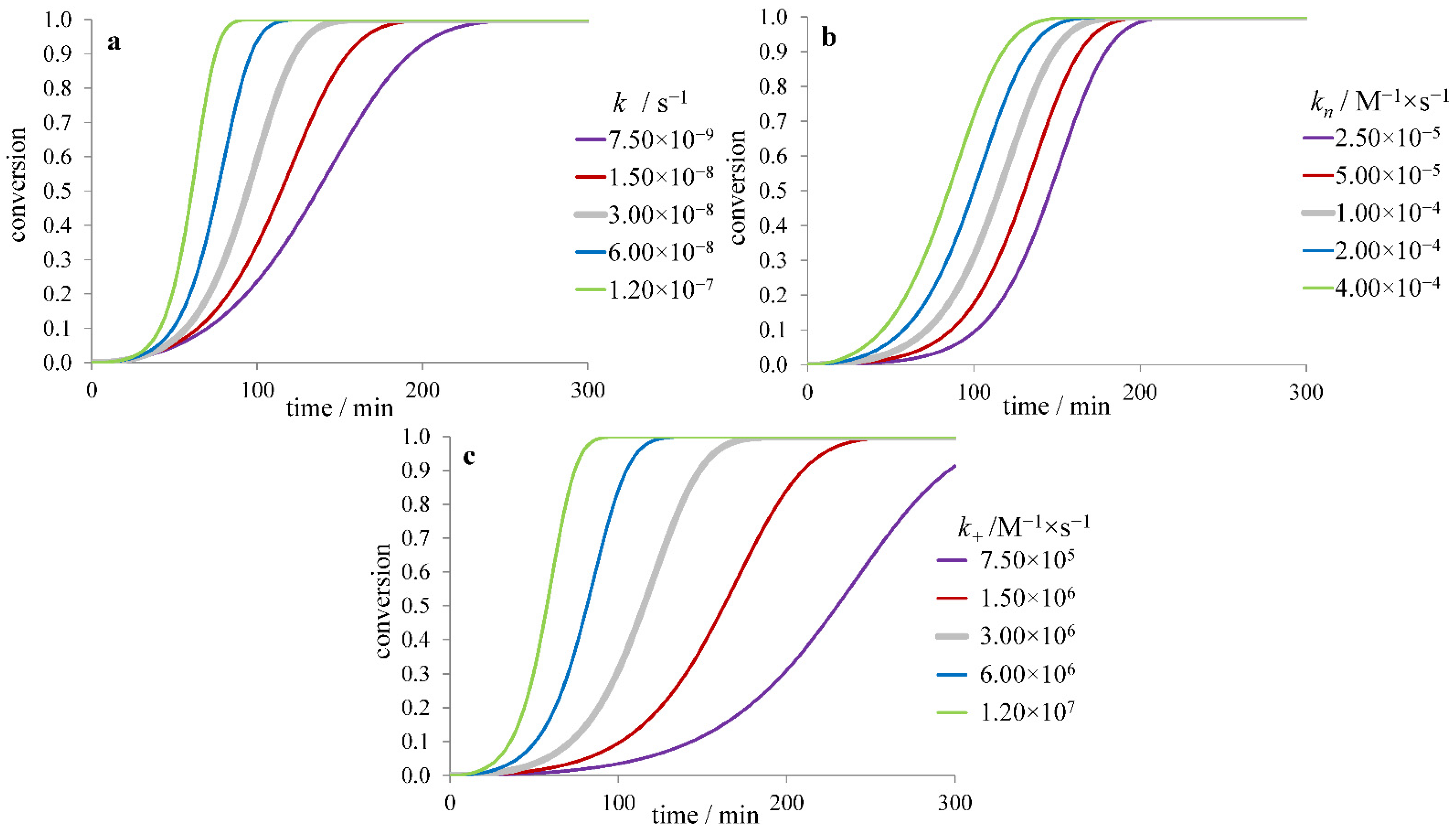
3.1. Inhibitors Influencing the Protein Monomer
3.1.1. The Effect of Inhibitor Binding to the Protein Monomer
3.1.2. The Effect of Protein Stabilizers and Denaturants
3.1.3. The Effect of Crowding Agents and Detergents
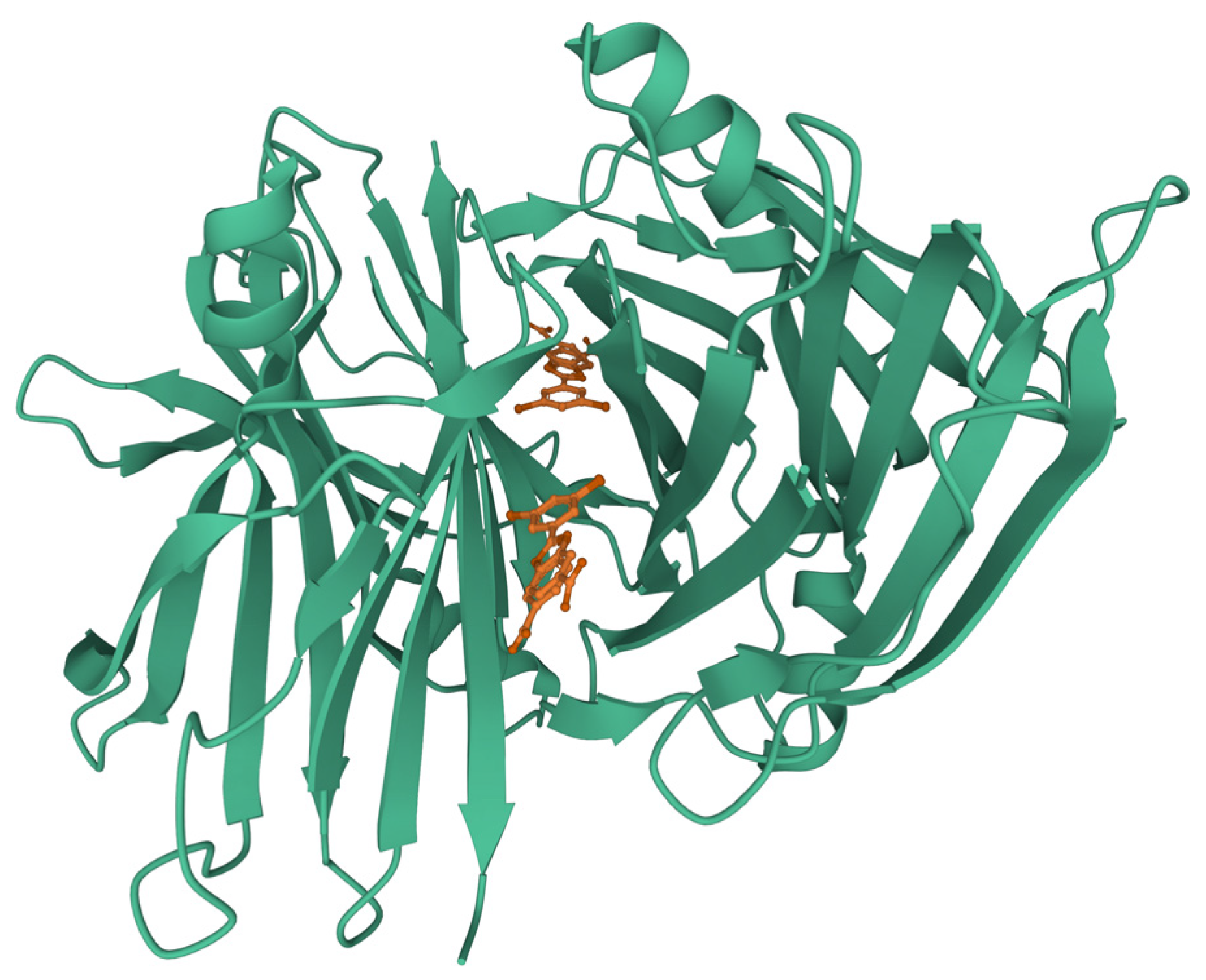
3.2. Inhibition of Fibril Growth by Binding to Aggregate Ends
3.3. Inhibition of Secondary Nucleation by Binding to the Fibril Surface
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wetzel, R. Amyloid. In Encyclopedia of Biological Chemistry; Elsevier: Amsterdam, The Netherlands, 2013; pp. 100–104. ISBN 978-0-12-378631-9. [Google Scholar]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid Nomenclature 2020: Update and Recommendations by the International Society of Amyloidosis (ISA) Nomenclature Committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef]
- Doig, A.J.; Derreumaux, P. Inhibition of Protein Aggregation and Amyloid Formation by Small Molecules. Curr. Opin. Struct. Biol. 2015, 30, 50–56. [Google Scholar] [CrossRef]
- Porat, Y.; Abramowitz, A.; Gazit, E. Inhibition of Amyloid Fibril Formation by Polyphenols: Structural Similarity and Aromatic Interactions as a Common Inhibition Mechanism. Chem. Biol. Drug Des. 2006, 67, 27–37. [Google Scholar] [CrossRef]
- Young, L.M.; Ashcroft, A.E.; Radford, S.E. Small Molecule Probes of Protein Aggregation. Curr. Opin. Chem. Biol. 2017, 39, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Chia, S.; Limbocker, R.; Mannini, B.; Ahn, M.; Perni, M.; Hansson, O.; Arosio, P.; Kumita, J.R.; Challa, P.K.; et al. Systematic Development of Small Molecules to Inhibit Specific Microscopic Steps of Aβ42 Aggregation in Alzheimer’s Disease. Proc. Natl. Acad. Sci. USA 2017, 114, E200–E208. [Google Scholar] [CrossRef]
- Tjernberg, L.O.; Näslund, J.; Lindqvist, F.; Johansson, J.; Karlström, A.R.; Thyberg, J.; Terenius, L.; Nordstedt, C. Arrest of -Amyloid Fibril Formation by a Pentapeptide Ligand. J. Biol. Chem. 1996, 271, 8545–8548. [Google Scholar] [CrossRef] [PubMed]
- Sievers, S.A.; Karanicolas, J.; Chang, H.W.; Zhao, A.; Jiang, L.; Zirafi, O.; Stevens, J.T.; Münch, J.; Baker, D.; Eisenberg, D. Structure-Based Design of Non-Natural Amino-Acid Inhibitors of Amyloid Fibril Formation. Nature 2011, 475, 96–100. [Google Scholar] [CrossRef]
- Frydman-Marom, A.; Rechter, M.; Shefler, I.; Bram, Y.; Shalev, D.E.; Gazit, E. Cognitive-Performance Recovery of Alzheimer’s Disease Model Mice by Modulation of Early Soluble Amyloidal Assemblies. Angew. Chem. Int. Ed. 2009, 48, 1981–1986. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.A.; Hu, C.J.; Griner, S.L.; Pan, H.; Bowler, J.T.; Abskharon, R.; Rosenberg, G.M.; Cheng, X.; Seidler, P.M.; Eisenberg, D.S. De Novo Designed Protein Inhibitors of Amyloid Aggregation and Seeding. Proc. Natl. Acad. Sci. USA 2022, 119, e2206240119. [Google Scholar] [CrossRef]
- Ikeda, K.; Suzuki, S.; Shigemitsu, Y.; Tenno, T.; Goda, N.; Oshima, A.; Hiroaki, H. Presence of Intrinsically Disordered Proteins Can Inhibit the Nucleation Phase of Amyloid Fibril Formation of Aβ(1–42) in Amino Acid Sequence Independent Manner. Sci. Rep. 2020, 10, 12334. [Google Scholar] [CrossRef]
- Cohen, S.I.A.; Arosio, P.; Presto, J.; Kurudenkandy, F.R.; Biverstål, H.; Dolfe, L.; Dunning, C.; Yang, X.; Frohm, B.; Vendruscolo, M.; et al. A Molecular Chaperone Breaks the Catalytic Cycle That Generates Toxic Aβ Oligomers. Nat. Struct. Mol. Biol. 2015, 22, 207–213. [Google Scholar] [CrossRef]
- Stanyon, H.F.; Viles, J.H. Human Serum Albumin Can Regulate Amyloid-β Peptide Fiber Growth in the Brain Interstitium. J. Biol. Chem. 2012, 287, 28163–28168. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, W.; Zhao, F.; Dong, Q.; Wang, Y.; Wei, W.; Jia, L.; Li, L.; Lu, F. Dual Effect of the Acidic Polysaccharose Ulvan on the Inhibition of Amyloid-β Protein Fibrillation and Disintegration of Mature Fibrils. ACS Appl. Mater. Interfaces 2020, 12, 41167–41176. [Google Scholar] [CrossRef]
- Liang, Y.; Ueno, M.; Zha, S.; Okimura, T.; Jiang, Z.; Yamaguchi, K.; Hatakeyama, T.; Oda, T. Sulfated Polysaccharide Ascophyllan Prevents Amyloid Fibril Formation of Human Insulin and Inhibits Amyloid-Induced Hemolysis and Cytotoxicity in PC12 Cells. Biosci. Biotechnol. Biochem. 2021, 85, 2281–2291. [Google Scholar] [CrossRef] [PubMed]
- Semenyuk, P.; Kurochkina, L.; Barinova, K.; Muronetz, V. Alpha-Synuclein Amyloid Aggregation Is Inhibited by Sulfated Aromatic Polymers and Pyridinium Polycation. Polymers 2020, 12, 517. [Google Scholar] [CrossRef]
- Holubová, M.; Štěpánek, P.; Hrubý, M. Polymer Materials as Promoters/Inhibitors of Amyloid Fibril Formation. Colloid Polym. Sci. 2021, 299, 343–362. [Google Scholar] [CrossRef]
- Yang, Z.; Ge, C.; Liu, J.; Chong, Y.; Gu, Z.; Jimenez-Cruz, C.A.; Chai, Z.; Zhou, R. Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides. Nanoscale 2015, 7, 18725–18737. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Lee, M. Fullerene Inhibits β-Amyloid Peptide Aggregation. Biochem. Biophys. Res. Commun. 2003, 303, 576–579. [Google Scholar] [CrossRef]
- Xie, L.; Luo, Y.; Lin, D.; Xi, W.; Yang, X.; Wei, G. The Molecular Mechanism of Fullerene-Inhibited Aggregation of Alzheimer’s β-Amyloid Peptide Fragment. Nanoscale 2014, 6, 9752–9762. [Google Scholar] [CrossRef]
- Brahmachari, S.; Paul, A.; Segal, D.; Gazit, E. Inhibition of Amyloid Oligomerization into Different Supramolecular Architectures by Small Molecules: Mechanistic Insights and Design Rules. Future Med. Chem. 2017, 9, 797–810. [Google Scholar] [CrossRef]
- Sharma, V.; Ghosh, K.S. Inhibition of Amyloid Fibrillation by Small Molecules and Nanomaterials: Strategic Development of Pharmaceuticals Against Amyloidosis. PPL 2019, 26, 315–323. [Google Scholar] [CrossRef]
- Rajan, R.; Ahmed, S.; Sharma, N.; Kumar, N.; Debas, A.; Matsumura, K. Review of the Current State of Protein Aggregation Inhibition from a Materials Chemistry Perspective: Special Focus on Polymeric Materials. Mater. Adv. 2021, 2, 1139–1176. [Google Scholar] [CrossRef]
- Mitra, A.; Sarkar, N. Sequence and Structure-Based Peptides as Potent Amyloid Inhibitors: A Review. Arch. Biochem. Biophys. 2020, 695, 108614. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.C.; Nam, E.; Lee, H.J.; Savelieff, M.G.; Lim, M.H. Towards an Understanding of Amyloid-β Oligomers: Characterization, Toxicity Mechanisms, and Inhibitors. Chem. Soc. Rev. 2017, 46, 310–323. [Google Scholar] [CrossRef]
- Boopathi, S.; Poma, A.B.; Garduño-Juárez, R. An Overview of Several Inhibitors for Alzheimer’s Disease: Characterization and Failure. IJMS 2021, 22, 10798. [Google Scholar] [CrossRef] [PubMed]
- Jokar, S.; Khazaei, S.; Behnammanesh, H.; Shamloo, A.; Erfani, M.; Beiki, D.; Bavi, O. Recent Advances in the Design and Applications of Amyloid-β Peptide Aggregation Inhibitors for Alzheimer’s Disease Therapy. Biophys. Rev. 2019, 11, 901–925. [Google Scholar] [CrossRef]
- Guo, X.; Liu, Z.; Zheng, Y.; Li, Y.; Li, L.; Liu, H.; Chen, Z.; Wu, L. Review on the Structures and Activities of Transthyretin Amyloidogenesis Inhibitors. DDDT 2020, 14, 1057–1081. [Google Scholar] [CrossRef]
- Mok, Y.-F.; Howlett, G.J.; Griffin, M.D.W. Sedimentation Velocity Analysis of the Size Distribution of Amyloid Oligomers and Fibrils. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 562, pp. 241–256. ISBN 978-0-12-802908-4. [Google Scholar]
- Jarrett, J.T.; Lansbury, P.T. Seeding “One-Dimensional Crystallization” of Amyloid: A Pathogenic Mechanism in Alzheimer’s Disease and Scrapie? Cell 1993, 73, 1055–1058. [Google Scholar] [CrossRef]
- Linse, S. Monomer-Dependent Secondary Nucleation in Amyloid Formation. Biophys. Rev. 2017, 9, 329–338. [Google Scholar] [CrossRef]
- Meisl, G.; Kirkegaard, J.B.; Arosio, P.; Michaels, T.C.T.; Vendruscolo, M.; Dobson, C.M.; Linse, S.; Knowles, T.P.J. Molecular Mechanisms of Protein Aggregation from Global Fitting of Kinetic Models. Nat. Protoc. 2016, 11, 252–272. [Google Scholar] [CrossRef] [PubMed]
- Limbocker, R.; Staats, R.; Chia, S.; Ruggeri, F.S.; Mannini, B.; Xu, C.K.; Perni, M.; Cascella, R.; Bigi, A.; Sasser, L.R.; et al. Squalamine and Its Derivatives Modulate the Aggregation of Amyloid-β and α-Synuclein and Suppress the Toxicity of Their Oligomers. Front. Neurosci. 2021, 15, 680026. [Google Scholar] [CrossRef]
- Irwin, R.; Faust, O.; Petrovic, I.; Wolf, S.G.; Hofmann, H.; Rosenzweig, R. Hsp40s Play Complementary Roles in the Prevention of Tau Amyloid Formation. eLife 2021, 10, e69601. [Google Scholar] [CrossRef]
- Zhong, X.; Kumar, R.; Wang, Y.; Biverstål, H.; Ingeborg Jegerschöld, C.; Koeck, J.B.P.; Johansson, J.; Abelein, A.; Chen, G. Amyloid Fibril Formation of Arctic Amyloid-β 1–42 Peptide Is Efficiently Inhibited by the BRICHOS Domain. ACS Chem. Biol. 2022, 17, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Belsare, K.D.; Wu, H.; Mondal, D.; Bond, A.; Castillo, E.; Jin, J.; Jo, H.; Roush, A.E.; Pilla, K.B.; Sali, A.; et al. Soluble TREM2 Inhibits Secondary Nucleation of Aβ Fibrillization and Enhances Cellular Uptake of Fibrillar Aβ. Proc. Natl. Acad. Sci. USA 2022, 119, e2114486119. [Google Scholar] [CrossRef]
- Linse, S.; Sormanni, P.; O’Connell, D.J. An Aggregation Inhibitor Specific to Oligomeric Intermediates of Aβ42 Derived from Phage Display Libraries of Stable, Small Proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2121966119. [Google Scholar] [CrossRef] [PubMed]
- Kalapothakis, J.M.D.; Morris, R.J.; Szavits-Nossan, J.; Eden, K.; Covill, S.; Tabor, S.; Gillam, J.; Barran, P.E.; Allen, R.J.; MacPhee, C.E. A Kinetic Study of Ovalbumin Fibril Formation: The Importance of Fragmentation and End-Joining. Biophys. J. 2015, 108, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, L.; Madurga, S.; Pukala, T.; Vilaseca, M.; López-Iglesias, C.; Robinson, C.V.; Giralt, E.; Carulla, N. Aβ40 and Aβ42 Amyloid Fibrils Exhibit Distinct Molecular Recycling Properties. J. Am. Chem. Soc. 2011, 133, 6505–6508. [Google Scholar] [CrossRef]
- Cohen, S.I.A.; Vendruscolo, M.; Welland, M.E.; Dobson, C.M.; Terentjev, E.M.; Knowles, T.P.J. Nucleated Polymerization with Secondary Pathways. I. Time Evolution of the Principal Moments. J. Chem. Phys. 2011, 135, 065105. [Google Scholar] [CrossRef]
- Giehm, L.; Otzen, D.E. Strategies to Increase the Reproducibility of Protein Fibrillization in Plate Reader Assays. Anal. Biochem. 2010, 400, 270–281. [Google Scholar] [CrossRef]
- Nicoud, L.; Lazzari, S.; Balderas Barragán, D.; Morbidelli, M. Fragmentation of Amyloid Fibrils Occurs in Preferential Positions Depending on the Environmental Conditions. J. Phys. Chem. B 2015, 119, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Huseby, C.J.; Bundschuh, R.; Kuret, J. The Role of Annealing and Fragmentation in Human Tau Aggregation Dynamics. J. Biol. Chem. 2019, 294, 4728–4737. [Google Scholar] [CrossRef]
- Powers, E.T.; Powers, D.L. Mechanisms of Protein Fibril Formation: Nucleated Polymerization with Competing Off-Pathway Aggregation. Biophys. J. 2008, 94, 379–391. [Google Scholar] [CrossRef]
- Hasecke, F.; Niyangoda, C.; Borjas, G.; Pan, J.; Matthews, G.; Muschol, M.; Hoyer, W. Protofibril–Fibril Interactions Inhibit Amyloid Fibril Assembly by Obstructing Secondary Nucleation. Angew. Chem. Int. Ed. 2021, 60, 3016–3021. [Google Scholar] [CrossRef]
- Uversky, V.N.; Fink, A.L. Conformational Constraints for Amyloid Fibrillation: The Importance of Being Unfolded. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2004, 1698, 131–153. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V. Amyloidogenesis of Natively Unfolded Proteins. CAR 2008, 5, 260–287. [Google Scholar] [CrossRef]
- Buell, A.K. Stability Matters, Too–The Thermodynamics of Amyloid Fibril Formation. Chem. Sci. 2022, 13, 10177–10192. [Google Scholar] [CrossRef]
- Hurshman, A.R.; White, J.T.; Powers, E.T.; Kelly, J.W. Transthyretin Aggregation under Partially Denaturing Conditions Is a Downhill Polymerization. Biochemistry 2004, 43, 7365–7381. [Google Scholar] [CrossRef] [PubMed]
- Faria, T.Q.; Almeida, Z.L.; Cruz, P.F.; Jesus, C.S.H.; Castanheira, P.; Brito, R.M.M. A Look into Amyloid Formation by Transthyretin: Aggregation Pathway and a Novel Kinetic Model. Phys. Chem. Chem. Phys. 2015, 17, 7255–7263. [Google Scholar] [CrossRef] [PubMed]
- Juárez, J.; Taboada, P.; Mosquera, V. Existence of Different Structural Intermediates on the Fibrillation Pathway of Human Serum Albumin. Biophys. J. 2009, 96, 2353–2370. [Google Scholar] [CrossRef]
- Holm, N.K.; Jespersen, S.K.; Thomassen, L.V.; Wolff, T.Y.; Sehgal, P.; Thomsen, L.A.; Christiansen, G.; Andersen, C.B.; Knudsen, A.D.; Otzen, D.E. Aggregation and Fibrillation of Bovine Serum Albumin. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2007, 1774, 1128–1138. [Google Scholar] [CrossRef]
- Thorn, D.C.; Ecroyd, H.; Sunde, M.; Poon, S.; Carver, J.A. Amyloid Fibril Formation by Bovine Milk α s2 -Casein Occurs under Physiological Conditions Yet Is Prevented by Its Natural Counterpart, αs1-Casein. Biochemistry 2008, 47, 3926–3936. [Google Scholar] [CrossRef] [PubMed]
- Thorn, D.C.; Meehan, S.; Sunde, M.; Rekas, A.; Gras, S.L.; MacPhee, C.E.; Dobson, C.M.; Wilson, M.R.; Carver, J.A. Amyloid Fibril Formation by Bovine Milk κ-Casein and Its Inhibition by the Molecular Chaperones αS-and β-Casein. Biochemistry 2005, 44, 17027–17036. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Taddei, N.; Baroni, F.; Capanni, C.; Stefani, M.; Ramponi, G.; Dobson, C.M. Kinetic Partitioning of Protein Folding and Aggregation. Nat. Struct. Biol. 2002, 9, 137–143. [Google Scholar] [CrossRef]
- Jain, S.; Udgaonkar, J.B. Evidence for Stepwise Formation of Amyloid Fibrils by the Mouse Prion Protein. J. Mol. Biol. 2008, 382, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Udgaonkar, J.B. Defining the Pathway of Worm-like Amyloid Fibril Formation by the Mouse Prion Protein by Delineation of the Productive and Unproductive Oligomerization Reactions. Biochemistry 2011, 50, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Rashno, F.; Khajeh, K.; Capitini, C.; Sajedi, R.H.; Shokri, M.M.; Chiti, F. Very Rapid Amyloid Fibril Formation by a Bacterial Lipase in the Absence of a Detectable Lag Phase. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2017, 1865, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Yagi, H.; Goto, Y.; Matsuzaki, K.; Hoshino, M. A Disulfide-Linked Amyloid-β Peptide Dimer Forms a Protofibril-like Oligomer through a Distinct Pathway from Amyloid Fibril Formation. Biochemistry 2010, 49, 7100–7107. [Google Scholar] [CrossRef]
- O’Nuallain, B.; Freir, D.B.; Nicoll, A.J.; Risse, E.; Ferguson, N.; Herron, C.E.; Collinge, J.; Walsh, D.M. Amyloid -Protein Dimers Rapidly Form Stable Synaptotoxic Protofibrils. J. Neurosci. 2010, 30, 14411–14419. [Google Scholar] [CrossRef]
- Calabrese, A.N.; Liu, Y.; Wang, T.; Musgrave, I.F.; Pukala, T.L.; Tabor, R.F.; Martin, L.L.; Carver, J.A.; Bowie, J.H. The Amyloid Fibril-Forming Properties of the Amphibian Antimicrobial Peptide Uperin 3.5. ChemBioChem 2016, 17, 239–246. [Google Scholar] [CrossRef]
- Ohhashi, Y.; Hasegawa, K.; Naiki, H.; Goto, Y. Optimum Amyloid Fibril Formation of a Peptide Fragment Suggests the Amyloidogenic Preference of Β2-Microglobulin under Physiological Conditions. J. Biol. Chem. 2004, 279, 10814–10821. [Google Scholar] [CrossRef]
- Rangachari, V.; Moore, B.D.; Reed, D.K.; Sonoda, L.K.; Bridges, A.W.; Conboy, E.; Hartigan, D.; Rosenberry, T.L. Amyloid-β(1−42) Rapidly Forms Protofibrils and Oligomers by Distinct Pathways in Low Concentrations of Sodium Dodecylsulfate. Biochemistry 2007, 46, 12451–12462. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.R.; Moss, M.A.; Reed, D.K.; Cratic-McDaniel, S.; Hoh, J.H.; Rosenberry, T.L. Amyloid-β Protofibrils Differ from Amyloid-β Aggregates Induced in Dilute Hexafluoroisopropanol in Stability and Morphology. J. Biol. Chem. 2005, 280, 2471–2480. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P.; Jones, S.; Serpell, L.C.; Sunde, M.; Radford, S.E. A Systematic Investigation into the Effect of Protein Destabilisation on Beta 2-Microglobulin Amyloid Formation. J. Mol. Biol. 2003, 330, 943–954. [Google Scholar] [CrossRef]
- Smith, D.P.; Woods, L.A.; Radford, S.E.; Ashcroft, A.E. Structure and Dynamics of Oligomeric Intermediates in Β2-Microglobulin Self-Assembly. Biophys. J. 2011, 101, 1238–1247. [Google Scholar] [CrossRef]
- Crespo, R.; Rocha, F.A.; Damas, A.M.; Martins, P.M. A Generic Crystallization-like Model That Describes the Kinetics of Amyloid Fibril Formation. J. Biol. Chem. 2012, 287, 30585–30594. [Google Scholar] [CrossRef]
- Khaibrakhmanova, D.; Nikiforova, A.; Li, Z.; Sedov, I. Effect of Ligands with Different Affinity on Albumin Fibril Formation. Int. J. Biol. Macromol. 2022, 204, 709–717. [Google Scholar] [CrossRef]
- Grüning, C.S.R.; Klinker, S.; Wolff, M.; Schneider, M.; Toksöz, K.; Klein, A.N.; Nagel-Steger, L.; Willbold, D.; Hoyer, W. The Off-Rate of Monomers Dissociating from Amyloid-β Protofibrils. J. Biol. Chem. 2013, 288, 37104–37111. [Google Scholar] [CrossRef]
- Heller, G.T.; Aprile, F.A.; Michaels, T.C.T.; Limbocker, R.; Perni, M.; Ruggeri, F.S.; Mannini, B.; Löhr, T.; Bonomi, M.; Camilloni, C.; et al. Small-Molecule Sequestration of Amyloid-β as a Drug Discovery Strategy for Alzheimer’s Disease. Sci. Adv. 2020, 6, eabb5924. [Google Scholar] [CrossRef]
- Sinha, S.; Lopes, D.H.J.; Du, Z.; Pang, E.S.; Shanmugam, A.; Lomakin, A.; Talbiersky, P.; Tennstaedt, A.; McDaniel, K.; Bakshi, R.; et al. Lysine-Specific Molecular Tweezers Are Broad-Spectrum Inhibitors of Assembly and Toxicity of Amyloid Proteins. J. Am. Chem. Soc. 2011, 133, 16958–16969. [Google Scholar] [CrossRef]
- Acharya, S.; Safaie, B.M.; Wongkongkathep, P.; Ivanova, M.I.; Attar, A.; Klärner, F.-G.; Schrader, T.; Loo, J.A.; Bitan, G.; Lapidus, L.J. Molecular Basis for Preventing α-Synuclein Aggregation by a Molecular Tweezer. J. Biol. Chem. 2014, 289, 10727–10737. [Google Scholar] [CrossRef]
- Hoyer, W.; Grönwall, C.; Jonsson, A.; Ståhl, S.; Härd, T. Stabilization of a β-Hairpin in Monomeric Alzheimer’s Amyloid-β Peptide Inhibits Amyloid Formation. Proc. Natl. Acad. Sci. USA 2008, 105, 5099–5104. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, H.; Härd, T.; Löfblom, J.; Ståhl, S. A Truncated and Dimeric Format of an Affibody Library on Bacteria Enables FACS-mediated Isolation of Amyloid-beta Aggregation Inhibitors with Subnanomolar Affinity. Biotechnol. J. 2015, 10, 1707–1718. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Gao, W.; Cui, Z.; Zhang, H.; Lu, F.; Liu, F. Ulvan Inhibits α-Synuclein Fibrillation and Disrupts the Mature Fibrils: In Vitro and in Vivo Studies. Int. J. Biol. Macromol. 2022, 211, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Valle-Delgado, J.J.; Alfonso-Prieto, M.; Groot, N.S.; Ventura, S.; Samitier, J.; Rovira, C.; Fernàndez-Busquets, X. Modulation of Aβ 42 Fìbrillogenesis by Glycosaminoglycan Structure. FASEB J. 2010, 24, 4250–4261. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Hou, W.; Sun, Y.; Gao, Z.; Zhu, S.; Jiang, Z. Chitosan Oligosaccharides Inhibit/Disaggregate Fibrils and Attenuate Amyloid β-Mediated Neurotoxicity. IJMS 2015, 16, 10526–10536. [Google Scholar] [CrossRef]
- Liu, H.; Ojha, B.; Morris, C.; Jiang, M.; Wojcikiewicz, E.P.; Rao, P.P.N.; Du, D. Positively Charged Chitosan and N -Trimethyl Chitosan Inhibit Aβ40 Fibrillogenesis. Biomacromolecules 2015, 16, 2363–2373. [Google Scholar] [CrossRef]
- Rekas, A.; Lo, V.; Gadd, G.E.; Cappai, R.; Yun, S.I. PAMAM Dendrimers as Potential Agents against Fibrillation of α -Synuclein, a Parkinson’s Disease-Related Protein. Macromol. Biosci. 2009, 9, 230–238. [Google Scholar] [CrossRef]
- Klajnert, B.; Cortijo-Arellano, M.; Cladera, J.; Bryszewska, M. Influence of Dendrimer’s Structure on Its Activity against Amyloid Fibril Formation. Biochem. Biophys. Res. Commun. 2006, 345, 21–28. [Google Scholar] [CrossRef]
- Gurzov, E.N.; Wang, B.; Pilkington, E.H.; Chen, P.; Kakinen, A.; Stanley, W.J.; Litwak, S.A.; Hanssen, E.G.; Davis, T.P.; Ding, F.; et al. Inhibition of HIAPP Amyloid Aggregation and Pancreatic β-Cell Toxicity by OH-Terminated PAMAM Dendrimer. Small 2016, 12, 1615–1626. [Google Scholar] [CrossRef]
- Klementieva, O.; Benseny-Cases, N.; Gella, A.; Appelhans, D.; Voit, B.; Cladera, J. Dense Shell Glycodendrimers as Potential Nontoxic Anti-Amyloidogenic Agents in Alzheimer’s Disease. Amyloid–Dendrimer Aggregates Morphology and Cell Toxicity. Biomacromolecules 2011, 12, 3903–3909. [Google Scholar] [CrossRef]
- Wang, M.; Kakinen, A.; Pilkington, E.H.; Davis, T.P.; Ke, P.C. Differential Effects of Silver and Iron Oxide Nanoparticles on IAPP Amyloid Aggregation. Biomater. Sci. 2017, 5, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Palmal, S.; Jana, N.R.; Jana, N.R. Inhibition of Amyloid Fibril Growth by Nanoparticle Coated with Histidine-Based Polymer. J. Phys. Chem. C 2014, 118, 21630–21638. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Akhavan, O.; Ghavami, M.; Rezaee, F.; Ghiasi, S.M.A. Graphene Oxide Strongly Inhibits Amyloid Beta Fibrillation. Nanoscale 2012, 4, 7322. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Luo, Y.; Derreumaux, P.; Wei, G. Induced β-Barrel Formation of the Alzheimer’s Aβ25–35 Oligomers on Carbon Nanotube Surfaces: Implication for Amyloid Fibril Inhibition. Biophys. J. 2009, 97, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, W.; Sang, J.; Jia, L.; Lu, F. Hydroxylated Single-Walled Carbon Nanotubes Inhibit Aβ 42 Fibrillogenesis, Disaggregate Mature Fibrils, and Protect against Aβ 42 -Induced Cytotoxicity. ACS Chem. Neurosci. 2019, 10, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Nedumpully-Govindan, P.; Gurzov, E.N.; Chen, P.; Pilkington, E.H.; Stanley, W.J.; Litwak, S.A.; Davis, T.P.; Ke, P.C.; Ding, F. Graphene Oxide Inhibits HIAPP Amyloid Fibrillation and Toxicity in Insulin-Producing NIT-1 Cells. Phys. Chem. Chem. Phys. 2016, 18, 94–100. [Google Scholar] [CrossRef]
- Ghaeidamini, M.; Bernson, D.; Sasanian, N.; Kumar, R.; Esbjörner, E.K. Graphene Oxide Sheets and Quantum Dots Inhibit α-Synuclein Amyloid Formation by Different Mechanisms. Nanoscale 2020, 12, 19450–19460. [Google Scholar] [CrossRef]
- Kim, D.; Yoo, J.M.; Hwang, H.; Lee, J.; Lee, S.H.; Yun, S.P.; Park, M.J.; Lee, M.; Choi, S.; Kwon, S.H.; et al. Graphene Quantum Dots Prevent α-Synucleinopathy in Parkinson’s Disease. Nat. Nanotech 2018, 13, 812–818. [Google Scholar] [CrossRef]
- Dumoulin, M.; Last, A.M.; Desmyter, A.; Decanniere, K.; Canet, D.; Larsson, G.; Spencer, A.; Archer, D.B.; Sasse, J.; Muyldermans, S.; et al. A Camelid Antibody Fragment Inhibits the Formation of Amyloid Fibrils by Human Lysozyme. Nature 2003, 424, 783–788. [Google Scholar] [CrossRef]
- Pagano, R.S.; López Medus, M.; Gómez, G.E.; Couto, P.M.; Labanda, M.S.; Landolfo, L.; D’Alessio, C.; Caramelo, J.J. Protein Fibrillation Lag Times During Kinetic Inhibition. Biophys. J. 2014, 107, 711–720. [Google Scholar] [CrossRef]
- Ashrafian, H.; Zadeh, E.H.; Tajbakhsh, M.; Majid, N.; Srivastava, G.N.; Khan, R.H. Discovery of a Tetracyclic Indole Alkaloid That Postpones Fibrillation of Hen Egg White Lysozyme Protein. Int. J. Biol. Macromol. 2021, 183, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F. Reduction of the Amyloidogenicity of a Protein by Specific Binding of Ligands to the Native Conformation. Protein Sci. 2001, 10, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Kamatari, Y.O.; Ono, F.; Shibata, H.; Fuse, T.; Elhelaly, A.E.; Fukuoka, M.; Kimura, T.; Hosokawa-Muto, J.; Ishikawa, T.; et al. A Designer Molecular Chaperone against Transmissible Spongiform Encephalopathy Slows Disease Progression in Mice and Macaques. Nat. Biomed. Eng. 2019, 3, 206–219. [Google Scholar] [CrossRef]
- Astolfi, A.; Spagnolli, G.; Biasini, E.; Barreca, M.L. The Compelling Demand for an Effective PrP C -Directed Therapy against Prion Diseases. ACS Med. Chem. Lett. 2020, 11, 2063–2067. [Google Scholar] [CrossRef] [PubMed]
- Brumshtein, B.; Esswein, S.R.; Salwinski, L.; Phillips, M.L.; Ly, A.T.; Cascio, D.; Sawaya, M.R.; Eisenberg, D.S. Inhibition by Small-Molecule Ligands of Formation of Amyloid Fibrils of an Immunoglobulin Light Chain Variable Domain. eLife 2015, 4, e10935. [Google Scholar] [CrossRef]
- Anand, B.G.; Prajapati, K.P.; Purohit, S.; Ansari, M.; Panigrahi, A.; Kaushik, B.; Behera, R.K.; Kar, K. Evidence of Anti-Amyloid Characteristics of Plumbagin via Inhibition of Protein Aggregation and Disassembly of Protein Fibrils. Biomacromolecules 2021, 22, 3692–3703. [Google Scholar] [CrossRef]
- Gomathi, K.; Haribabu, J.; Saranya, S.; Gayathri, D.; Jeyalakshmi, K.; Sendilvelan, S.; Echeverria, C.; Karvembu, R. Effective Inhibition of Insulin Amyloid Fibril Aggregation by Nickel(II) Complexes Containing Heterocyclic Thiosemicarbazones. Eur. Biophys. J. 2021, 50, 1069–1081. [Google Scholar] [CrossRef]
- Siddiqi, M.K.; Alam, P.; Malik, S.; Majid, N.; Chaturvedi, S.K.; Rajan, S.; Ajmal, M.R.; Khan, M.V.; Uversky, V.N.; Khan, R.H. Stabilizing Proteins to Prevent Conformational Changes Required for Amyloid Fibril Formation. J. Cell. Biochem. 2019, 120, 2642–2656. [Google Scholar] [CrossRef]
- Prasanthan, P.; Kishore, N. Unusual Human Serum Albumin Fibrillation Inhibition by Ketoprofen and Fenoprofen: Mechanistic Insights. J. Mol. Recognit. 2021, 34. [Google Scholar] [CrossRef]
- Nettleton, E.J.; Sunde, M.; Lai, Z.; Kelly, J.W.; Dobson, C.M.; Robinson, C.V. Protein Subunit Interactions and Structural Integrity of Amyloidogenic Transthyretins: Evidence from Electrospray Mass Spectrometry. J. Mol. Biol. 1998, 281, 553–564. [Google Scholar] [CrossRef]
- Adamski-Werner, S.L.; Palaninathan, S.K.; Sacchettini, J.C.; Kelly, J.W. Diflunisal Analogues Stabilize the Native State of Transthyretin. Potent Inhibition of Amyloidogenesis. J. Med. Chem. 2004, 47, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Raghu, P.; Reddy, G.B.; Sivakumar, B. Inhibition of Transthyretin Amyloid Fibril Formation by 2,4-Dinitrophenol through Tetramer Stabilization. Arch. Biochem. Biophys. 2002, 400, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Iakovleva, I.; Begum, A.; Brännström, K.; Wijsekera, A.; Nilsson, L.; Zhang, J.; Andersson, P.L.; Sauer-Eriksson, A.E.; Olofsson, A. Tetrabromobisphenol A Is an Efficient Stabilizer of the Transthyretin Tetramer. PLoS ONE 2016, 11, e0153529. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, L.; Tonali, N.; Nencetti, S.; Orlandini, E. Natural Compounds as Inhibitors of Transthyretin Amyloidosis and Neuroprotective Agents: Analysis of Structural Data for Future Drug Design. J. Enzym. Inhib. Med. Chem. 2020, 35, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Reum Han, A.; Hee Jeon, E.; Woo Kim, K.; Ki Lee, S.; Ohn, C.; Jean Park, S.; Sook Kang, N.; Koo, T.-S.; Bum Hong, K.; Choi, S. Synthesis and Biological Evaluation of Quinolone Derivatives as Transthyretin Amyloidogenesis Inhibitors and Fluorescence Sensors. Bioorg. Med. Chem. 2022, 53, 116550. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, F.; Kim, J.H.; Chiellini, G. Transthyretin Stabilization: An Emerging Strategy for the Treatment of Alzheimer’s Disease? IJMS 2020, 21, 8672. [Google Scholar] [CrossRef]
- Palaninathan, S.K.; Mohamedmohaideen, N.N.; Orlandini, E.; Ortore, G.; Nencetti, S.; Lapucci, A.; Rossello, A.; Freundlich, J.S.; Sacchettini, J.C. Novel Transthyretin Amyloid Fibril Formation Inhibitors: Synthesis, Biological Evaluation, and X-Ray Structural Analysis. PLoS ONE 2009, 4, e6290. [Google Scholar] [CrossRef]
- Baures, P.W.; Peterson, S.A.; Kelly, J.W. Discovering Transthyretin Amyloid Fibril Inhibitors by Limited Screening. Bioorg. Med. Chem. 1998, 6, 1389–1401. [Google Scholar] [CrossRef]
- Ciccone, L.; Nencetti, S.; Camodeca, C.; Ortore, G.; Cuffaro, D.; Socci, S.; Orlandini, E. Synthesis and Evaluation of Monoaryl Derivatives as Transthyretin Fibril Formation Inhibitors. Pharm. Chem. J. 2022, 56, 38–47. [Google Scholar] [CrossRef]
- Bulawa, C.E.; Connelly, S.; DeVit, M.; Wang, L.; Weigel, C.; Fleming, J.A.; Packman, J.; Powers, E.T.; Wiseman, R.L.; Foss, T.R.; et al. Tafamidis, a Potent and Selective Transthyretin Kinetic Stabilizer That Inhibits the Amyloid Cascade. Proc. Natl. Acad. Sci. USA 2012, 109, 9629–9634. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, Z.; Wang, Y.; Fei, Z.; Cao, J. Changing the Activities and Structures of Bovine Serum Albumin Bound to Graphene Oxide. Appl. Surf. Sci. 2018, 427, 1019–1029. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, Y.; Wang, Q.; Wu, X. Inhibition of Lysozyme Fibrillation by Functional Groups in Graphene Oxide Quantum Dots. Chem. Phys. Lett. 2022, 801, 139749. [Google Scholar] [CrossRef]
- Gregory, W.E.; Sharma, B.; Hu, L.; Raghavendra, A.J.; Podila, R. Interfacial Charge Transfer with Exfoliated Graphene Inhibits Fibril Formation in Lysozyme Amyloid. Biointerphases 2020, 15, 031010. [Google Scholar] [CrossRef] [PubMed]
- Griebenow, K.; Klibanov, A.M. On Protein Denaturation in Aqueous−Organic Mixtures but Not in Pure Organic Solvents. J. Am. Chem. Soc. 1996, 118, 11695–11700. [Google Scholar] [CrossRef]
- Magsumov, T.; Ziying, L.; Sedov, I. Comparative Study of the Protein Denaturing Ability of Different Organic Cosolvents. Int. J. Biol. Macromol. 2020, 160, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Jungwirth, P.; Cremer, P.S. Beyond Hofmeister. Nat. Chem 2014, 6, 261–263. [Google Scholar] [CrossRef]
- Butler, S.L.; Falke, J.J. Effects of Protein Stabilizing Agents on Thermal Backbone Motions: A Disulfide Trapping Study. Biochemistry 1996, 35, 10595–10600. [Google Scholar] [CrossRef]
- Ueda, T.; Nagata, M.; Imoto, T. Aggregation and Chemical Reaction in Hen Lysozyme Caused by Heating at PH 6 Are Depressed by Osmolytes, Sucrose and Trehalose. J. Biochem. 2001, 130, 491–496. [Google Scholar] [CrossRef]
- White, D.A.; Buell, A.K.; Knowles, T.P.J.; Welland, M.E.; Dobson, C.M. Protein Aggregation in Crowded Environments. J. Am. Chem. Soc. 2010, 132, 5170–5175. [Google Scholar] [CrossRef]
- Arora, A.; Ha, C.; Park, C.B. Inhibition of Insulin Amyloid Formation by Small Stress Molecules. FEBS Lett. 2004, 564, 121–125. [Google Scholar] [CrossRef]
- Rahamtullah; Ahmad, A.; Mishra, R. Polyol and Sugar Osmolytes Stabilize the Molten Globule State of α-Lactalbumin and Inhibit Amyloid Fibril Formation. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2022, 1870, 140853. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Pasha, J.M.; Deep, S. Effect of the Sugar and Polyol Additives on the Aggregation Kinetics of BSA in the Presence of N-Cetyl-N,N,N-Trimethyl Ammonium Bromide. J. Colloid Interface Sci. 2010, 350, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Abe, Y.; Ohkuri, T.; Mishima, T.; Monji, A.; Kanba, S.; Ueda, T. Mechanism for Retardation of Amyloid Fibril Formation by Sugars in Vλ6 Protein: Retardation of Amyloid by Sugar. Protein Sci. 2013, 22, 467–474. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Shu, W.; Devlin, G.L.; Meehan, S.; Auer, S.; Dobson, C.M.; Welland, M.E. Kinetics and Thermodynamics of Amyloid Formation from Direct Measurements of Fluctuations in Fibril Mass. Proc. Natl. Acad. Sci. USA 2007, 104, 10016–10021. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.; Khurana, R.; Coats, A.; Frokjaer, S.; Brange, J.; Vyas, S.; Uversky, V.N.; Fink, A.L. Effect of Environmental Factors on the Kinetics of Insulin Fibril Formation: Elucidation of the Molecular Mechanism. Biochemistry 2001, 40, 6036–6046. [Google Scholar] [CrossRef] [PubMed]
- Macchi, F.; Eisenkolb, M.; Kiefer, H.; Otzen, D.E. The Effect of Osmolytes on Protein Fibrillation. IJMS 2012, 13, 3801–3819. [Google Scholar] [CrossRef]
- Fung, J.; Darabie, A.A.; McLaurin, J. Contribution of Simple Saccharides to the Stabilization of Amyloid Structure. Biochem. Biophys. Res. Commun. 2005, 328, 1067–1072. [Google Scholar] [CrossRef]
- Liu, R.; Barkhordarian, H.; Emadi, S.; Park, C.; Sierks, M. Trehalose Differentially Inhibits Aggregation and Neurotoxicity of Beta-Amyloid 40 and 42. Neurobiol. Dis. 2005, 20, 74–81. [Google Scholar] [CrossRef]
- Uversky, V.N.; Li, J.; Fink, A.L. Trimethylamine- N -Oxide-Induced Folding of α-Synuclein. FEBS Lett. 2001, 509, 31–35. [Google Scholar] [CrossRef]
- Yang, D.-S.; Yip, C.M.; Huang, T.H.J.; Chakrabartty, A.; Fraser, P.E. Manipulating the Amyloid-β Aggregation Pathway with Chemical Chaperones. J. Biol. Chem. 1999, 274, 32970–32974. [Google Scholar] [CrossRef]
- Scaramozzino, F.; Peterson, D.W.; Farmer, P.; Gerig, J.T.; Graves, D.J.; Lew, J. TMAO Promotes Fibrillization and Microtubule Assembly Activity in the C-Terminal Repeat Region of Tau. Biochemistry 2006, 45, 3684–3691. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Muresan, A.; Lee, K.Y.C.; Murphy, R.M. Urea Modulation of β-Amyloid Fibril Growth: Experimental Studies and Kinetic Models. Protein Sci. 2008, 13, 2888–2898. [Google Scholar] [CrossRef] [PubMed]
- Weiffert, T.; Meisl, G.; Curk, S.; Cukalevski, R.; Šarić, A.; Knowles, T.P.J.; Linse, S. Influence of Denaturants on Amyloid Β42 Aggregation Kinetics. Front. Neurosci. 2022, 16, 943355. [Google Scholar] [CrossRef]
- Tiiman, A.; Krishtal, J.; Palumaa, P.; Tõugu, V. In Vitro Fibrillization of Alzheimer’s Amyloid-β Peptide (1-42). AIP Adv. 2015, 5, 092401. [Google Scholar] [CrossRef]
- van Gils, J.H.M.; van Dijk, E.; Peduzzo, A.; Hofmann, A.; Vettore, N.; Schützmann, M.P.; Groth, G.; Mouhib, H.; Otzen, D.E.; Buell, A.K.; et al. The Hydrophobic Effect Characterises the Thermodynamic Signature of Amyloid Fibril Growth. PLoS Comput. Biol. 2020, 16, e1007767. [Google Scholar] [CrossRef]
- Levine, Z.A.; Larini, L.; LaPointe, N.E.; Feinstein, S.C.; Shea, J.-E. Regulation and Aggregation of Intrinsically Disordered Peptides. Proc. Natl. Acad. Sci. USA 2015, 112, 2758–2763. [Google Scholar] [CrossRef] [PubMed]
- Fezoui, Y.; Teplow, D.B. Kinetic Studies of Amyloid β-Protein Fibril Assembly. J. Biol. Chem. 2002, 277, 36948–36954. [Google Scholar] [CrossRef]
- Munishkina, L.A.; Phelan, C.; Uversky, V.N.; Fink, A.L. Conformational Behavior and Aggregation of α-Synuclein in Organic Solvents: Modeling the Effects of Membranes. Biochemistry 2003, 42, 2720–2730. [Google Scholar] [CrossRef]
- Anderson, V.L.; Webb, W.W.; Eliezer, D. Interplay between Desolvation and Secondary Structure in Mediating Cosolvent and Temperature Induced Alpha-Synuclein Aggregation. Phys. Biol. 2012, 9, 056005. [Google Scholar] [CrossRef]
- Chen, S. Solubilization and Disaggregation of Polyglutamine Peptides. Protein Sci. 2001, 10, 887–891. [Google Scholar] [CrossRef]
- Zagorski, M.G.; Yang, J.; Shao, H.; Ma, K.; Zeng, H.; Hong, A. [13] Methodological and Chemical Factors Affecting Amyloid β Peptide Amyloidogenicity. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 309, pp. 189–204. ISBN 978-0-12-182210-1. [Google Scholar]
- Marinelli, P.; Castillo, V.; Ventura, S. Trifluoroethanol Modulates Amyloid Formation by the All α-Helical URN1 FF Domain. IJMS 2013, 14, 17830–17844. [Google Scholar] [CrossRef] [PubMed]
- Vernaglia, B.A.; Huang, J.; Clark, E.D. Guanidine Hydrochloride Can Induce Amyloid Fibril Formation from Hen Egg-White Lysozyme. Biomacromolecules 2004, 5, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.J.; Knowles, T.P.J.; Tartaglia, G.G.; Fitzpatrick, A.W.; Devlin, G.L.; Shammas, S.L.; Waudby, C.A.; Mossuto, M.F.; Meehan, S.; Gras, S.L.; et al. Metastability of Native Proteins and the Phenomenon of Amyloid Formation. J. Am. Chem. Soc. 2011, 133, 14160–14163. [Google Scholar] [CrossRef] [PubMed]
- Hamada, D.; Dobson, C.M. A Kinetic Study of β-Lactoglobulin Amyloid Fibril Formation Promoted by Urea. Protein Sci. 2009, 11, 2417–2426. [Google Scholar] [CrossRef] [PubMed]
- Kayser, J.J.; Arnold, P.; Steffen-Heins, A.; Schwarz, K.; Keppler, J.K. Functional Ethanol-Induced Fibrils: Influence of Solvents and Temperature on Amyloid-like Aggregation of Beta-Lactoglobulin. J. Food Eng. 2020, 270, 109764. [Google Scholar] [CrossRef]
- Chiti, F.; Webster, P.; Taddei, N.; Clark, A.; Stefani, M.; Ramponi, G.; Dobson, C.M. Designing Conditions for in Vitro Formation of Amyloid Protofilaments and Fibrils. Proc. Natl. Acad. Sci. USA 1999, 96, 3590–3594. [Google Scholar] [CrossRef]
- Žerovnik, E.; Škarabot, M.; Škerget, K.; Giannini, S.; Žerovnik, E.; Škarabot, M.; Škerget, K.; Giannini, S.; Stoka, V.; Jenko-Kokalj, S.; et al. Amyloid Fibril Formation by Human Stefin B: Influence of PH and TFE on Fibril Growth and Morphology. Amyloid 2007, 14, 237–247. [Google Scholar] [CrossRef]
- Muta, H.; Lee, Y.-H.; Kardos, J.; Lin, Y.; Yagi, H.; Goto, Y. Supersaturation-Limited Amyloid Fibrillation of Insulin Revealed by Ultrasonication. J. Biol. Chem. 2014, 289, 18228–18238. [Google Scholar] [CrossRef]
- Bernson, D.; Mecinovic, A.; Abed, M.T.; Limé, F.; Jageland, P.; Palmlöf, M.; Esbjörner, E.K. Amyloid Formation of Bovine Insulin Is Retarded in Moderately Acidic PH and by Addition of Short-Chain Alcohols. Eur. Biophys. J. 2020, 49, 145–153. [Google Scholar] [CrossRef]
- Dzwolak, W.; Grudzielanek, S.; Smirnovas, V.; Ravindra, R.; Nicolini, C.; Jansen, R.; Loksztejn, A.; Porowski, S.; Winter, R. Ethanol-Perturbed Amyloidogenic Self-Assembly of Insulin: Looking for Origins of Amyloid Strains. Biochemistry 2005, 44, 8948–8958. [Google Scholar] [CrossRef]
- Ahmad, A.; Millett, I.S.; Doniach, S.; Uversky, V.N.; Fink, A.L. Partially Folded Intermediates in Insulin Fibrillation. Biochemistry 2003, 42, 11404–11416. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.; Frokjaer, S.; Brange, J.; Uversky, V.N.; Fink, A.L. Probing the Mechanism of Insulin Fibril Formation with Insulin Mutants. Biochemistry 2001, 40, 8397–8409. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Mondal, S.; Bagchi, B. Effect of Ethanol on Insulin Dimer Dissociation. J. Chem. Phys. 2019, 150, 084902. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Xie, J.; Wei, L.; Li, W. Macromolecular Crowding Modulates the Kinetics and Morphology of Amyloid Self-Assembly by β-Lactoglobulin. Int. J. Biol. Macromol. 2013, 53, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Holubová, M.; Lobaz, V.; Loukotová, L.; Rabyk, M.; Hromádková, J.; Trhlíková, O.; Pechrová, Z.; Groborz, O.; Štěpánek, P.; Hrubý, M. Does Polysaccharide Glycogen Behave as a Promoter of Amyloid Fibril Formation at Physiologically Relevant Concentrations? Soft. Matter. 2021, 17, 1628–1641. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.F.; Bird, S.; Shaw, M.; Jean, L.; Vaux, D.J. Combined Effects of Agitation, Macromolecular Crowding, and Interfaces on Amyloidogenesis. J. Biol. Chem. 2012, 287, 38006–38019. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Cooper, M.E.; Bower, K.S.; Li, J.; Fink, A.L. Accelerated α-Synuclein Fibrillation in Crowded Milieu. FEBS Lett. 2002, 515, 99–103. [Google Scholar] [CrossRef]
- Horvath, I.; Kumar, R.; Wittung-Stafshede, P. Macromolecular Crowding Modulates α-Synuclein Amyloid Fiber Growth. Biophys. J. 2021, 120, 3374–3381. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Fan, J.-B.; Zhou, Z.; Zhou, B.-R.; Meng, S.-R.; Hu, J.-Y.; Chen, J.; Liang, Y. The Contrasting Effect of Macromolecular Crowding on Amyloid Fibril Formation. PLoS ONE 2012, 7, e36288. [Google Scholar] [CrossRef]
- Ellis, R.J.; Minton, A.P. Protein Aggregation in Crowded Environments. Biol. Chem. 2006, 387, 485–497. [Google Scholar] [CrossRef]
- Munishkina, L.A.; Ahmad, A.; Fink, A.L.; Uversky, V.N. Guiding Protein Aggregation with Macromolecular Crowding. Biochemistry 2008, 47, 8993–9006. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Jao, S.; Ma, K.; Zagorski, M.G. Solution Structures of Micelle-Bound Amyloid β-(1-40) and β-(1-42) Peptides of Alzheimer’s Disease 1 1Edited by P. E. Wright. J. Mol. Biol. 1999, 285, 755–773. [Google Scholar] [CrossRef] [PubMed]
- Shabestari, M.H.; Meeuwenoord, N.J.; Filippov, D.V.; Huber, M. Interaction of the Amyloid β Peptide with Sodium Dodecyl Sulfate as a Membrane-Mimicking Detergent. J. Biol. Phys. 2016, 42, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Hasegawa, K.; Matsuzaki, K.; Naiki, H.; Yanagisawa, K. Environment- and Mutation-Dependent Aggregation Behavior of Alzheimer Amyloid β-Protein: Aβ Aggregation in Membrane-Mimicking Environment. J. Neurochem. 2004, 90, 62–69. [Google Scholar] [CrossRef]
- Rangachari, V.; Reed, D.K.; Moore, B.D.; Rosenberry, T.L. Secondary Structure and Interfacial Aggregation of Amyloid-β(1−40) on Sodium Dodecyl Sulfate Micelles. Biochemistry 2006, 45, 8639–8648. [Google Scholar] [CrossRef]
- Khan, J.M.; Malik, A.; Ahmed, A.; Alghamdi, O.H.A.; Ahmed, M. SDS Induces Cross Beta-Sheet Amyloid as Well as Alpha-Helical Structure in Conconavalin A. J. Mol. Liq. 2020, 319, 114154. [Google Scholar] [CrossRef]
- Pertinhez, T.A.; Bouchard, M.; Smith, R.A.G.; Dobson, C.M.; Smith, L.J. Stimulation and Inhibition of Fibril Formation by a Peptide in the Presence of Different Concentrations of SDS. FEBS Lett. 2002, 529, 193–197. [Google Scholar] [CrossRef]
- Leffers, K.-W.; Wille, H.; Stöhr, J.; Junger, E.; Prusiner, S.B.; Riesner, D. Assembly of Natural and Recombinant Prion Protein into Fibrils. Biol. Chem. 2005, 386, 569–580. [Google Scholar] [CrossRef]
- Xiong, L.-W.; Raymond, L.D.; Hayes, S.F.; Raymond, G.J.; Caughey, B. Conformational Change, Aggregation and Fibril Formation Induced by Detergent Treatments of Cellular Prion Protein: Detergent-Induced Changes in PrP. J. Neurochem. 2008, 79, 669–678. [Google Scholar] [CrossRef]
- Ryan, T.M.; Howlett, G.J.; Bailey, M.F. Fluorescence Detection of a Lipid-Induced Tetrameric Intermediate in Amyloid Fibril Formation by Apolipoprotein C-II. J. Biol. Chem. 2008, 283, 35118–35128. [Google Scholar] [CrossRef]
- Ghanta, J.; Shen, C.-L.; Kiessling, L.L.; Murphy, R.M. A Strategy for Designing Inhibitors of β-Amyloid Toxicity. J. Biol. Chem. 1996, 271, 29525–29528. [Google Scholar] [CrossRef] [PubMed]
- Chafekar, S.M.; Malda, H.; Merkx, M.; Meijer, E.W.; Viertl, D.; Lashuel, H.A.; Baas, F.; Scheper, W. Branched KLVFF Tetramers Strongly Potentiate Inhibition of β-Amyloid Aggregation. ChemBioChem 2007, 8, 1857–1864. [Google Scholar] [CrossRef]
- Zhang, G.; Leibowitz, M.J.; Sinko, P.J.; Stein, S. Multiple-Peptide Conjugates for Binding β-Amyloid Plaques of Alzheimer’s Disease. Bioconj. Chem. 2003, 14, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Formaggio, F.; Bettio, A.; Moretto, V.; Crisma, M.; Toniolo, C.; Broxterman, Q.B. Disruption of the ?-Sheet Structure of a Protected Pentapeptide, Related to the ?-Amyloid Sequence 17-21, Induced by a Single, Helicogenic C?-Tetrasubstituted ?-Amino Acid. J. Pept. Sci. 2003, 9, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Etienne, M.A.; Aucoin, J.P.; Fu, Y.; McCarley, R.L.; Hammer, R.P. Stoichiometric Inhibition of Amyloid β-Protein Aggregation with Peptides Containing Alternating α,α-Disubstituted Amino Acids. J. Am. Chem. Soc. 2006, 128, 3522–3523. [Google Scholar] [CrossRef]
- Soto, C.; Kindy, M.S.; Baumann, M.; Frangione, B. Inhibition of Alzheimer’s Amyloidosis by Peptides That Prevent β-Sheet Conformation. Biochem. Biophys. Res. Commun. 1996, 226, 672–680. [Google Scholar] [CrossRef]
- Jagota, S.; Rajadas, J. Synthesis of D-Amino Acid Peptides and Their Effect on Beta-Amyloid Aggregation and Toxicity in Transgenic Caenorhabditis Elegans. Med. Chem. Res. 2013, 22, 3991–4000. [Google Scholar] [CrossRef]
- Horsley, J.R.; Jovcevski, B.; Wegener, K.L.; Yu, J.; Pukala, T.L.; Abell, A.D. Rationally Designed Peptide-Based Inhibitor of Aβ42 Fibril Formation and Toxicity: A Potential Therapeutic Strategy for Alzheimer’s Disease. Biochem. J. 2020, 477, 2039–2054. [Google Scholar] [CrossRef]
- Adessi, C.; Frossard, M.-J.; Boissard, C.; Fraga, S.; Bieler, S.; Ruckle, T.; Vilbois, F.; Robinson, S.M.; Mutter, M.; Banks, W.A.; et al. Pharmacological Profiles of Peptide Drug Candidates for the Treatment of Alzheimer’s Disease. J. Biol. Chem. 2003, 278, 13905–13911. [Google Scholar] [CrossRef]
- Gordon, D.J.; Sciarretta, K.L.; Meredith, S.C. Inhibition of β-Amyloid(40) Fibrillogenesis and Disassembly of β-Amyloid(40) Fibrils by Short β-Amyloid Congeners Containing N -Methyl Amino Acids at Alternate Residues. Biochemistry 2001, 40, 8237–8245. [Google Scholar] [CrossRef]
- Taylor, M.; Moore, S.; Mayes, J.; Parkin, E.; Beeg, M.; Canovi, M.; Gobbi, M.; Mann, D.M.A.; Allsop, D. Development of a Proteolytically Stable Retro-Inverso Peptide Inhibitor of β-Amyloid Oligomerization as a Potential Novel Treatment for Alzheimer’s Disease. Biochemistry 2010, 49, 3261–3272. [Google Scholar] [CrossRef] [PubMed]
- Parthsarathy, V.; McClean, P.L.; Hölscher, C.; Taylor, M.; Tinker, C.; Jones, G.; Kolosov, O.; Salvati, E.; Gregori, M.; Masserini, M.; et al. A Novel Retro-Inverso Peptide Inhibitor Reduces Amyloid Deposition, Oxidation and Inflammation and Stimulates Neurogenesis in the APPswe/PS1ΔE9 Mouse Model of Alzheimer’s Disease. PLoS ONE 2013, 8, e54769. [Google Scholar] [CrossRef]
- Goyal, D.; Shuaib, S.; Mann, S.; Goyal, B. Rationally Designed Peptides and Peptidomimetics as Inhibitors of Amyloid-β (Aβ) Aggregation: Potential Therapeutics of Alzheimer’s Disease. ACS Comb. Sci. 2017, 19, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Kaffy, J.; Brinet, D.; Soulier, J.-L.; Correia, I.; Tonali, N.; Fera, K.F.; Iacone, Y.; Hoffmann, A.R.F.; Khemtémourian, L.; Crousse, B.; et al. Designed Glycopeptidomimetics Disrupt Protein–Protein Interactions Mediating Amyloid β-Peptide Aggregation and Restore Neuroblastoma Cell Viability. J. Med. Chem. 2016, 59, 2025–2040. [Google Scholar] [CrossRef] [PubMed]
- Shvadchak, V.V.; Afitska, K.; Yushchenko, D.A. Inhibition of α-Synuclein Amyloid Fibril Elongation by Blocking Fibril Ends. Angew. Chem. Int. Ed. 2018, 57, 5690–5694. [Google Scholar] [CrossRef]
- Agerschou, E.D.; Borgmann, V.; Wördehoff, M.M.; Hoyer, W. Inhibitor and Substrate Cooperate to Inhibit Amyloid Fibril Elongation of α-Synuclein. Chem. Sci. 2020, 11, 11331–11337. [Google Scholar] [CrossRef]
- Bove-Fenderson, E.; Urano, R.; Straub, J.E.; Harris, D.A. Cellular Prion Protein Targets Amyloid-β Fibril Ends via Its C-Terminal Domain to Prevent Elongation. J. Biol. Chem. 2017, 292, 16858–16871. [Google Scholar] [CrossRef]
- Spatharas, P.M.; Nasi, G.I.; Tsiolaki, P.L.; Theodoropoulou, M.K.; Papandreou, N.C.; Hoenger, A.; Trougakos, I.P.; Iconomidou, V.A. Clusterin in Alzheimer’s Disease: An Amyloidogenic Inhibitor of Amyloid Formation? Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2022, 1868, 166384. [Google Scholar] [CrossRef]
- Bett, C.K.; Serem, W.K.; Fontenot, K.R.; Hammer, R.P.; Garno, J.C. Effects of Peptides Derived from Terminal Modifications of the Aβ Central Hydrophobic Core on Aβ Fibrillization. ACS Chem. Neurosci. 2010, 1, 661–678. [Google Scholar] [CrossRef][Green Version]
- Paul, A.; Frenkel-Pinter, M.; Escobar Alvarez, D.; Milordini, G.; Gazit, E.; Zacco, E.; Segal, D. Tryptophan-Galactosylamine Conjugates Inhibit and Disaggregate Amyloid Fibrils of Aβ42 and HIAPP Peptides While Reducing Their Toxicity. Commun. Biol. 2020, 3, 484. [Google Scholar] [CrossRef]
- Kobayashi, H.; Murata, M.; Kawanishi, S.; Oikawa, S. Polyphenols with Anti-Amyloid β Aggregation Show Potential Risk of Toxicity Via Pro-Oxidant Properties. IJMS 2020, 21, 3561. [Google Scholar] [CrossRef] [PubMed]
- Sanders, H.M.; Jovcevski, B.; Marty, M.T.; Pukala, T.L. Structural and Mechanistic Insights into Amyloid-β and A-synuclein Fibril Formation and Polyphenol Inhibitor Efficacy in Phospholipid Bilayers. FEBS J. 2022, 289, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhan, C.; Dong, X.; Wei, G. Molecular Mechanisms of Resveratrol and EGCG in the Inhibition of Aβ 42 Aggregation and Disruption of Aβ 42 Protofibril: Similarities and Differences. Phys. Chem. Chem. Phys. 2021, 23, 18843–18854. [Google Scholar] [CrossRef] [PubMed]
- Cieślik-Boczula, K.; Trombik, P. Resveratrol Modulates Fibrillogenesis in a Poly-l-lysine Peptide. Int. J. Biol. Macromol. 2019, 125, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Seidler, P.M.; Murray, K.A.; Boyer, D.R.; Ge, P.; Sawaya, M.R.; Hu, C.J.; Cheng, X.; Abskharon, R.; Pan, H.; DeTure, M.A.; et al. Structure-Based Discovery of Small Molecules That Disaggregate Alzheimer’s Disease Tissue Derived Tau Fibrils in Vitro. Nat. Commun. 2022, 13, 5451. [Google Scholar] [CrossRef]
- Sternke-Hoffmann, R.; Peduzzo, A.; Bolakhrif, N.; Haas, R.; Buell, A.K. The Aggregation Conditions Define Whether EGCG Is an Inhibitor or Enhancer of α-Synuclein Amyloid Fibril Formation. IJMS 2020, 21, 1995. [Google Scholar] [CrossRef]
- Madhuranthakam, C.M.R.; Shakeri, A.; Rao, P.P.N. Modeling the Inhibition Kinetics of Curcumin, Orange G, and Resveratrol with Amyloid-β Peptide. ACS Omega 2021, 6, 8680–8686. [Google Scholar] [CrossRef]
- Radbakhsh, S.; Barreto, G.E.; Bland, A.R.; Sahebkar, A. Curcumin: A Small Molecule with Big Functionality against Amyloid Aggregation in Neurodegenerative Diseases and Type 2 Diabetes. BioFactors 2021, 47, 570–586. [Google Scholar] [CrossRef]
- Sun, J.; Jiang, G.; Shigemori, H. Inhibitory Activity on Amyloid Aggregation of Rosmarinic Acid and Its Substructures From Isodon japonicus. Nat. Prod. Commun. 2019, 14, 1934578X1984303. [Google Scholar] [CrossRef]
- Ogawa, K.; Ishii, A.; Shindo, A.; Hongo, K.; Mizobata, T.; Sogon, T.; Kawata, Y. Spearmint Extract Containing Rosmarinic Acid Suppresses Amyloid Fibril Formation of Proteins Associated with Dementia. Nutrients 2020, 12, 3480. [Google Scholar] [CrossRef]
- Jahić Mujkić, A.; Tušek Žnidarič, M.; Berbić, S.; Žerovnik, E. Synergy of the Inhibitory Action of Polyphenols Plus Vitamin C on Amyloid Fibril Formation: Case Study of Human Stefin B. Antioxidants 2021, 10, 1471. [Google Scholar] [CrossRef] [PubMed]
- Abioye, R.O.; Okagu, O.D.; Udenigwe, C.C. Inhibition of Islet Amyloid Polypeptide Fibrillation by Structurally Diverse Phenolic Compounds and Fibril Disaggregation Potential of Rutin and Quercetin. J. Agric. Food Chem. 2022, 70, 392–402. [Google Scholar] [CrossRef]
- Bhatia, N.K.; Modi, P.; Sharma, S.; Deep, S. Quercetin and Baicalein Act as Potent Antiamyloidogenic and Fibril Destabilizing Agents for SOD1 Fibrils. ACS Chem. Neurosci. 2020, 11, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Howlett, D.R.; George, A.R.; Owen, D.E.; Ward, R.V.; Markwell, R.E. Common Structural Features Determine the Effectiveness of Carvedilol, Daunomycin and Rolitetracycline as Inhibitors of Alzheimer Beta-Amyloid Fibril Formation. Biochem. J. 1999, 343, 419–423. [Google Scholar] [CrossRef]
- Meena, V.K.; Kumar, V.; Karalia, S.; Dangi, R.S.; Sundd, M. Structural and Mechanistic Insights into Modulation of α-Synuclein Fibril Formation by Aloin and Emodin. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2022, 1866, 130151. [Google Scholar] [CrossRef]
- Han, F.; Jiang, B.; Lü, M.-H.; Wang, Z.-P.; Liu, W.; Zhang, Y.-X.; Xu, J. Hybrids of Polyphenolic Acids and Xanthone, the Potential Preventive and Therapeutic Effects on PD: Design, Synthesis, in Vitro Anti-Aggregation of α-Synuclein, and Disaggregation against the Existed α-Synuclein Oligomer and Fibril. Bioorg. Med. Chem. 2022, 66, 116818. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, Z.; Zhang, L.; Sui, R.; Khan, S. Exploring the Inhibitory Effects of Liquiritigenin against Tau Fibrillation and Related Neurotoxicity as a Model of Preventive Care in Alzheimer’s Disease. Int. J. Biol. Macromol. 2021, 183, 1184–1190. [Google Scholar] [CrossRef]
- Siposova, K.; Kozar, T.; Huntosova, V.; Tomkova, S.; Musatov, A. Inhibition of Amyloid Fibril Formation and Disassembly of Pre-Formed Fibrils by Natural Polyphenol Rottlerin. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2019, 1867, 259–274. [Google Scholar] [CrossRef]
- Yang, F.; Lim, G.P.; Begum, A.N.; Ubeda, O.J.; Simmons, M.R.; Ambegaokar, S.S.; Chen, P.P.; Kayed, R.; Glabe, C.G.; Frautschy, S.A.; et al. Curcumin Inhibits Formation of Amyloid β Oligomers and Fibrils, Binds Plaques, and Reduces Amyloid in Vivo. J. Biol. Chem. 2005, 280, 5892–5901. [Google Scholar] [CrossRef]
- Sasanian, N.; Bernson, D.; Horvath, I.; Wittung-Stafshede, P.; Esbjörner, E.K. Redox-Dependent Copper Ion Modulation of Amyloid-β (1-42) Aggregation In Vitro. Biomolecules 2020, 10, 924. [Google Scholar] [CrossRef]
- Abelein, A.; Gräslund, A.; Danielsson, J. Zinc as Chaperone-Mimicking Agent for Retardation of Amyloid β Peptide Fibril Formation. Proc. Natl. Acad. Sci. USA 2015, 112, 5407–5412. [Google Scholar] [CrossRef] [PubMed]
- Michaels, T.C.T.; Šarić, A.; Meisl, G.; Heller, G.T.; Curk, S.; Arosio, P.; Linse, S.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P.J. Thermodynamic and Kinetic Design Principles for Amyloid-Aggregation Inhibitors. Proc. Natl. Acad. Sci. USA 2020, 117, 24251–24257. [Google Scholar] [CrossRef] [PubMed]
- Linse, S.; Scheidt, T.; Bernfur, K.; Vendruscolo, M.; Dobson, C.M.; Cohen, S.I.A.; Sileikis, E.; Lundqvist, M.; Qian, F.; O’Malley, T.; et al. Kinetic Fingerprints Differentiate the Mechanisms of Action of Anti-Aβ Antibodies. Nat. Struct. Mol. Biol. 2020, 27, 1125–1133. [Google Scholar] [CrossRef]
- Törner, R.; Kupreichyk, T.; Gremer, L.; Debled, E.C.; Fenel, D.; Schemmert, S.; Gans, P.; Willbold, D.; Schoehn, G.; Hoyer, W.; et al. Structural Basis for the Inhibition of IAPP Fibril Formation by the Co-Chaperonin Prefoldin. Nat. Commun. 2022, 13, 2363. [Google Scholar] [CrossRef]
- Ghadami, S.A.; Chia, S.; Ruggeri, F.S.; Meisl, G.; Bemporad, F.; Habchi, J.; Cascella, R.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P.J.; et al. Transthyretin Inhibits Primary and Secondary Nucleations of Amyloid-β Peptide Aggregation and Reduces the Toxicity of Its Oligomers. Biomacromolecules 2020, 21, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Staats, R.; Michaels, T.C.T.; Flagmeier, P.; Chia, S.; Horne, R.I.; Habchi, J.; Linse, S.; Knowles, T.P.J.; Dobson, C.M.; Vendruscolo, M. Screening of Small Molecules Using the Inhibition of Oligomer Formation in α-Synuclein Aggregation as a Selection Parameter. Commun. Chem. 2020, 3, 191. [Google Scholar] [CrossRef]
- Groenning, M. Binding Mode of Thioflavin T and Other Molecular Probes in the Context of Amyloid Fibrils—Current Status. J. Chem. Biol. 2010, 3, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Lin, T.Y.; Chang, D.; Guo, Z. Thioflavin T as an Amyloid Dye: Fibril Quantification, Optimal Concentration and Effect on Aggregation. R. Soc. Open Sci. 2017, 4, 160696. [Google Scholar] [CrossRef]
- Yamamoto, N.; Akai, T.; Inoue, R.; Sugiyama, M.; Tamura, A.; Chatani, E. Structural Insights into the Inhibition of Amyloid Fibril Formation by Fibrinogen via Interaction with Prefibrillar Intermediates. Biochemistry 2019, 58, 2769–2781. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedov, I.; Khaibrakhmanova, D. Molecular Mechanisms of Inhibition of Protein Amyloid Fibril Formation: Evidence and Perspectives Based on Kinetic Models. Int. J. Mol. Sci. 2022, 23, 13428. https://doi.org/10.3390/ijms232113428
Sedov I, Khaibrakhmanova D. Molecular Mechanisms of Inhibition of Protein Amyloid Fibril Formation: Evidence and Perspectives Based on Kinetic Models. International Journal of Molecular Sciences. 2022; 23(21):13428. https://doi.org/10.3390/ijms232113428
Chicago/Turabian StyleSedov, Igor, and Diliara Khaibrakhmanova. 2022. "Molecular Mechanisms of Inhibition of Protein Amyloid Fibril Formation: Evidence and Perspectives Based on Kinetic Models" International Journal of Molecular Sciences 23, no. 21: 13428. https://doi.org/10.3390/ijms232113428
APA StyleSedov, I., & Khaibrakhmanova, D. (2022). Molecular Mechanisms of Inhibition of Protein Amyloid Fibril Formation: Evidence and Perspectives Based on Kinetic Models. International Journal of Molecular Sciences, 23(21), 13428. https://doi.org/10.3390/ijms232113428






