COVID-19 Molecular Pathophysiology: Acetylation of Repurposing Drugs
Abstract
1. Introduction
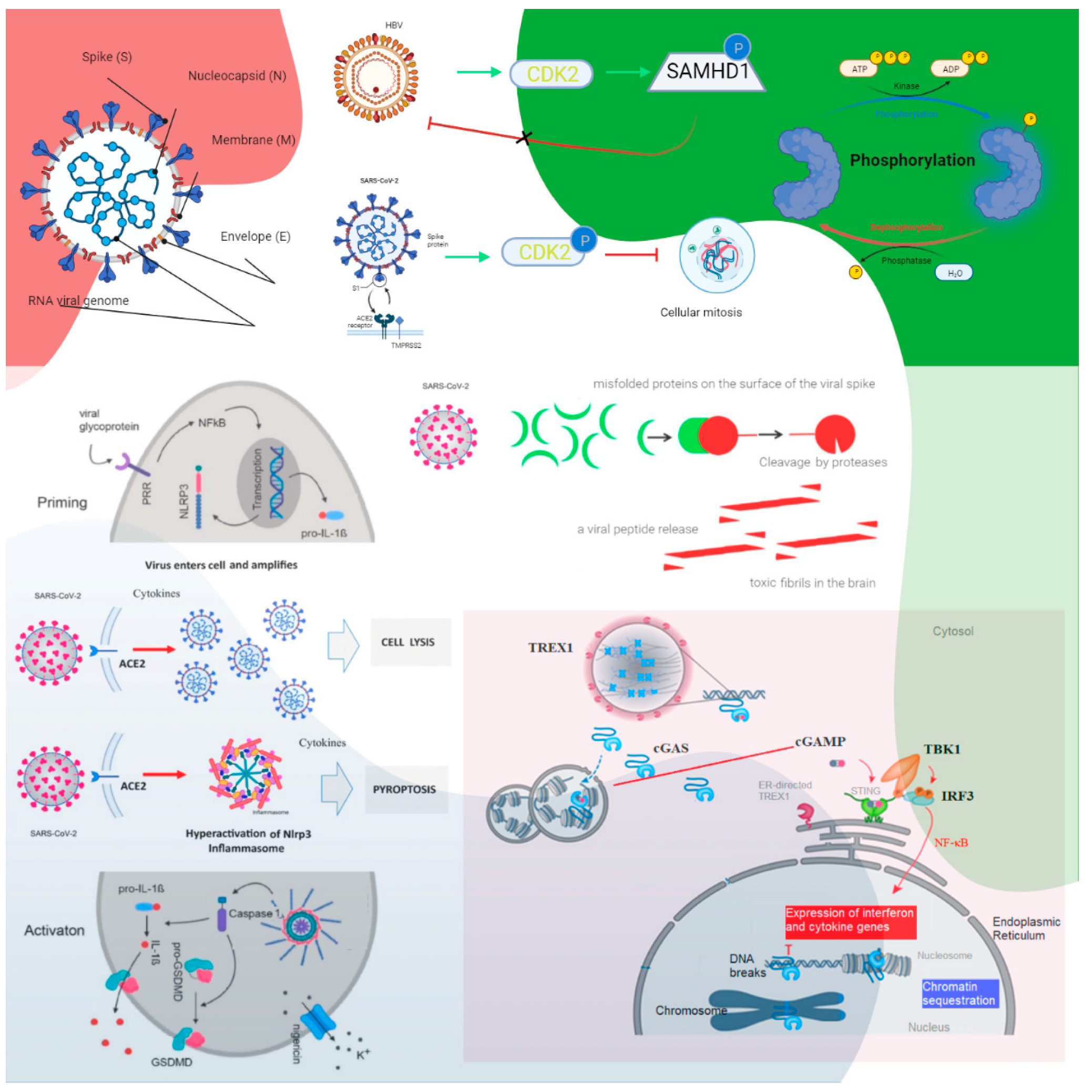
2. Molecular Pathophysiology
2.1. SAMHD1 Tetramerization Yields the Catalytically Active Tetramer: SARS-CoV-2 Might Use CDK2 to Phosphorylate SAMHD1
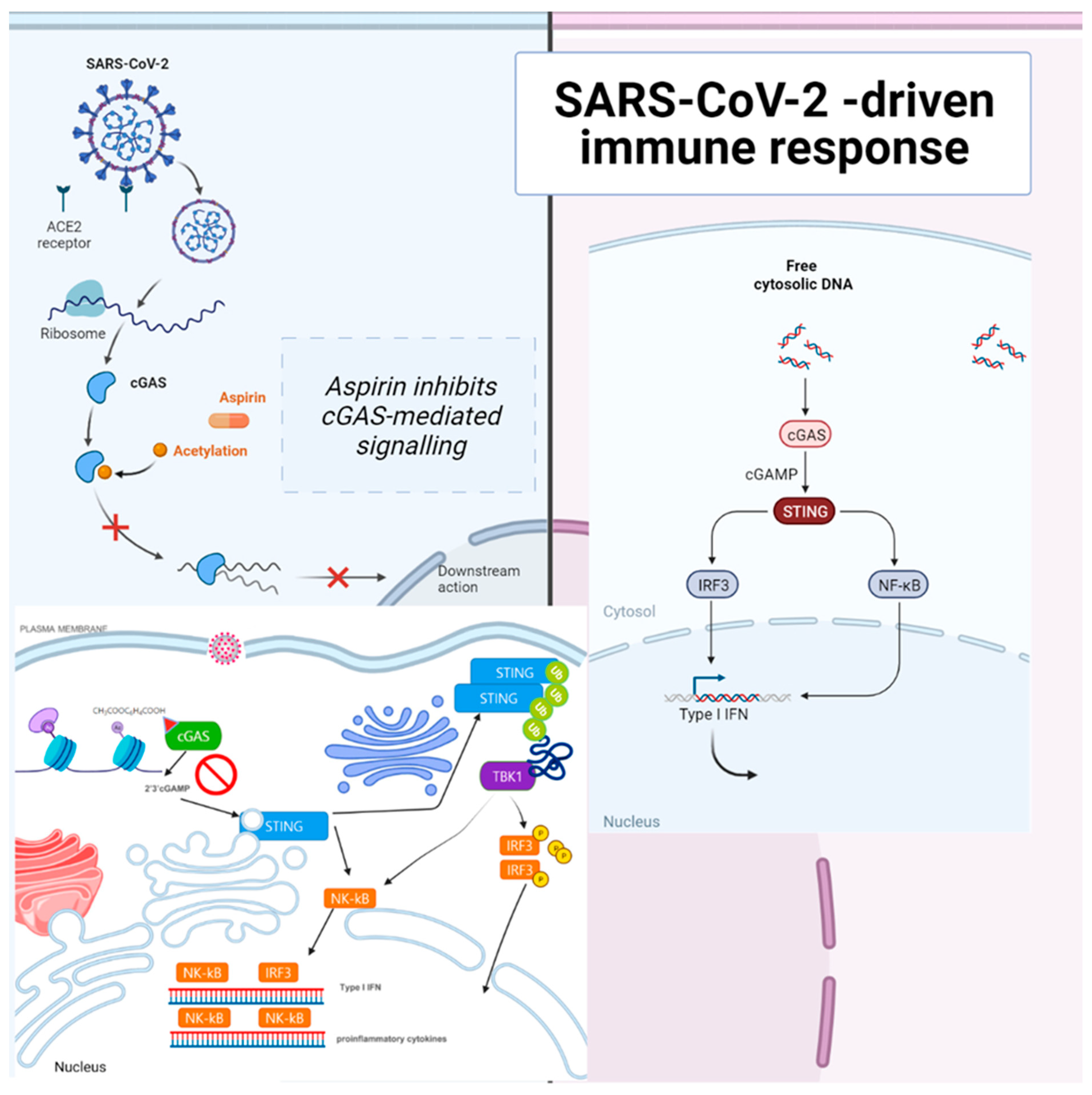
2.2. cGAS–STING Signaling
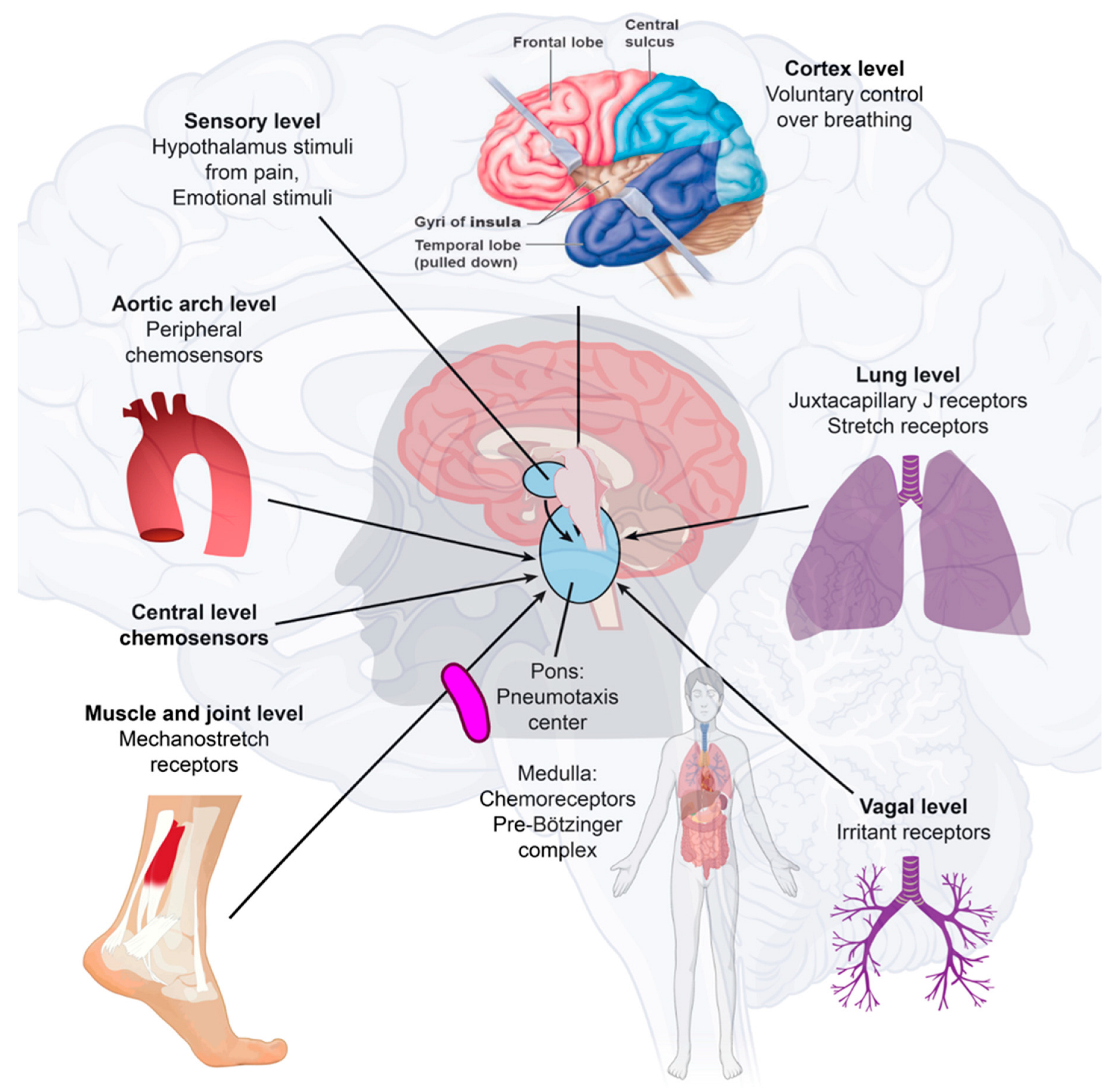
2.3. Immunological-Induced Engram Pathway
3. Acetylation and Molecular Treatment Pathway
3.1. Aspirin Inhibited cGAS and Did Not Affect STING Directly
3.2. Aspirin Treatment Decreased Mortality
3.3. The Acetylation Properties of Dapsone Competitively Anticatalyze COVID-19 Exacerbations
3.4. Only Dapsone Treats SARS-CoV-2 Exacerbated Acute Respiratory Distress Syndrome
3.5. Dapsone Treatment Mechanism
3.6. Repurposing Drugs That Do Not Treat COVID-19 Type-1 Interferonopathy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACE | Angiotensin-converting enzyme |
| AD | Alzheimer’s disease |
| AESI | Adverse events of special interest |
| AGS | Aicardi–Goutières syndrome |
| AMI | Acute myocardial infarction |
| AMI I/R injury | AMI ischemia–reperfusion injury |
| ARDS | Acute respiratory distress syndrome |
| ASC | Apoptosis-associated speck-like protein containing a CARD |
| ATF4 | Activated parkin via protein kinase RNA-like endoplasmic reticulum kinase-activating transcription factor 4 |
| BDNF | Brain-derived neurotrophic factor |
| BiPAP | Bilevel positive airway pressure |
| BV-2 | A type of microglial cell derived from C57/BL6 mice |
| CAPS | Cryopyrin-associated periodic syndromes |
| CARD | Caspase activation and recruitment domain |
| CCNE2 | Essential for the control of the cell cycle at the late G1 and early S phases; belongs to the cyclin family |
| CCR2 | C–C motif chemokine receptor 2 |
| CH | Clonal hematopoiesis, hematopoietic stem and progenitor cells |
| CI | Confidence interval |
| CK-MB | Creatine kinase-MB fraction |
| COPD | Chronic obstructive pulmonary disease |
| COX-2 | Cyclooxygenase 2 |
| CRP | C-reactive protein |
| CRS | Cytokine release syndrome |
| Cyclin E2 | Cyclin E2 is a protein that in humans is encoded by the CCNE2 gene |
| DDS | 4,4′-Diaminodiphenyl sulfone (dapsone) |
| DIC | Disseminated intravascular coagulation |
| ECG | Electrocardiogram |
| G6PDH | Glucose-6-phosphate dehydrogenase |
| HLA | Human leukocyte antigen |
| HLA-DRB1 | Major histocompatibility complex, class II, DR beta 1 |
| ICU | Intensive care unit |
| IFN | Interferon |
| IFNAR2 | Interferon-alpha and beta receptor subunit 2 |
| IL | Interleukin |
| IL-1β | Interleukin-1 beta |
| IMV | intensive mechanical ventilation |
| IRF3 | Interferon regulatory factor 3 |
| JNK | Jun N-terminal kinases |
| LDH | Lactate dehydrogenase |
| LDL | Low-density lipoprotein |
| LL | Lepromatous leprosy |
| MADDS | Monoacetyldapsone |
| MAPK | Mitogen-activated protein kinase |
| MCI | Mild cognitive impairment |
| MHC | Major histocompatibility complex |
| MIS-C/A | Multisystem inflammation syndrome in children and adults |
| MPO | Myeloperoxidase |
| NOD2 | Nucleotide-binding oligomerization domain containing 2 |
| mRNA | Messenger RNA |
| mtDNA | Mitochondrial DNA |
| NACHT | Domain conserved in NAIP, CIITA, HET-E, and TP1 |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLRP3 | NOD-, LRR-, and pyrin domain-containing protein 3 |
| PAI-1 | Plasminogen activator inhibitor-1 |
| PAMPs | Pathogen-associated molecular patterns |
| PBMCs | Human peripheral blood mononuclear cells |
| PD | Parkinson’s disease |
| PEDF | Pigment epithelium-derived factor |
| PEDFR/iPLA2 | PEDF/calcium-independent phospholipase A2 |
| Phospho-p65 | Anti-phospho-NFkB p65 (Ser536) monoclonal antibody (T.849.2) |
| Phospho-IκBα | Phospho-IκBα (Ser32/36) (5A5) mouse mAb #9246 |
| PTGS2 | Prostaglandin synthase 2 |
| PTM | Multiple post-translational modification |
| ROS | Reactive oxygen species |
| S | Full-length prefusion spike glycoprotein of SARS-CoV-2 |
| S1 | SARS-CoV-2 spike protein subunit 1 |
| SCLS | Systemic capillary leak syndrome |
| RCT | Randomized controlled trial |
| SOD | Superoxide dismutase |
| TGF-β | Transforming growth factor-beta |
| THP-1 | A spontaneously immortalized monocyte-like cell line |
| TNF | Tumor necrosis factor |
| TLR | Toll-like receptor |
| TMPRSS2 | Transmembrane protease serine subtype 2 |
| TTS | Thrombosis with thrombocytopenia syndrome |
| TYK2 | Tyrosine kinase 2 |
References
- Rodrigues, T.S.; de Sá, K.S.G.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R.; et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2020, 218, e20201707. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.P.; Cassel, S.L. Inflammasome-Mediated Autoinflammatory Disorders. Postgrad. Med. 2010, 122, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.; Fujiogi, M.; Koutsogiannaki, S. COVID-19 pathophysiology: A review. Clin. Immunol. 2020, 215, 108427. [Google Scholar] [CrossRef] [PubMed]
- Saris, A.; Reijnders, T.D.Y.; Nossent, E.J.; Schuurman, A.R.; Verhoeff, J.; van Asten, S.; Bontkes, H.; Blok, S.; Duitman, J.; Bogaard, H.-J.; et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax 2021, 76, 1010–1019. [Google Scholar] [CrossRef]
- Wauters, E.; Van Mol, P.; Garg, A.D.; Jansen, S.; Van Herck, Y.; Vanderbeke, L.; Bassez, A.; Boeckx, B.; Malengier-Devlies, B.; Timmerman, A.; et al. Discriminating Mild from Critical COVID-19 by Innate and Adaptive Immune Single-cell Profiling of Bronchoalveolar Lavages. BioRxiv 2021, 31, 272–290. [Google Scholar] [CrossRef]
- Zarrilli, G.; Angerilli, V.; Businello, G.; Sbaraglia, M.; Traverso, G.; Fortarezza, F.; Rizzo, S.; De Gaspari, M.; Basso, C.; Calabrese, F.; et al. The Immunopathological and Histological Landscape of COVID-19-Mediated Lung Injury. Int. J. Mol. Sci. 2021, 22, 974. [Google Scholar] [CrossRef]
- Li, D.; Ren, W.; Jiang, Z.; Zhu, L. Regulation of the NLRP3 inflammasome and macrophage pyroptosis by the p38 MAPK signaling pathway in a mouse model of acute lung injury. Mol. Med. Rep. 2018, 18, 4399–4409. [Google Scholar] [CrossRef]
- Middleton, E.A.; Rondina, M.T.; Schwertz, H.; Zimmerman, G. Amicus or Adversary Revisited: Platelets in Acute Lung Injury and Acute Respiratory Distress Syndrome. Am. J. Respir. Cell Mol. Biol. 2018, 59, 18–35. [Google Scholar] [CrossRef]
- Gerard, L.; Lecocq, M.; Bouzin, C.; Hoton, D.; Schmit, G.; Pereira, J.P.; Montiel, V.; Plante-Bordeneuve, T.; Laterre, P.-F.; Pilette, C. Increased Angiotensin-Converting Enzyme 2 and Loss of Alveolar Type II Cells in COVID-19–related Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2021, 204, 1024–1034. [Google Scholar] [CrossRef]
- Rando, H.M.; MacLean, A.L.; Lee, A.J.; Ray, S.; Bansal, V.; Skelly, A.N.; Sell, E.; Dziak, J.J.; Shinholster, L.; McGowan, L.D.; et al. Pathogenesis, Symptomatology, and Transmission of SARS-CoV-2 through Analysis of Viral Genomics and Structure. mSystems 2021, 6, e00095-21. [Google Scholar] [CrossRef]
- Kanneganti, T. Intracellular innate immune receptors: Life inside the cell. Immunol. Rev. 2020, 297, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Kanneganti, T.-D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Bork, P.; Ponting, C.P.; Hofmann, K. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997, 6, 249–253. [Google Scholar] [CrossRef]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012, 13, 223–228, Erratum in Nat. Immunol. 2013, 14, 877. [Google Scholar] [CrossRef]
- Coquel, F.; Silva, M.-J.; Técher, H.; Zadorozhny, K.; Sharma, S.; Nieminuszczy, J.; Mettling, C.; Dardillac, E.; Barthe, A.; Schmitz, A.-L.; et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 2018, 557, 57–61. [Google Scholar] [CrossRef] [PubMed]
- White, T.E.; Brandariz-Nuñez, A.; Casuso, J.C.V.; Amie, S.; Nguyen, L.A.; Kim, B.; Tuzova, M.; Diaz-Griffero, F. The Retroviral Restriction Ability of SAMHD1, but Not Its Deoxynucleotide Triphosphohydrolase Activity, Is Regulated by Phosphorylation. Cell Host Microbe 2013, 13, 441–451. [Google Scholar] [CrossRef]
- Tang, C.; Ji, X.; Wu, L.; Xiong, Y. Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of Thr-592. J. Biol. Chem. 2015, 290, 26352–26359. [Google Scholar] [CrossRef]
- Ji, X.; Wu, Y.; Yan, J.; Mehrens, J.; Yang, H.; DeLucia, M.; Hao, C.; Gronenborn, A.M.; Skowronski, J.; Ahn, J.; et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 2013, 20, 1304–1309. [Google Scholar] [CrossRef]
- Yu, C.H.; Bhattacharya, A.; Persaud, M.; Taylor, A.B.; Wang, Z.; Bulnes-Ramos, A.; Xu, J.; Selyutina, A.; Martinez-Lopez, A.; Cano, K.; et al. Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification. Nat. Commun. 2021, 12, 731. [Google Scholar] [CrossRef]
- Oo, A.; Zandi, K.; Shepard, C.; Bassit, L.C.; Musall, K.; Goh, S.L.; Cho, Y.J.; Kim, D.H.; Schinazi, R.F.; Kim, B. Elimination of Aicardi–Goutières syndrome protein SAMHD1 activates cellular innate immunity and suppresses SARS-CoV-2 replication. J. Biol. Chem. 2022, 298, 101635. [Google Scholar] [CrossRef]
- Crow, Y.J.; Manel, N. Aicardi–Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015, 15, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.I.; Bond, J.; Asipu, A.; Brunette, R.L.; Manfield, I.W.; Carr, I.M.; Fuller, J.C.; Jackson, R.M.; Lamb, T.; Briggs, T.A.; et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 2009, 41, 829–832. [Google Scholar] [CrossRef]
- Krasemann, S.; Haferkamp, U.; Pfefferle, S.; Woo, M.S.; Heinrich, F.; Schweizer, M.; Appelt-Menzel, A.; Cubukova, A.; Barenberg, J.; Leu, J.; et al. The blood-brain barrier is dysregulated in COVID-19 and serves as a CNS entry route for SARS-CoV-2. Stem Cell Rep. 2022, 17, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Sergi, C. SAMHD1 as the potential link between SARS-CoV-2 infection and neurological complications. Front. Neurol. 2020, 11, 562913. [Google Scholar] [CrossRef] [PubMed]
- Maelfait, J.; Bridgeman, A.; Benlahrech, A.; Cursi, C.; Rehwinkel, J. Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell Rep. 2016, 16, 1492–1501. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Yu, Q.; Chu, L.; Cui, Y.; Ding, M.; Wang, Q.; Wang, H.; Chen, Y.; Liu, X.; Wang, C. Nuclear cGAS Functions Non-canonically to Enhance Antiviral Immunity via Recruiting Methyltransferase Prmt5. Cell Rep. 2020, 33, 108490. [Google Scholar] [CrossRef]
- de Oliveira Mann, C.C.; Hopfner, K.-P. Nuclear cGAS: Guard or prisoner? EMBO J. 2021, 40, e108293. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H.; Bohr, V.A. Signaling by cGAS–STING in Neurodegeneration, Neuroinflammation, and Aging. Trends Neurosci. 2021, 44, 83–96. [Google Scholar] [CrossRef]
- Ren, H.; Ma, C.; Peng, H.; Zhang, B.; Zhou, L.; Su, Y.; Gao, X.; Huang, H. Micronucleus production, activation of DNA damage response and cGAS-STING signaling in syncytia induced by SARS-CoV-2 infection. Biol. Direct 2021, 16, 20. [Google Scholar] [CrossRef]
- Su, J.; Shen, S.; Hu, Y.; Chen, S.; Cheng, L.; Cai, Y.; Wei, W.; Wang, Y.; Rui, Y.; Yu, X.-F. SARS-CoV-2 ORF3a inhibits cGAS-STING-mediated autophagy flux and antiviral function. J. Med. Virol. 2022, 1–9. [Google Scholar] [CrossRef]
- Han, L.; Zheng, Y.; Deng, J.; Nan, M.-L.; Xiao, Y.; Zhuang, M.-W.; Zhang, J.; Wang, W.; Gao, C.; Wang, P.-H. SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy. J. Med. Virol. 2022, 94, 5174–5188. [Google Scholar] [CrossRef]
- Domizio, J.D.; Gulen, M.F.; Saidoune, F.; Thacker, V.V.; Yatim, A.; Sharma, K.; Nass, T.; Guenova, E.; Schaller, M.; Conrad, C.; et al. The cGAS–STING pathway drives type I IFN immunopathology in COVID-19. Nature 2022, 603, 145–151. [Google Scholar] [CrossRef]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brünink, S.; Greuel, S.; et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Matschke, J.; Lütgehetmann, M.; Hagel, C.; Sperhake, J.P.; Schröder, A.S.; Edler, C.; Mushumba, H.; Fitzek, A.; Allweiss, L.; Dandri, M.; et al. Neuropathology of patients with COVID-19 in Germany: A post-mortem case series. Lancet Neurol. 2020, 19, 919–929. [Google Scholar] [CrossRef]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; Weizman, O.E.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021, 218, e20202135. [Google Scholar] [CrossRef] [PubMed]
- Bauer, L.; Brigitta, M.; de Vrij, F.M.S.; Kushner, S.A.; Harschnitz, O.; van Riel, D. The neuroinvasiveness, neurotropism, and neurovirulence of SARS-CoV-2. Trends Neurosci. 2022, 45, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Cajanding, R.J.M. Silent Hypoxia in COVID-19 Pneumonia: State of Knowledge, Pathophysiology, Mechanisms, and Management. AACN Adv. Crit. Care 2022, 33, 143–153. [Google Scholar] [CrossRef]
- Finsterer, J.; Scorza, F.A. Clinical and Pathophysiologic Spectrum of Neuro-COVID. Mol. Neurobiol. 2021, 58, 3787–3791. [Google Scholar] [CrossRef]
- Schwabenland, M.; Salié, H.; Tanevski, J.; Killmer, S.; Lago, M.S.; Schlaak, A.E.; Mayer, L.; Matschke, J.; Püschel, K.; Fitzek, A.; et al. Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia-T-cell interactions. Immunity 2021, 54, 1594–1610.e11. [Google Scholar] [CrossRef]
- Kanwar, B.; Lee, C.J.; Lee, J.-H. Specific Treatment Exists for SARS-CoV-2 ARDS. Vaccines 2021, 9, 635. [Google Scholar] [CrossRef]
- Lee, M.-H.; Perl, D.P.; Nair, G.; Li, W.; Maric, D.; Murray, H.; Dodd, S.J.; Koretsky, A.P.; Watts, J.A.; Cheung, V.; et al. Microvascular Injury in the Brains of Patients with COVID-19. N. Engl. J. Med. 2020, 384, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Stratton, C.W.; Tang, Y.; Lu, H. Pathogenesis-directed therapy of 2019 novel coronavirus disease. J. Med Virol. 2020, 93, 1320–1342. [Google Scholar] [CrossRef] [PubMed]
- Koren, T.; Yifa, R.; Amer, M.; Krot, M.; Boshnak, N.; Ben-Shaanan, T.L.; Azulay-Debby, H.; Zalayat, I.; Avishai, E.; Hajjo, H.; et al. Insular cortex neurons encode and retrieve specific immune responses. Cell 2021, 184, 5902–5915.e17. [Google Scholar] [CrossRef] [PubMed]
- Gogolla, N. The brain remembers where and how inflammation struck. Cell 2021, 184, 5851–5853. [Google Scholar] [CrossRef] [PubMed]
- Koike, K.; Yoo, S.-J.; Bleymehl, K.; Omura, M.; Zapiec, B.; Pyrski, M.; Blum, T.; Khan, M.; Bai, Z.; Leinders-Zufall, T.; et al. Danger Perception and Stress Response through an Olfactory Sensor for the Bacterial Metabolite Hydrogen Sulfide. Neuron 2021, 109, 2469–2484.e7. [Google Scholar] [CrossRef]
- Norden, D.M.; Trojanowski, P.J.; Villanueva, E.; Navarro, E.; Godbout, J.P. Sequential activation of microglia and astrocyte cytokine expression precedes increased iba-1 or GFAP immunoreactivity following systemic immune challenge. Glia 2015, 64, 300–316. [Google Scholar] [CrossRef]
- O’Neil, S.M.; Hans, E.E.; Jiang, S.; Wangler, L.M.; Godbout, J.P. Astrocyte immunosenescence and deficits in interleukin 10 signaling in the aged brain disrupt the regulation of microglia following innate immune activation. Glia 2022, 70, 913–934. [Google Scholar] [CrossRef]
- Little, P. Non-steroidal anti-inflammatory drugs and COVID-19. BMJ 2020, 368, m1185. [Google Scholar] [CrossRef]
- Dai, J.; Huang, Y.-J.; He, X.; Zhao, M.; Wang, X.; Liu, Z.-S.; Xue, W.; Cai, H.; Zhan, X.-Y.; Huang, S.-Y.; et al. Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell 2019, 176, 1447–1460.e14. [Google Scholar] [CrossRef]
- Abu Esba, L.C.; Alqahtani, R.A.; Thomas, A.; Shamas, N.; Alswaidan, L.; Mardawi, G. Ibuprofen and NSAID Use in COVID-19 Infected Patients Is Not Associated with Worse Outcomes: A Prospective Cohort Study. Infect. Dis. Ther. 2021, 10, 253–268. [Google Scholar] [CrossRef]
- Bruce, E.; Barlow-Pay, F.; Short, R.; Vilches-Moraga, A.; Price, A.; McGovern, A.; Braude, P.; Stechman, M.J.; Moug, S.; McCarthy, K.; et al. Prior Routine Use of Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) and Important Outcomes in Hospitalised Patients with COVID-19. J. Clin. Med. 2020, 9, 2586. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.H.; Rahnavard, A.; Gomberg-Maitland, M.; Chatterjee, R.; Patodi, P.; Yamane, D.P.; Levine, A.R.; Davison, D.; Hawkins, K.; Jackson, A.M.; et al. Association of Early Aspirin Use with in-Hospital Mortality in Patients with Moderate COVID-19. JAMA Netw. Open 2022, 5, e223890. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-M.; Sansores-Garcia, L.; Chen, X.-M.; Matijevic-Aleksic, N.; Du, M.; Wu, K.K. Suppression of inducible cyclooxygenase 2 gene transcription by aspirin and sodium salicylate. Proc. Natl. Acad. Sci. USA 1999, 96, 5292–5297. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Pfeffer, L.M. The Role of Nuclear Factor κB in the Interferon Response. J. Interf. Cytokine Res. 2011, 31, 553–559. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, L.; Hong, Z.; Lv, Z.; Mao, Z.; Tang, Y.; Kong, X.; Li, S.; Cui, Y.; Liu, H.; et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017, 13, e1006264. [Google Scholar] [CrossRef]
- Esparza-Ibarra, E.L.; Ayala-Luján, J.L.; Mendoza-Almanza, B.; González-Curiel, I.; Godina-González, S.; Hernández-Barrales, M.; Mendoza-Almanza, G. The Role of Platelet in Severe and Fatal Forms of COVID-19. Curr. Mol. Med. 2022, 22, 572–583. [Google Scholar] [CrossRef]
- Whyte, C.S.; Morrow, G.B.; Mitchell, J.L.; Chowdary, P.; Mutch, N.J. Fibrinolytic abnormalities in acute respiratory distress syndrome (ARDS) and versatility of thrombolytic drugs to treat COVID-19. J. Thromb. Haemost. 2020, 18, 1548–1555. [Google Scholar] [CrossRef]
- Willems, L.; Nagy, M.; Cate, H.T.; Spronk, H.; Groh, L.; Leentjens, J.; Janssen, N.; Netea, M.; Thijssen, D.; Hannink, G.; et al. Sustained inflammation, coagulation activation and elevated endothelin-1 levels without macrovascular dysfunction at 3 months after COVID-19. Thromb. Res. 2022, 209, 106–114. [Google Scholar] [CrossRef]
- Koupenova, M.; Kehrel, B.E.; Corkrey, H.A.; Freedman, J.E. Thrombosis and platelets: An update. Eur. Heart J. 2017, 38, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Toner, P.; McAuley, D.F.; Shyamsundar, M. Aspirin as a potential treatment in sepsis or acute respiratory distress syndrome. Crit. Care 2015, 19, 374. [Google Scholar] [CrossRef] [PubMed]
- Scheuch, G.; Canisius, S.; Nocker, K.; Hofmann, T.; Naumann, R.; Pleschka, S.; Ludwig, S.; Welte, T.; Planz, O. Targeting intracellular signaling as an antiviral strategy: Aerosolized LASAG for the treatment of influenza in hospitalized patients. Emerg. Microbes Infect. 2018, 7, 21. [Google Scholar] [CrossRef]
- The OpenSAFELY Collaborative; Wong, A.Y.S.; MacKenna, B.; Morton, C.E.; Schultze, A.; Walker, A.J.; Bhaskaran, K.; Brown, J.P.; Rentsch, C.T.; Williamson, E.; et al. OpenSAFELY: Do adults prescribed non-steroidal anti-inflammatory drugs have an increased risk of death from COVID-19? medRxiv 2020, 2020.08.12.20171405. [Google Scholar] [CrossRef]
- Rinott, E.; Kozer, E.; Shapira, Y.; Bar-Haim, A.; Youngster, I. Ibuprofen use and clinical outcomes in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1259.e5–1259.e7. [Google Scholar] [CrossRef]
- Imam, Z.; Odish, F.; Gill, I.; O’Connor, D.; Armstrong, J.; Vanood, A.; Ibironke, O.; Hanna, A.; Ranski, A.; Halalau, A. Older age and comorbidity are independent mortality predictors in a large cohort of 1305 COVID-19 patients in Michigan, United States. J. Intern. Med. 2020, 288, 469–476. [Google Scholar] [CrossRef]
- Lund, L.C.; Kristensen, K.B.; Reilev, M.; Christensen, S.; Thomsen, R.W.; Christiansen, C.F.; Støvring, H.; Johansen, N.B.; Brun, N.C.; Hallas, J.; et al. Adverse outcomes and mortality in users of non-steroidal anti-inflammatory drugs who tested positive for SARS-CoV-2: A Danish nationwide cohort study. PLoS Med. 2020, 17, e1003308. [Google Scholar] [CrossRef]
- Osborne, T.F.; Veigulis, Z.P.; Arreola, D.M.; Mahajan, S.M.; Röösli, E.; Curtin, C.M. Association of mortality and aspirin prescription for COVID-19 patients at the Veterans Health Administration. PLoS ONE 2021, 16, e0246825. [Google Scholar] [CrossRef]
- Gatti, G.; Hossein, J.; Malena, M.; Cruciani, M.; Bassetti, M. Penetration of dapsone into cerebrospinal fluid of patients with AIDS. J. Antimicrob. Chemother. 1997, 40, 113–115. [Google Scholar] [CrossRef][Green Version]
- Rich, J.D.; Mirochnick, M. Dapsone penetrates cerebrospinal fluid during Pneumocystis carinii pneumonia prophylaxis. Diagn. Microbiol. Infect. Dis. 1996, 24, 77–79. [Google Scholar] [CrossRef]
- Ellard, G.A.; Gammon, P.T.; Helmy, H.S.; Rees, R.J.W. Dapsone Acetylation and the Treatment of Leprosy. Nature 1972, 239, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; An, H.; Sohn, M.-G.; Kivela, P.; Oh, S. 4,4’-Diaminodiphenyl Sulfone (DDS) as An Inflammasome Competitor. Int. J. Mol. Sci. 2020, 21, 5953. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Choi, S.-H.; Lee, C.J.; Oh, S.-S. Recovery of Dementia Syndrome following Treatment of Brain Inflammation. Dement. Geriatr. Cogn. Disord. Extra 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, C.J.; Park, J.; Lee, S.J.; Choi, S.-H. The Neuroinflammasome in Alzheimer’s Disease and Cerebral Stroke. Dement. Geriatr. Cogn. Disord. Extra 2021, 11, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, B.A.; Khattak, A.; Balentine, J.; Lee, J.H.; Kast, R.E. Benefits of Using Dapsone in Patients Hospitalized with COVID-19. Vaccines 2022, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Khattak, A.; Kanwar, B.; Sergi, C.; Lee, C.J.; Balentine, J.; Lee, J.-H.; Park, J.; Lee, S.J.; Choi, S.-H. Commentary for the Elderly in the Pandemic Era. Dement. Geriatr. Cogn. Disord. Extra 2021, 11, 168–171. [Google Scholar] [CrossRef]
- Zhang, F.-R.; Liu, H.; Irwanto, A.; Fu, X.-A.; Li, Y.; Yu, G.-Q.; Yu, Y.-X.; Chen, M.-F.; Low, H.-Q.; Li, J.-H.; et al. HLA-B*13:01 and the Dapsone Hypersensitivity Syndrome. N. Engl. J. Med. 2013, 369, 1620–1628. [Google Scholar] [CrossRef]
- Tempark, T.; Satapornpong, P.; Rerknimitr, P.; Nakkam, N.; Saksit, N.; Wattanakrai, P.; Jantararoungtong, T.; Koomdee, N.; Mahakkanukrauh, A.; Tassaneeyakul, W.; et al. Dapsone-induced severe cutaneous adverse drug reactions are strongly linked with HLA-B*13: 01 allele in the Thai population. Pharm. Genom. 2017, 27, 429–437. [Google Scholar] [CrossRef]
- Watanabe, H.; Watanabe, Y.; Tashiro, Y.; Mushiroda, T.; Ozeki, T.; Hashizume, H.; Sueki, H.; Yamamoto, T.; Utsunomiya-Tate, N.; Gouda, H.; et al. A docking model of dapsone bound to HLA-B*13:01 explains the risk of dapsone hypersensitivity syndrome. J. Dermatol. Sci. 2017, 88, 320–329. [Google Scholar] [CrossRef]
- Chakraborty, A.; Panda, A.K.; Ghosh, R.; Biswas, A. DNA minor groove binding of a well known anti-mycobacterial drug dapsone: A spectroscopic, viscometric and molecular docking study. Arch. Biochem. Biophys. 2019, 665, 107–113. [Google Scholar] [CrossRef]
- Zhao, Q.; Alhilali, K.; Alzahrani, A.; Almutairi, M.; Amjad, J.; Liu, H.; Sun, Y.; Sun, L.; Zhang, H.; Meng, X.; et al. Dapsone-and nitroso dapsone-specific activation of T cells from hypersensitive patients expressing the risk allele HLA-B* 13: 01. Allergy 2019, 74, 1533–1548. [Google Scholar] [CrossRef] [PubMed]
- Leite, M.D.M.; Gonzalez-Galarza, F.F.; da Silva, B.C.C.; Middleton, D.; dos Santos, E.J.M. Predictive immunogenetic markers in COVID-19. Hum. Immunol. 2021, 82, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-T.; Wang, C.-W.; Lu, C.-W.; Chen, C.-B.; Lee, H.-E.; Hung, S.-I.; Choon, S.-E.; Yang, C.-H.; Liu, M.-T.; Chen, T.J.; et al. The Function of HLA-B*13:01 Involved in the Pathomechanism of Dapsone-Induced Severe Cutaneous Adverse Reactions. J. Investig. Dermatol. 2018, 138, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Colella, M.P.; Orsi, F.A.; Alves, E.C.F.; Delmoro, G.F.; Yamaguti-Hayakawa, G.G.; de Paula, E.V.; Annichino-Bizzacchi, J.M. A retrospective analysis of 122 immune thrombocytopenia patients treated with dapsone: Efficacy, safety and factors associated with treatment response. J. Thromb. Haemost. 2021, 19, 2275–2286. [Google Scholar] [CrossRef]
- Cho, S.C.; Rhim, J.H.; Choi, H.R.; Son, Y.H.; Lee, S.J.; Song, K.-Y.; Park, S.C. Protective effect of 4,4’-diaminodiphenylsulfone against paraquat-induced mouse lung injury. Exp. Mol. Med. 2011, 43, 525–537. [Google Scholar] [CrossRef]
- Mahale, A.; Kumar, R.; Sarode, L.P.; Gakare, S.; Prakash, A.; Ugale, R.R. Dapsone prolong delayed excitotoxic neuronal cell death by interacting with proapoptotic/survival signaling proteins. J. Stroke Cerebrovasc. Dis. 2020, 29, 104848. [Google Scholar] [CrossRef]
- Zhan, R.; Zhao, M.; Zhou, T.; Chen, Y.; Yu, W.; Zhao, L.; Zhang, T.; Wang, H.; Yang, H.; Jin, Y.; et al. Dapsone protects brain microvascular integrity from high-fat diet induced LDL oxidation. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Rashidian, A.; Rashki, A.; Abdollahi, A.; Haddadi, N.S.; Chamanara, M.; Mumtaz, F.; Dehpour, A.R. Dapsone reduced acetic acid-induced inflammatory response in rat colon tissue through inhibition of NF-kB signaling pathway. Immunopharm. Immunotoxicol 2019, 41, 607–613. [Google Scholar] [CrossRef]
- Erttmann, S.F.; Swacha, P.; Aung, K.M.; Brindefalk, B.; Jiang, H.; Härtlova, A.; Uhlin, B.E.; Wai, S.N.; Gekara, N.O. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity 2022, 55, 847–861.e10. [Google Scholar] [CrossRef]
- Roy, E.R.; Chiu, G.; Li, S.; Propson, N.E.; Kanchi, R.; Wang, B.; Coarfa, C.; Zheng, H.; Cao, W. Concerted type I interferon signaling in microglia and neural cells promotes memory impairment associated with amyloid β plaques. Immunity 2022, 55, 879–894.e6. [Google Scholar] [CrossRef]
- Lee, J.H.; Kanwar, B.; Lee, C.J.; Sergi, C.; Coleman, M.D. Dapsone is an anticatalysis for Alzheimer’s disease exacerbation. iScience 2022, 25, 104274. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.C.; Vigod, S.; Bortolussi-Courval, É.; Hanula, R.; Boulware, D.R.; Lenze, E.J.; Reiersen, A.M.; McDonald, E.G. Fluvoxamine for Outpatient Management of COVID-19 to Prevent Hospitalization: A Systematic Review and Meta-analysis. JAMA Netw. Open 2022, 5, e226269. [Google Scholar] [CrossRef] [PubMed]
- Reis, G.; Moreira-Silva, E.A.D.S.; Silva, D.C.M.; Thabane, L.; Milagres, A.C.; Ferreira, T.S.; dos Santos, C.V.Q.; Campos, V.H.d.S.; Nogueira, A.M.R.; de Almeida, A.P.F.G.; et al. Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: The TOGETHER randomised, platform clinical trial. Lancet Glob. Health 2021, 10, e42–e51. [Google Scholar] [CrossRef]
- Bramante, C.T.; Huling, J.D.; Tignanelli, C.J.; Buse, J.B.; Liebovitz, D.M.; Nicklas, J.M.; Cohen, K.; Puskarich, M.A.; Belani, H.K.; Proper, J.L.; et al. Randomized Trial of Metformin, Ivermectin, and Fluvoxamine for COVID-19. N. Engl. J. Med. 2022, 387, 599–610. [Google Scholar] [CrossRef]
- Nguyen, L.C.; Yang, D.; Nicolaescu, V.; Best, T.J.; Gula, H.; Saxena, D.; Gabbard, J.D.; Chen, S.N.; Ohtsuki, T.; Friesen, J.B.; et al. Cannabidiol inhibits SARS-CoV-2 replication through induction of the host ER stress and innate immune responses. Sci. Adv. 2022, 8, eabi6110. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, Y.-J.; Dobbs, N.; Sakai, T.; Liou, J.; Miner, J.J.; Yan, N. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 2019, 216, 867–883. [Google Scholar] [CrossRef]
- Corley, M.J.; Pang, A.P.; Dody, K.; Mudd, P.A.; Patterson, B.K.; Seethamraju, H.; Bram, Y.; Peluso, M.J.; Torres, L.; Iyer, N.S.; et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J. Leukoc. Biol. 2021, 110, 21–26. [Google Scholar] [CrossRef]
- Marié, I.J.; Chang, H.-M.; Levy, D.E. HDAC stimulates gene expression through BRD4 availability in response to IFN and in interferonopathies. J. Exp. Med. 2018, 215, 3194–3212. [Google Scholar] [CrossRef]
- Feser, J.; Tyler, J. Chromatin structure as a mediator of aging. FEBS Lett. 2011, 585, 2041–2048. [Google Scholar] [CrossRef]
- Bracken, A.P.; Kleine-Kohlbrecher, D.; Dietrich, N.; Pasini, D.; Gargiulo, G.; Beekman, C.; Theilgaard-Mönch, K.; Minucci, S.; Porse, B.T.; Marine, J.-C.; et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007, 21, 525–530. [Google Scholar] [CrossRef]
- Kamada, R.; Yang, W.; Zhang, Y.; Patel, M.C.; Yang, Y.; Ouda, R.; Dey, A.; Wakabayashi, Y.; Sakaguchi, K.; Fujita, T.; et al. Interferon stimulation creates chromatin marks and establishes transcriptional memory. Proc. Natl. Acad. Sci. USA 2018, 115, E9162–E9171. [Google Scholar] [CrossRef] [PubMed]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270. [Google Scholar] [CrossRef]
- Shirvaliloo, M. The unfavorable clinical outcome of COVID-19 in smokers is mediated by H3K4me3, H3K9me3 and H3K27me3 histone marks. Epigenomics 2022, 14, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wadgaonkar, P.; Xu, L.; Thakur, C.; Fu, Y.; Bi, Z.; Qiu, Y.; Almutairy, B.; Zhang, W.; Stemmer, P.; et al. Environmentally-induced mdig contributes to the severity of COVID-19 through fostering expression of SARS-CoV-2 receptor NRPs and glycan metabolism. Theranostics 2021, 11, 7970–7983. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.-W.; Shie, J.-J.; Chen, C.-H.; Wu, C.-Y.; Hsu, T.-L.; Wong, C.-H. O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc. Natl. Acad. Sci. USA 2018, 115, 7302–7307. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Zhou, L. EZH2-mediated H3K27me3 inhibits ACE2 expression. Biochem. Biophys. Res. Commun. 2020, 526, 947–952. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Antón-Plágaro, C.; Williamson, M.K.; Shoemark, D.K.; Simón-Gracia, L.; Klein, K.; Bauer, M.; Hollandi, R.; Greber, U.F.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Ogunlade, B.O.; Lazartigues, E.; Filipeanu, C.M. Angiotensin type 1 receptor-dependent internalization of SARS-CoV-2 by angiotensin-converting enzyme 2. Hypertension 2021, 77, e42–e43. [Google Scholar] [CrossRef]
- Yeung, M.L.; Teng, J.L.L.; Jia, L.; Zhang, C.; Huang, C.; Cai, J.-P.; Zhou, R.; Chan, K.-H.; Zhao, H.; Zhu, L.; et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell 2021, 184, 2212–2228.e12. [Google Scholar] [CrossRef]
- Kumar, V.; Behera, R.; Lohite, K.; Karnik, S.; Kundu, G.C. p38 Kinase Is Crucial for Osteopontin-Induced Furin Expression That Supports Cervical Cancer Progression. Cancer Res. 2010, 70, 10381–10391. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Furushima, D.; Niki, T.; Matsuba, T.; Maeda, Y.; Takahashi, A.; Hattori, T.; Ashino, Y. High Levels of the Cleaved form of Galectin-9 and Osteopontin in the Plasma Are Associated with Inflammatory Markers That Reflect the Severity of COVID-19 Pneumonia. Int. J. Mol. Sci. 2021, 22, 4978. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.; Schack, L.; Kläning, E.; Sørensen, E.S. Osteopontin Is Cleaved at Multiple Sites Close to Its Integrin-binding Motifs in Milk and Is a Novel Substrate for Plasmin and Cathepsin D. J. Biol. Chem. 2010, 285, 7929–7937. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-M.; Yang, W.-L.; Yang, F.-Y.; Zhang, L.; Huang, W.-J.; Hou, W.; Fan, C.-F.; Jin, R.-H.; Feng, Y.-M.; Wang, Y.-C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Ricke-Hoch, M.; Stelling, E.; Lasswitz, L.; Gunesch, A.P.; Kasten, M.; Zapatero-Belinchón, F.J.; Brogden, G.; Gerold, G.; Pietschmann, T.; Montiel, V.; et al. Impaired immune response mediated by prostaglandin E2 promotes severe COVID-19 disease. PLoS ONE 2021, 16, e0255335. [Google Scholar] [CrossRef]
- Sena, K.; Furue, K.; Setoguchi, F.; Noguchi, K. Altered expression of SARS-CoV-2 entry and processing genes by Porphyromonas gingivalis-derived lipopolysaccharide, inflammatory cytokines and prostaglandin E2 in human gingival fibroblasts. Arch. Oral Biol. 2021, 129, 105201. [Google Scholar] [CrossRef]
- Kasela, S.; Daniloski, Z.; Bollepalli, S.; Jordan, T.X.; tenOever, B.R.; Sanjana, N.E.; Lappalainen, T. Integrative approach identifies SLC6A20 and CXCR6 as putative causal genes for the COVID-19 GWAS signal in the 3p21. 31 locus. Genome Biol. 2021, 22, 242. [Google Scholar] [CrossRef]
- Vuille-Dit-Bille, R.N.; Camargo, S.M.; Emmenegger, L.; Sasse, T.; Kummer, E.; Jando, J.; Hamie, Q.M.; Meier, C.F.; Hunziker, S.; Forras-Kaufmann, Z.; et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 2015, 47, 693–705. [Google Scholar] [CrossRef]
- Labor, M.; Vrbica, Ž.; Gudelj, I.; Labor, S.; Jurić, I.; Plavec, D. Exhaled Breath Temperature as a Novel Marker of Future Development of COPD: Results of a Follow-Up Study in Smokers. COPD J. Chronic Obstr. Pulm. Dis. 2016, 13, 741–749. [Google Scholar] [CrossRef]
- Patra, T.; Meyer, K.; Geerling, L.; Isbell, T.S.; Hoft, D.F.; Brien, J.; Pinto, A.K.; Ray, R.B.; Ray, R. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020, 16, e1009128. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Fang, J.; Yu, F.; Heng, D.; Fan, Y.; Xu, J.; Peng, B.; Liu, W.; Han, S.; et al. Decreased Methylation Level of H3K27me3 Increases Seizure Susceptibility. Mol. Neurobiol. 2017, 54, 7343–7352. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Gack, M.U. Distinct and Orchestrated Functions of RNA Sensors in Innate Immunity. Immunity 2020, 53, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lee, J.-H.; Parker, Z.M.; Acharya, D.; Chiang, J.J.; van Gent, M.; Riedl, W.; Davis-Gardner, M.E.; Wies, E.; Chiang, C.; et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021, 6, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Sato, S.; Sotoyama, Y.; Orba, Y.; Sawa, H.; Yamauchi, H.; Sasaki, M.; Takaoka, A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021, 22, 820–828. [Google Scholar] [CrossRef]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef]
- Kissling, E.; Hooiveld, M.; Sandonis Martín, V.; Martínez-Baz, I.; William, N.; Vilcu, A.M.; Mazagatos, C.; Domegan, L.; de Lusignan, S.; Meijer, A.; et al. Vaccine effectiveness against symptomatic SARS-CoV-2 infection in adults aged 65 years and older in primary care: I-MOVE-COVID-19 project, Europe, December 2020 to May 2021. Eurosurveillance 2021, 26, 2100670. [Google Scholar] [CrossRef]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 2022, 399, 924–944. [Google Scholar] [CrossRef]
- Williams, A.H.; Zhan, C.-G. Generalized Methodology for the Quick Prediction of Variant SARS-CoV-2 Spike Protein Binding Affinities with Human Angiotensin-Converting Enzyme II. J. Phys. Chem. B 2022, 126, 2353–2360. [Google Scholar] [CrossRef]
| Dapsone-Administered (+)/Nonprescribed (–) | Decreased FIO2 | Others | Row Totals | Dapsone-Administered (+) Group | Decreased FIO2 + No Progressive ARDS | Progressive ARDS | Row Totals |
|---|---|---|---|---|---|---|---|
| Dapsone (+) onset *1 | 7 | 1 | 8 | Dapsone (+) onset *2 + Aggravated *3 | 17 | 3 | 20 |
| Dapsone (–) onset | 8 | 12 | 20 | Dapsone (+) severe *4 | 0 | 2 | 2 |
| Totals | 15 | 13 | 28 | Total | 17 | 5 | 22 |
| Factor | Receptor Mediator | Role in COVID-19 | Histone Marks in 1 mdig+ Epithelial Cells | Ref. |
|---|---|---|---|---|
| Aging | ACE2 | Age-dependent DNA methylation was observed close to the transcription start site of the ACE2 gene in airway epithelial cells. Histone modifications and the levels of histone proteins change during aging, dramatically influencing chromatin compaction and gene expression. | ↓ 3 H3K9me3 ↓3 H3K27me3 ↑3 H3K36me3 ↑3 H4K20me3 | [99] [100] [101] [102] |
| DNA methylation | *↑ methylation of IFN-related genes. **↓ Inflammatory genes hypomethylated. | [97] | ||
| Smoking | Epigenetic mechanisms alter the common transcriptional bridge between smoking and COVID-19 by trimethylation of particular lysine (K) residues at H3 and H4 histones. | ↑ H3K4me3 ↓ H3K9me3 ↓ H3K27me3 ↓ H4K20me3 | [103] | |
| ACE2 | The primary receptor mediating SARS-CoV-2 entry, the expression of ACE2 is maintained, if not upregulated, due to the arsenic-induced impaired activity of 2 EZH2. | ↓ 3 H3K27me3 | [104] [105] [106] | |
| 4 NRP1 | They are highly expressed in the respiratory tract epithelial cells; NRP1 binds the S1 segment of the SARS-CoV-2 spike protein following its cleavage by furin. | ↑ 3 H3K4me3 ↓ 3 H3K9me3 | [104] [107] | |
| NRP2 | Similar to NRP1. | ↑ 3 H3K4me3 ↓ 3 H3K9me3 ↓ 3 H3K27me3 | [104] [108] | |
| 5 AT1R | It facilitates SARS-CoV-2 entry through receptor-mediated endocytosis of ACE2-S complex following the interaction of viral spike protein with soluble ACE2. | ↓ 3 H3K9me3 | [104] [109] [110] | |
| 6 CTSD | Potentially facilitates SARS-CoV-2 entry through positive regulation of furin through osteopontin. | ↓ 3 H3K9me3 ↓ 3 H3K27me3 | [104] [111,112,113] | |
| 7 CTSL | Elevated in the serum of COVID-19 patients, CTSL mediates viral entry by participating in the cleavage of the viral spike protein. | ↑ 3 H3K4me3 ↓ 3 H3K9me3 ↓ 3 H3K27me3 | [104] [114] | |
| 8 PTGER2-4 | Upregulation of PGE2 receptors might potentiate the positive regulatory effect of PGE2 on ACE2 and TMPRSS2, facilitating SARS-CoV-2 entry. | ↑ 3 H3K4me3 | [104] [115,116] | |
| 9 SLC6A20/SIT1 | Positively regulated by ACE2, SLC6A20/SIT1 is suspected of interacting with ACE2 and enhancing its activity reciprocally. | ↓ 3 H3K27me3 | [104] [117,118] | |
| IL-6 | Present in high levels in the serum of COVID-19 patients, IL-6 is speculated to enhance viral entry by activating the AT1R signaling cascade. | ↓ 3 H3K27me3 | [110,119,120,121] | |
| 10 ISG | 11 HDAC | An ISG expression signature is a hallmark of interferonopathies and other autoimmune diseases. The combined inhibition of HDAC1/2 and 12BRD4 resolved the aberrant ISG expression detected in cells derived from patients with two inherited interferonopathies [98]. | ↑ 3 H3K36me3 | [101] |
| 13MDA5 | MDA5 preferentially binds negative-strand SARS-CoV-2 RNA. Active viral replication is required for triggering MDA5 activation. Type I/III IFN induction by SARS-CoV-2 relies on MDA5. | In study | [122] [123] | |
| 14 RIG-I | RIG-I binds preferentially blunt-ended short dsRNA bearing a 5’ triphosphate moiety such as the 15IAV (sub)genomic panhandle structure and exerts IFN-independent antiviral activity by competing with the viral polymerase for binding to the 3’ untranslated region (UTR) of the SARS-CoV-2 genomic RNA. Both MDA5 and RIG-I may contribute to SARS-CoV-2 restriction in a temporal manner. | In study | [124] [125] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.H.; Kanwar, B.; Khattak, A.; Balentine, J.; Nguyen, N.H.; Kast, R.E.; Lee, C.J.; Bourbeau, J.; Altschuler, E.L.; Sergi, C.M.; et al. COVID-19 Molecular Pathophysiology: Acetylation of Repurposing Drugs. Int. J. Mol. Sci. 2022, 23, 13260. https://doi.org/10.3390/ijms232113260
Lee JH, Kanwar B, Khattak A, Balentine J, Nguyen NH, Kast RE, Lee CJ, Bourbeau J, Altschuler EL, Sergi CM, et al. COVID-19 Molecular Pathophysiology: Acetylation of Repurposing Drugs. International Journal of Molecular Sciences. 2022; 23(21):13260. https://doi.org/10.3390/ijms232113260
Chicago/Turabian StyleLee, Jong Hoon, Badar Kanwar, Asif Khattak, Jenny Balentine, Ngoc Huy Nguyen, Richard E. Kast, Chul Joong Lee, Jean Bourbeau, Eric L. Altschuler, Consolato M. Sergi, and et al. 2022. "COVID-19 Molecular Pathophysiology: Acetylation of Repurposing Drugs" International Journal of Molecular Sciences 23, no. 21: 13260. https://doi.org/10.3390/ijms232113260
APA StyleLee, J. H., Kanwar, B., Khattak, A., Balentine, J., Nguyen, N. H., Kast, R. E., Lee, C. J., Bourbeau, J., Altschuler, E. L., Sergi, C. M., Nguyen, T. N. M., Oh, S., Sohn, M.-G., & Coleman, M. (2022). COVID-19 Molecular Pathophysiology: Acetylation of Repurposing Drugs. International Journal of Molecular Sciences, 23(21), 13260. https://doi.org/10.3390/ijms232113260









