Genome-Wide Characterization of Chrysanthemum indicum Nuclear Factor Y, Subunit C Gene Family Reveals the Roles of CiNF-YCs in Flowering Regulation
Abstract
1. Introduction
2. Results
2.1. Identification of the NF-YC Family Members
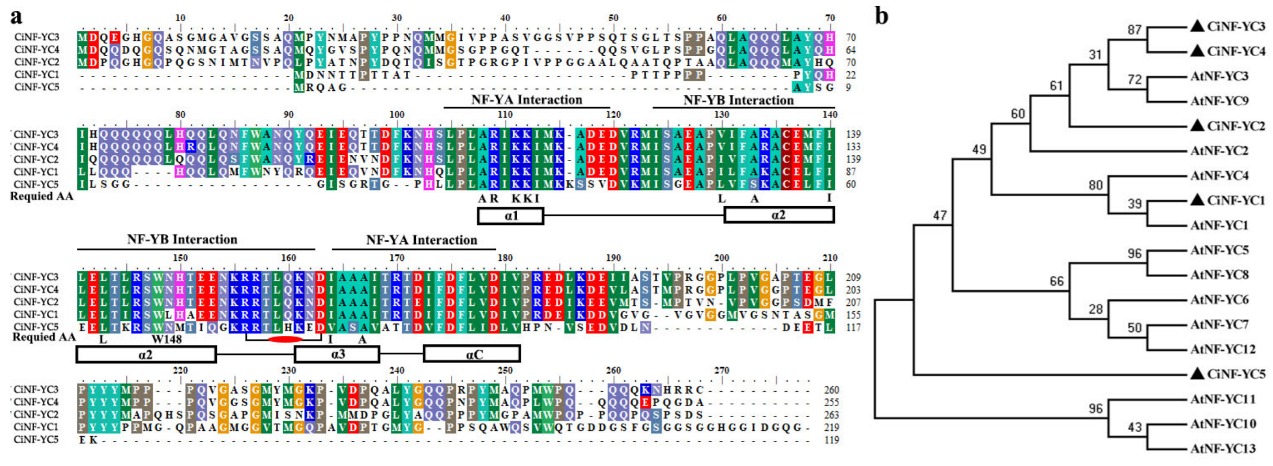
2.2. Multiple Alignment and Phylogenetic Analysis of NF-YC Proteins in Wild Chrysanthemum
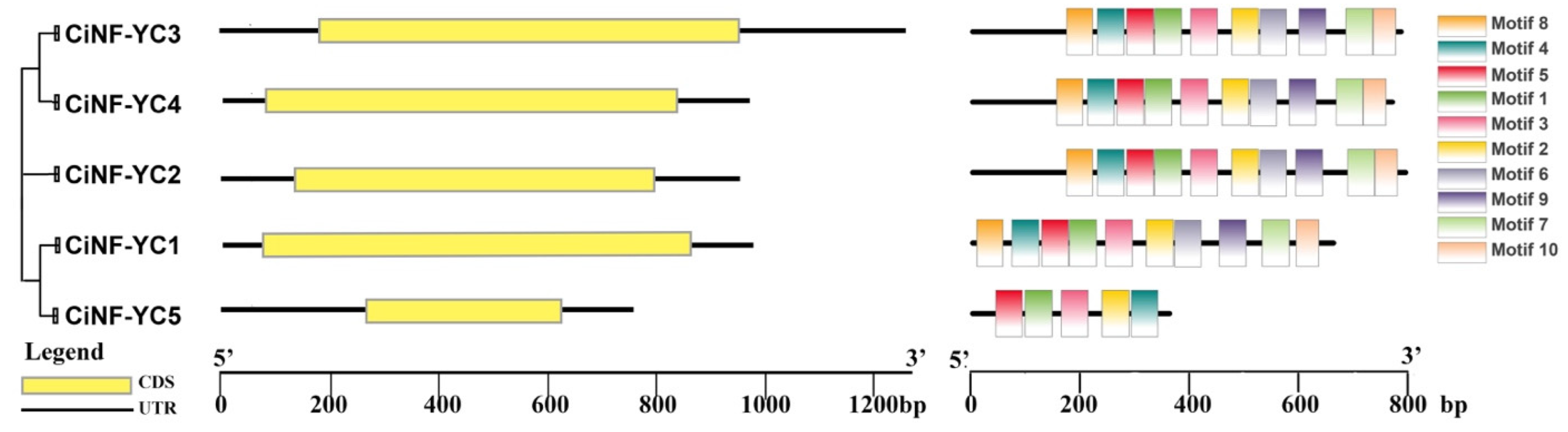
2.3. Gene Structure and Conserved Motif Analyses of NF-YC in Wild Chrysanthemum
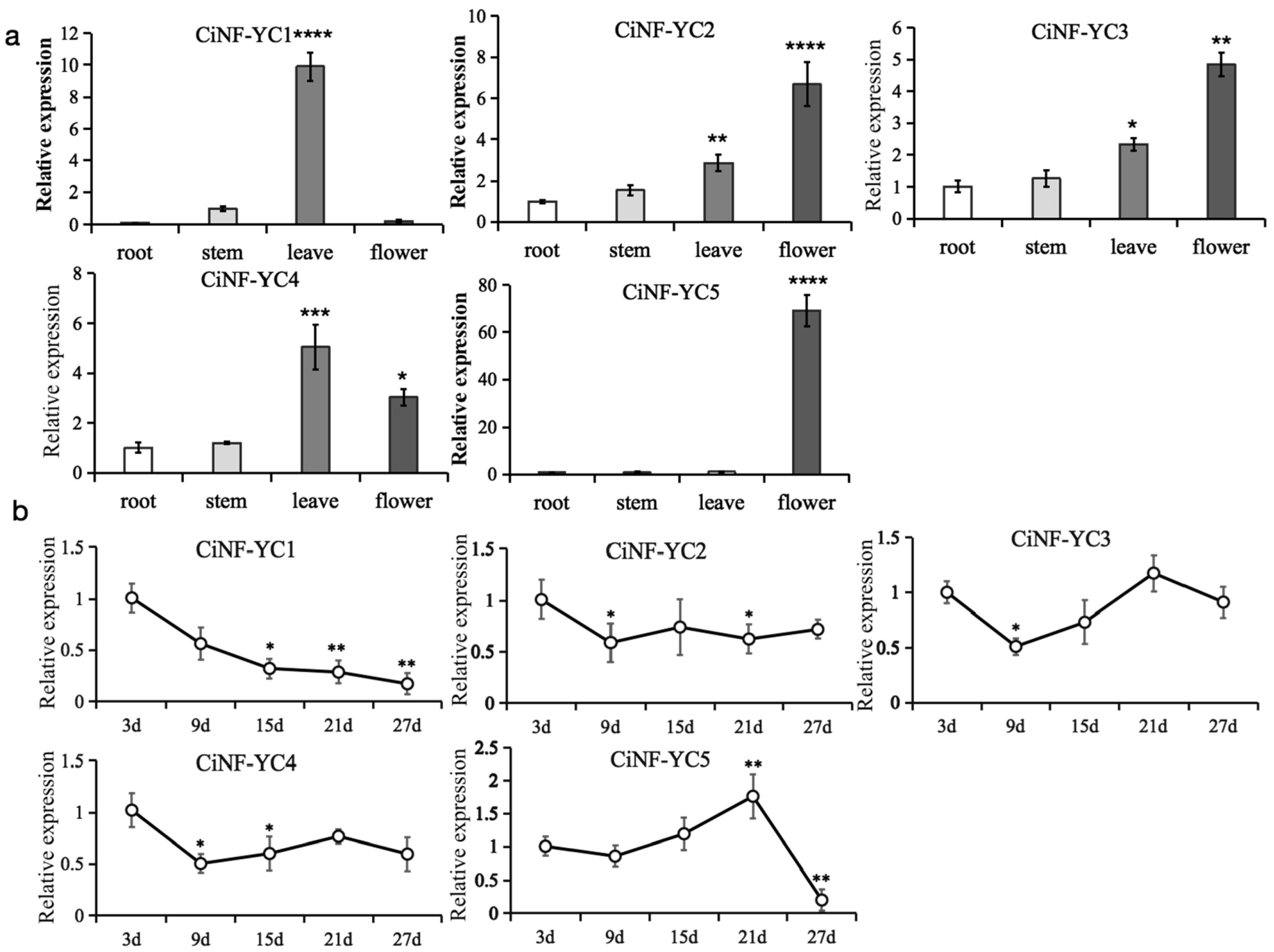
2.4. Expression Profiles of CiNF-YC Genes
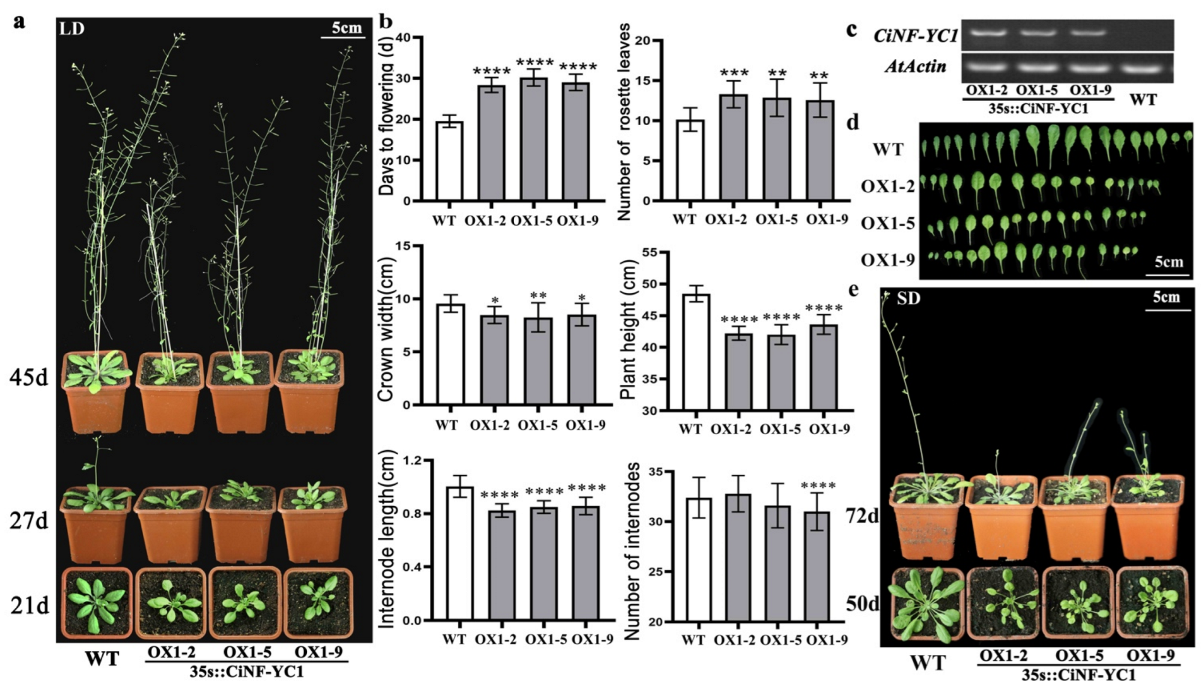
2.5. Modified Flowering Time of CiNF-YC1 Overexpressing Arabidopsis
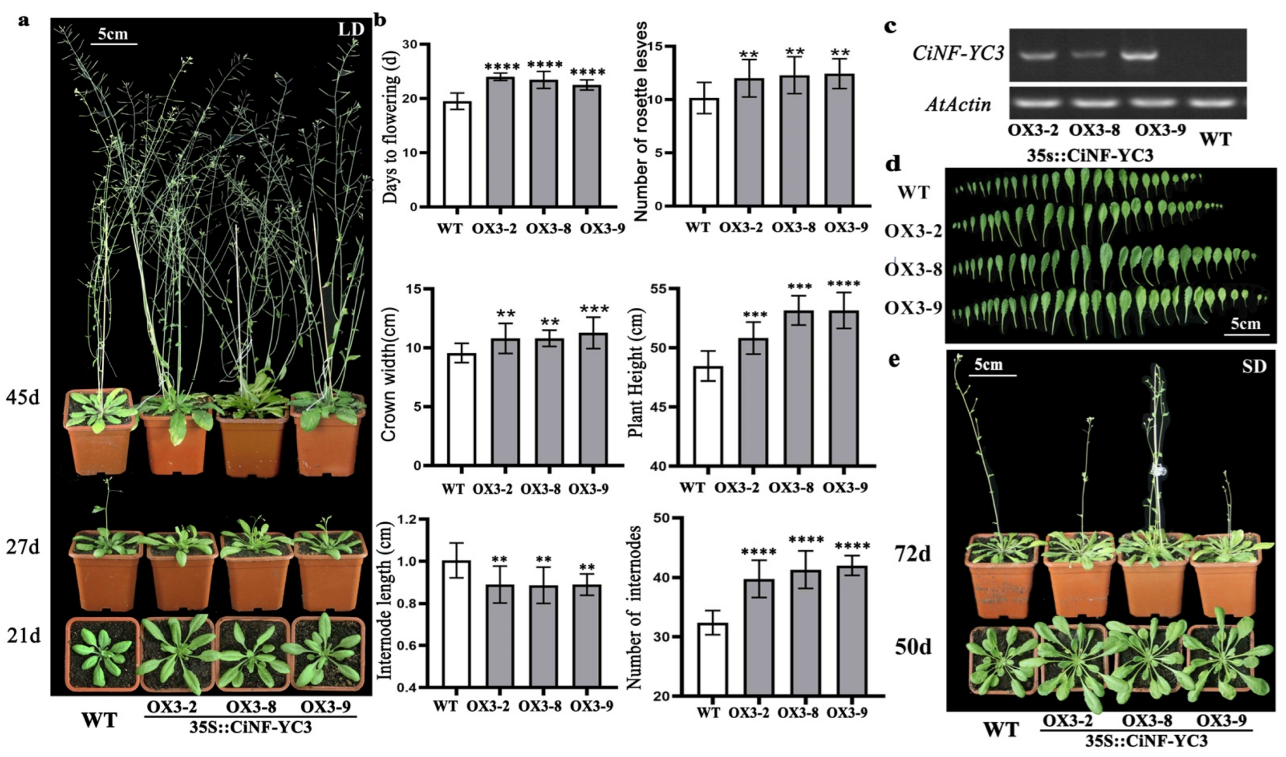
2.6. Effects of CiNF-YC3 on Flowering and Growth of Arabidopsis
2.7. Modified Expression of Genes Involved in Flowering in CiNF-YC1 and CiNF-YC3 Overexpressor Lines
3. Discussion
3.1. Characterization of the NF-YC Gene Family in Chrysanthemum Indicum
3.2. Expression Patterns of CiNF-YC Genes in Various Developmental Stages and Tissues
3.3. Function of CiNF-YC Genes in Flowering
3.4. Possible Flowering Pathways That Involved by CiNF-YC1 and CiNF-YC3
4. Materials and Methods
4.1. Identification, Alignments, and Phylogenetic Analysis of NF-YCs in Wild Chrysanthemum
4.2. Gene Structure and Motif Analyses of NF-YCs in Wild Chrysanthemum
4.3. Plant Materials and Culture Conditions
4.4. RNA Isolation and RT-qPCR Analysis
4.5. Generation of CiNF-YC1 and CiNF-YC3 Overexpressing Arabidopsis
4.6. Functional Validation of CiNF-YC1 and CiNF-YC3 in Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Srikanth, A.; Schmid, M. Regulation of Flowering Time: All Roads Lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of Flowering in Arabidopsis. Cell 2010, 141, 550.e1–550.e2. [Google Scholar] [CrossRef] [PubMed]
- Amasino, R.M.; Michaels, S.D. The Timing of Flowering. Plant Physiol. 2010, 154, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, V.; Fornara, F. Y Flowering? Regulation and Activity of CONSTANS and CCT-Domain Proteins in Arabidopsis and Crop Species. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 655–660. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, C.; Xu, Y.; Wang, T.; Chen, Y.; Lü, J.; Zhang, L.; Jiang, C.-Z.; Hong, B.; Gao, J. Control of Chrysanthemum Flowering through Integration with an Aging Pathway. Nat. Commun. 2017, 8, 829. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y Subunits NF-YB2 and NF-YB3 Play Additive Roles in the Promotion of Flowering by Inductive Long-Day Photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Hwang, Y.-H.; Kim, S.K.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.-K. Functional Conservation of Rice OsNF-YB/YC and Arabidopsis AtNF-YB/YC Proteins in the Regulation of Flowering Time. Plant Cell Rep. 2016, 35, 857–865. [Google Scholar] [CrossRef]
- Shen, C.; Liu, H.; Guan, Z.; Yan, J.; Zheng, T.; Yan, W.; Wu, C.; Zhang, Q.; Yin, P.; Xing, Y. Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering. Plant Cell 2020, 32, 3469–3484. [Google Scholar] [CrossRef]
- Nardone, V.; Chaves-Sanjuan, A.; Nardini, M. Structural Determinants for NF-Y/DNA Interaction at the CCAAT Box. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 571–580. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC Structure Gives Insight into DNA Binding and Transcription Regulation by CCAAT Factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R.M.; et al. NF-Y Plays Essential Roles in Flavonoid Biosynthesis by Modulating Histone Modifications in Tomato. New Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-BHLH144 Complex Directly Activates Wx to Regulate Grain Quality in Rice (Oryza Sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.M.; Li, Z.X.; Ran, Q.J.; Li, P.; Peng, Z.H.; Zhang, J.R. ZmNF-YB16 Overexpression Improves Drought Resistance and Yield by Enhancing Photosynthesis and the Antioxidant Capacity of Maize Plants. Front. Recent Dev. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Ma, Y.; Wang, X.F.; Zhang, D.P. Overexpression of the Transcription Factor NF-YC9 Confers Abscisic Acid Hypersensitivity in Arabidopsis. Plant Mol. Biol. 2017, 95, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Tan, H.; Hong, S.; Liang, Y.; Zuo, J. Arabidopsis Transcription Factor Genes NF-YA1, 5, 6, and 9 Play Redundant Roles in Male Gametogenesis, Embryogenesis, and Seed Development. Mol. Plant 2013, 6, 188–201. [Google Scholar] [CrossRef]
- Combier, J.P.; Frugier, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Verni, T.; Ott, T.; Gamas, P.; Crespi, M.; Niebel, A.; et al. MtHAP2-1 Is a Key Transcriptional Regulator of Symbiotic Nodule Development Regulated by MicroRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef]
- Han, X.; Tang, S.; An, Y.; Zheng, D.C.; Xia, X.L.; Yin, W.L. Overexpression of the Poplar NF-YB7 Transcription Factor Confers Drought Tolerance and Improves Water-Use Efficiency in Arabidopsis. J. Exp. Bot. 2013, 64, 4589–4601. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, Y.; Zhuo, C.; Lu, S.; Guo, Z. Overexpression of a NF-YC Transcription Factor from Bermudagrass Confers Tolerance to Drought and Salinity in Transgenic Rice. Plant Biotechnol. J. 2014, 13, 482–491. [Google Scholar] [CrossRef]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef]
- Hackenberg, D.; Keetman, U.; Grimm, B. Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering. Int. J. Mol. Sci. 2012, 13, 3458–3477. [Google Scholar] [CrossRef]
- Kim, S.K.; Park, H.Y.; Jang, Y.H.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.K. OsNF-YC2 and OsNF-YC4 Proteins Inhibit Flowering under Long-Day Conditions in Rice. Planta 2016, 243, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F. NF-YC3, NF-YC4 and NF-YC9 Are Required for CONSTANS-Mediated, Photoperiod-Dependent Flowering in Arabidopsis thaliana. Plant J. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, K.; Yang, X.; Guo, B.; Xue, Y.; Miao, D.; Huang, S.; An, X. Comprehensive Analyses of Four PtoNF-YC Genes from Populus Tomentosa and Impacts on Flowering Timing. Int. J. Mol. Sci. 2022, 23, 3116. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, Y.; Hu, Y.; Zhou, L.; Li, Y.; Hou, X. Temporal-Specific Interaction of NF-YC and CURLY LEAF during the Floral Transition Regulates Flowering. Plant Physiol. 2018, 177, 105–114. [Google Scholar] [CrossRef]
- Mei, X.; Li, P.; Wang, L.; Liu, C.; Zhou, L.; Cai, Y. Molecular and Functional Characterization of ZmNF-YC14 in Transgenic Arabidopsis. J. Plant Biol. 2018, 61, 410–423. [Google Scholar] [CrossRef]
- Van Lieshout, N.; van Kaauwen, M.; Kodde, L.; Arens, P.; Smulders, M.J.M.; Visser, R.G.F.; Finkers, R. De Novo Whole-Genome Assembly of Chrysanthemum Makinoi, a Key Wild Chrysanthemum. G3 Genes Genomes Genet. 2022, 12, jkab358. [Google Scholar] [CrossRef]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-Wide Analysis of Tomato NF-Y Factors and Their Role in Fruit Ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef]
- Quach, T.N.; Nguyen, H.T.M.H.T.; Valliyodan, B.; Joshi, T.; Xu, D.; Nguyen, H.T. Genome-Wide Expression Analysis of Soybean NF-Y Genes Reveals Potential Function in Development and Drought Response. Mol. Genet. Genom. 2015, 290, 1095–1115. [Google Scholar] [CrossRef]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.P. Genome-Wide Identification and Expression Analysis of the NF-Y Family of Transcription Factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef]
- Malviya, N.; Jaiswal, P.; Yadav, D. Genome-Wide Characterization of Nuclear Factor Y (NF-Y) Gene Family of Sorghum [Sorghum bicolor (L.) Moench]: A Bioinformatics Approach. Physiol. Mol. Biol. Plants 2016, 22, 33–49. [Google Scholar] [CrossRef]
- Li, J.; Gao, K.; Khan, W.U.; Yang, X.; Yang, X.; Zhao, T.; Chen, Z.; An, X. Genome-wide analysis of the poplar NF-Y gene family and its. expression in floral bud development of Populus tomentosa. Trees 2020, 34, 285–296. [Google Scholar] [CrossRef]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum IV, W.E.; Tayrose, G.; Holt, B.F. Tissue-Specific Expression Patterns of Arabidopsis NF-Y Transcription Factors Suggest Potential for Extensive Combinatorial Complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef]
- Huang, S.; Hu, L.; Xu, D.; Li, W.; Xu, Z.; Li, L.; Zhou, Y.; Diao, X.; Jia, G.; Ma, Y.; et al. Transcription Factor SiNF-YA5 from Foxtail Millet (Setaria italica) Conferred Tolerance to High-Salt Stress through ABA-Independent Pathway in Transgenic Arabidopsis. Acta Agron. Sin. 2016, 42, 1787–1797. [Google Scholar] [CrossRef]
- Cao, S.; Kumimoto, R.W.; Siriwardana, C.L.; Risinger, J.R.; Holt, B.F. Identification and Characterization of NF-Y Transcription Factor Families in the Monocot Model Plant Brachypodium distachyon. PLoS ONE 2011, 6, e21805. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Han, M.; Lv, Q.; Bao, F.; He, Y. Identification and Expression Profile Analysis of NUCLEAR FACTOR-Y Families in Physcomitrella patens. Front. Plant Sci. 2015, 6, 642. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Wang, Y.; Zhu, J.; Zhang, Y.; Hou, H. Genomic Organization, Phylogenetic Comparison, and Differential Expression of the Nuclear Factor-Y Gene Family in Apple (Malus domestica). Plants 2020, 10, 16. [Google Scholar] [CrossRef]
- Wan, Q.; Luo, L.; Zhang, X.; Lv, Y.; Zhu, S.; Kong, L.; Wan, Y.; Liu, F.; Zhang, K. Genome-Wide Identification and Abiotic Stress Response Pattern Analysis of NF-Y Gene Family in Peanut (Arachis hypogaea L.). Trop. Plant Biol. 2021, 14, 329–344. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.; Liang, Z. Genome-Wide Identification and Characterization of the NF-Y Gene Family in Grape (Vitis vinifera L.). BMC Genom. 2016, 17, 605. [Google Scholar] [CrossRef]
- Rípodas, C.; Castaingts, M.; Clúa, J.; Blanco, F.; Zanetti, M.E. Annotation, Phylogeny and Expression Analysis of the Nuclear Factor Y Gene Families in Common Bean (Phaseolus vulgaris). Front. Plant Sci. 2015, 5, 761. [Google Scholar] [CrossRef]
- Panzade, K.P.; Kale, S.S.; Manoj, M.L.; Kothawale, S.P.; Damse, D.N. Genome-Wide Analysis and Expression Profile of Nuclear Factor Y (NF-Y) Gene Family in Z. jujuba. Appl. Biochem. Biotechnol. 2022, 194, 1373–1389. [Google Scholar] [CrossRef]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, G.; Liu, W.; Dong, X.; Zhang, A. Genome-Wide Analysis of the NF-Y Gene Family in Peach (Prunus persica L.). BMC Genom. 2019, 20, 612. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.L.S.; Martins, C.P.S.; Sousa, A.O.; Camillo, L.R.; Araújo, C.P.; Alcantara, G.M.; Camargo, D.S.; Cidade, L.C.; de Almeida, A.F.; Costa, M.G.C. Genome-Wide Characterization and Expression Analysis of Citrus NUCLEAR FACTOR-Y (NF-Y) Transcription Factors Identified a Novel NF-YA Gene Involved in Drought-Stress Response and Tolerance. PLoS ONE 2018, 13, e0199187. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhu, J.; Yang, Y. Genome-Wide Identification and Expression Analysis of NF-Y Transcription Factor Families in Watermelon (Citrullus lanatus). J. Plant Growth Regul. 2017, 36, 590–607. [Google Scholar] [CrossRef]
- Wei, Q.; Wen, S.; Lan, C.; Yu, Y.; Chen, G. Genome-Wide Identification and Expression Profile Analysis of the NF-Y Transcription Factor Gene Family in Petunia hybrida. Plants 2020, 9, 336. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, H.; Zhang, L.; Xu, F.; Shi, L.; Wang, S.; Hong, J.; Ding, G. Identification and Comprehensive Analysis of the Nuclear Factor-Y Family Genes Reveal Their Multiple Roles in Response to Nutrient Deficiencies in Brassica napus. Int. J. Mol. Sci. 2021, 22, 10354. [Google Scholar] [CrossRef]
- Song, C.; Liu, Y.; Song, A.; Dong, G.; Zhao, H.; Sun, W.; Ramakrishnan, S.; Wang, Y.; Wang, S.; Li, T.; et al. The Chrysanthemum nankingense Genome Provides Insights into the Evolution and Diversification of Chrysanthemum Flowers and Medicinal Traits. Mol. Plant 2018, 11, 1482–1491. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Zhu, J.; Ma, W.; Li, Z.; Bi, Z.; Sun, C.; Bai, J.; Zhang, J.; Liu, Y. Genome-Wide Identification and Analysis of the NF-Y Gene Family in Potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 739989. [Google Scholar] [CrossRef]
- Feng, Z.J.; He, G.H.; Zheng, W.J.; Lu, P.P.; Chen, M.; Gong, Y.M.; Ma, Y.Z.; Xu, Z.S. Foxtail millet NF-Y Families: Genome-Wide Survey and Evolution Analyses Identified Two Functional Genes Important in Abiotic Stresses. Front. Plant Sci. 2015, 6, 1142. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 Represents a Functionally Specialized Subunit of the CCAAT Binding Transcription Factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef]
- Koralewski, T.E.; Krutovsky, K.V. Evolution of Exon-Intron Structure and Alternative Splicing. PLoS ONE 2011, 6, e18055. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Chen, C.; Hussain, N.; Wang, Y.; Li, M.; Liu, L.; Qin, M.; Ma, N.; Gao, J.; Sun, X. An Ethylene-Inhibited NF-YC Transcription Factor RhNF-YC9 Regulates Petal Expansion in Rose. Hortic. Plant J. 2020, 6, 419–427. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 Module Integrates GA and ABA Signalling to Regulate Seed Germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Hu, Y.; Yang, Y.; Cai, J.; Liu, H.; Zhang, C.; Liu, X.; Hou, X. Arabidopsis NF-YCs Play Dual Roles in Repressing Brassinosteroid Biosynthesis and Signaling during Light-Regulated Hypocotyl Elongation. Plant Cell 2021, 33, 2360–2374. [Google Scholar] [CrossRef]
- Mei, X.; Nan, J.; Zhao, Z.; Yao, S.; Wang, W.; Yang, Y.; Bai, Y.; Dong, E.; Liu, C.; Cai, Y. Maize Transcription Factor ZmNF-YC13 Regulates Plant Architecture. J. Exp. Bot. 2021, 72, 4757–4772. [Google Scholar] [CrossRef]
- Xiong, Y.; Ren, Y.; Li, W.; Wu, F.; Yang, W.; Huang, X.; Yao, J. NF-YC12 Is a Key Multi-Functional Regulator of Accumulation of Seed Storage Substances in Rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [CrossRef]
- Gao, R.; Wang, Y.; Gruber, M.Y.; Hannoufa, A. MiR156/SPL10 Modulates Lateral Root Development, Branching and Leaf Morphology in Arabidopsis by Silencing AGAMOUS-LIKE 79. Front. Plant Sci. 2018, 8, 2226. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists Integrating Various Biological Data Handling Tools with a User-Friendly Interface. bioRxiv 2018. [Google Scholar] [CrossRef]

| Gene Name | Genome ID | NCBI ID | mRNA Length (bp) | Peptide Residues (aa) | MW (kDa) | pI |
|---|---|---|---|---|---|---|
| CiNF-YC1 | CHR00079409-RA | IABW01058102.1 | 659 | 220 | 23.89 | 4.82 |
| CiNF-YC2 | CHR00063225-RA | IABW01127607.1 | 796 | 264 | 29.44 | 5.27 |
| CiNF-YC3 | CHR00082598-RA | IABW01051895.1 | 786 | 261 | 29.13 | 6.89 |
| CiNF-YC4 | CHR00018499-RA | IABW01098859.1 | 768 | 256 | 28.66 | 5.04 |
| CiNF-YC5 | CHR00020456-RA | IABW01033460.1 | 360 | 120 | 13.10 | 6.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yao, Y.; Wen, S.; Bin, J.; Tan, Q.; Lou, J.; Xie, L.; Zeng, R.; Guo, H.; Zhang, Z.; et al. Genome-Wide Characterization of Chrysanthemum indicum Nuclear Factor Y, Subunit C Gene Family Reveals the Roles of CiNF-YCs in Flowering Regulation. Int. J. Mol. Sci. 2022, 23, 12812. https://doi.org/10.3390/ijms232112812
Wang X, Yao Y, Wen S, Bin J, Tan Q, Lou J, Xie L, Zeng R, Guo H, Zhang Z, et al. Genome-Wide Characterization of Chrysanthemum indicum Nuclear Factor Y, Subunit C Gene Family Reveals the Roles of CiNF-YCs in Flowering Regulation. International Journal of Molecular Sciences. 2022; 23(21):12812. https://doi.org/10.3390/ijms232112812
Chicago/Turabian StyleWang, Xueting, Yao Yao, Shiyun Wen, Jing Bin, Qinghua Tan, Jinpeng Lou, Li Xie, Ruizhen Zeng, Herong Guo, Zhisheng Zhang, and et al. 2022. "Genome-Wide Characterization of Chrysanthemum indicum Nuclear Factor Y, Subunit C Gene Family Reveals the Roles of CiNF-YCs in Flowering Regulation" International Journal of Molecular Sciences 23, no. 21: 12812. https://doi.org/10.3390/ijms232112812
APA StyleWang, X., Yao, Y., Wen, S., Bin, J., Tan, Q., Lou, J., Xie, L., Zeng, R., Guo, H., Zhang, Z., & Wei, Q. (2022). Genome-Wide Characterization of Chrysanthemum indicum Nuclear Factor Y, Subunit C Gene Family Reveals the Roles of CiNF-YCs in Flowering Regulation. International Journal of Molecular Sciences, 23(21), 12812. https://doi.org/10.3390/ijms232112812






