Restriction of Flaviviruses by an Interferon-Stimulated Gene SHFL/C19orf66
Abstract
1. Introduction
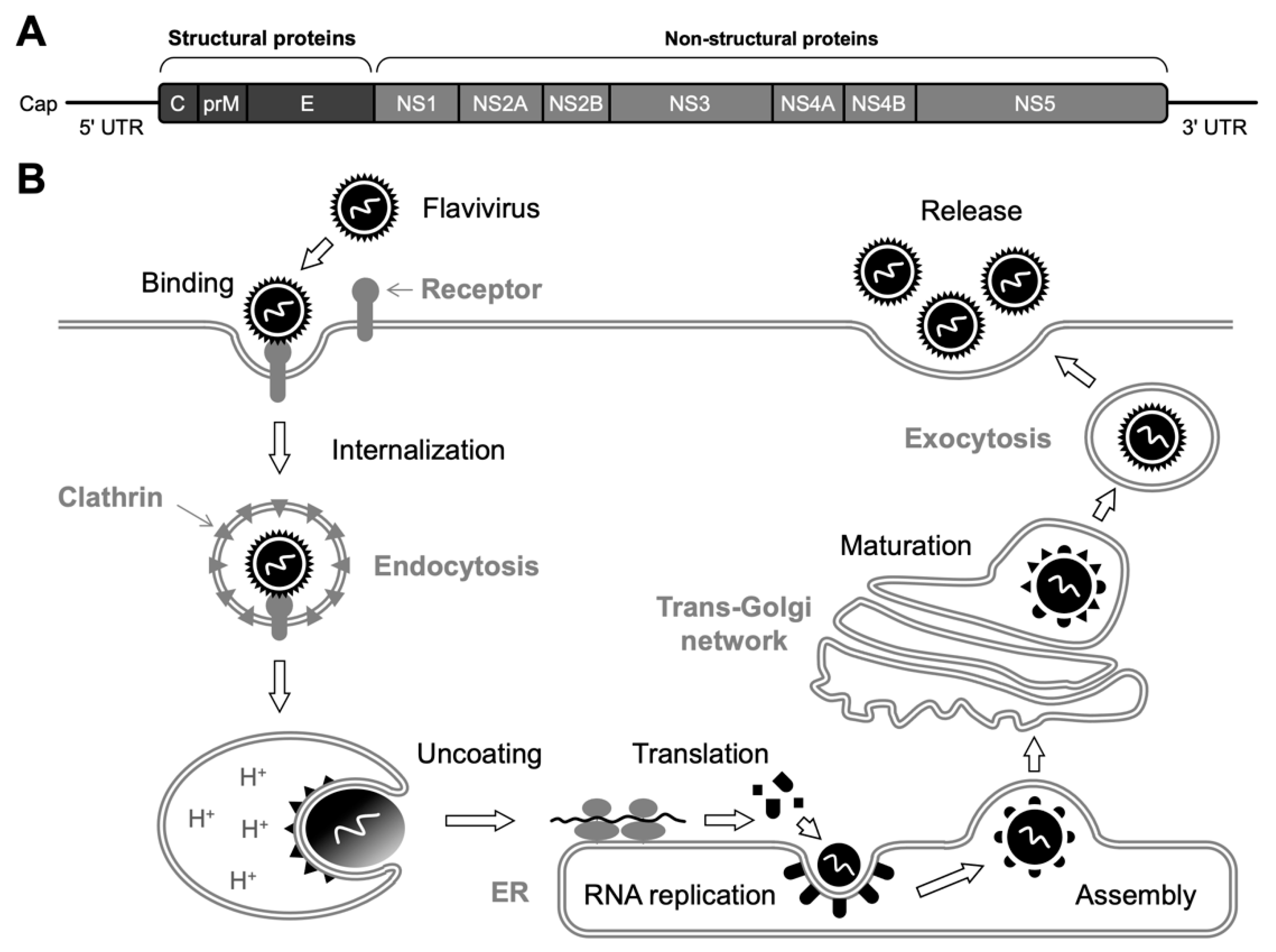
2. Flaviviruses
3. Identification of SHFL as a Novel Cellular Inhibitor against Flaviviruses
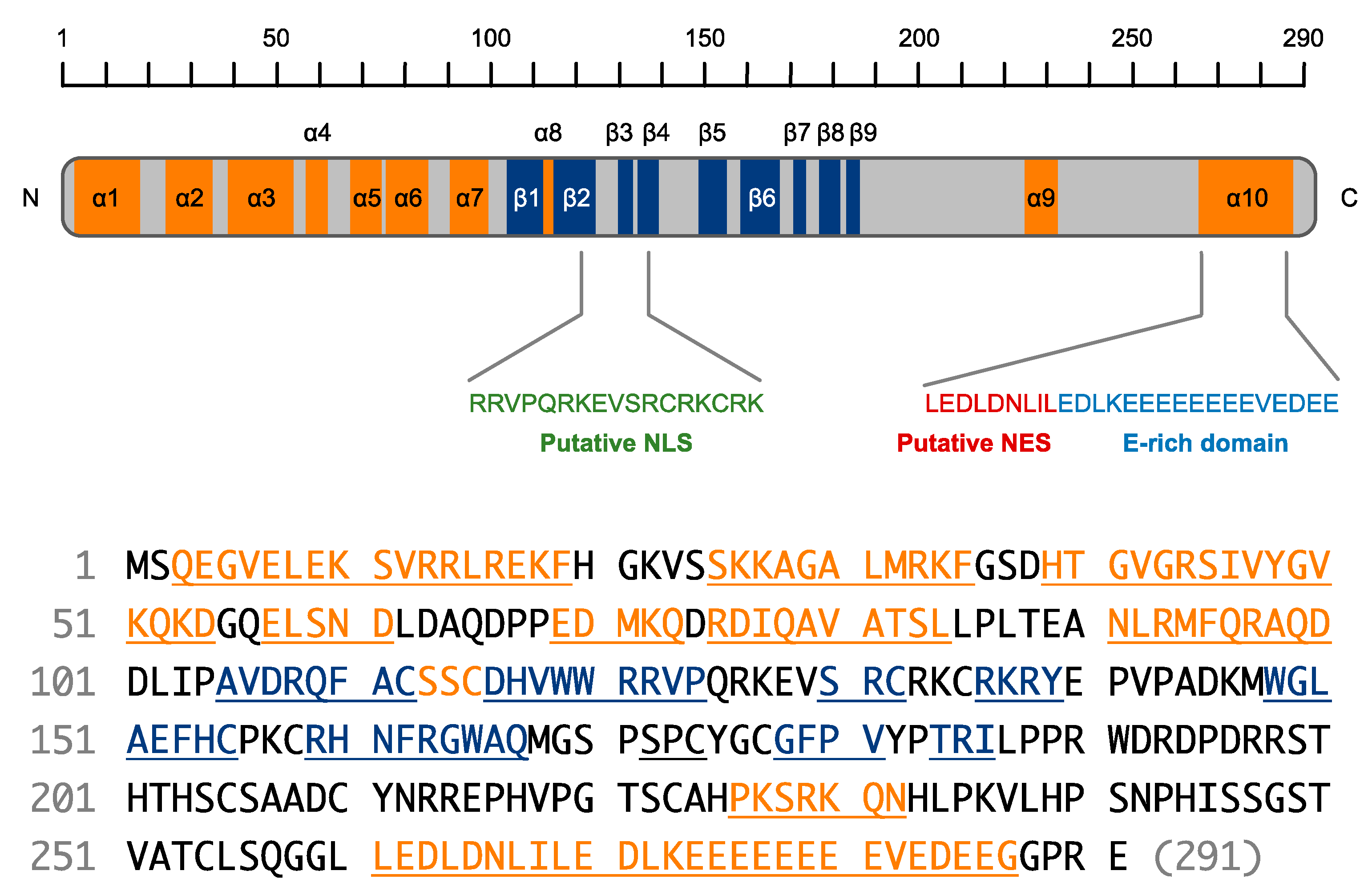
4. Domain Features of SHFL
5. Molecular Aspects of SHFL in the Inhibition of Flaviviruses
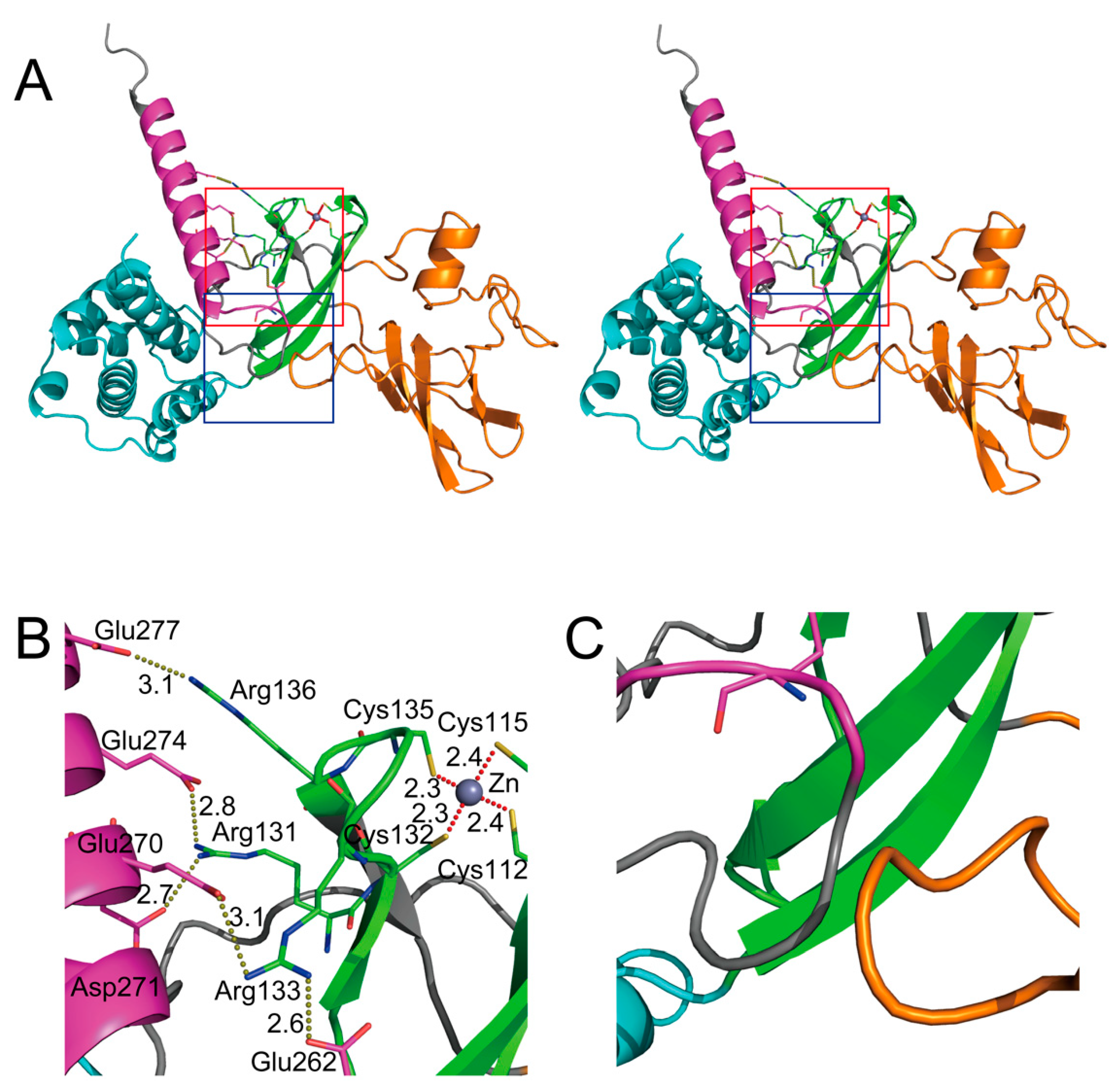
6. Structural Interpretation of Key Domains in SHFL—Seeing Is Believing
7. Anti-Flavivirus Activity of SHFL In Vivo
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-stimulated genes: What do they all do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef] [PubMed]
- HGNC. Symbol Report for SHFL. Available online: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:2160 (accessed on 15 July 2020).
- Berthoux, L. The restrictome of flaviviruses. Virol. Sin. 2020, 35, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Recent advances in antiviral interferon-stimulated gene biology. F1000Research 2018, 7, 309. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, W.; Muller, M. Shiftless, a critical piece of the innate immune response to viral infection. Viruses 2022, 14, 1338. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Cortese, M.; Acosta, E.G.; Bartenschlager, R. Rewiring cellular networks by members of the flaviviridae family. Nat. Rev. Microbiol. 2018, 16, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Wikan, N.; Smith, D.R. Zika virus: History of a newly emerging arbovirus. Lancet Infect. Dis. 2016, 16, e119–e126. [Google Scholar] [CrossRef]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus entry receptors: An update. Viruses 2013, 6, 69–88. [Google Scholar] [CrossRef]
- Bollati, M.; Alvarez, K.; Assenberg, R.; Baronti, C.; Canard, B.; Cook, S.; Coutard, B.; Decroly, E.; de Lamballerie, X.; Gould, E.A.; et al. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antivir. Res. 2010, 87, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lok, S.-M.; Yu, I.-M.; Zhang, Y.; Kuhn, R.J.; Chen, J.; Rossmann, M.G. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 2008, 319, 1830–1834. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Chin, W.-X.; Han, Q.; Ichiyama, K.; Lee, C.H.; Eyo, Z.W.; Ebina, H.; Takahashi, H.; Takahashi, C.; Tan, B.H.; et al. Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLoS Pathog. 2016, 12, e1005357. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.; Cherry, S. Virus-host interactions: From unbiased genetic screens to function. Annu. Rev. Virol. 2015, 2, 497–524. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/55337 (accessed on 28 September 2022).
- Wang, X.; Xuan, Y.; Han, Y.; Ding, X.; Ye, K.; Yang, F.; Gao, P.; Goff, S.P.; Gao, G. Regulation of HIV-1 gag-pol expression by shiftless, an inhibitor of programmed-1 ribosomal frameshifting. Cell 2019, 176, 625–635.e14. [Google Scholar] [CrossRef] [PubMed]
- JPred. Available online: http://www.compbio.dundee.ac.uk/jpred4/index.html (accessed on 28 September 2022).
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Balinsky, C.A.; Schmeisser, H.; Wells, A.I.; Ganesan, S.; Jin, T.; Singh, K.; Zoon, K.C. IRAV (FLJ11286), an interferon-stimulated gene with antiviral activity against dengue virus, interacts with MOV10. J. Virol. 2017, 91, e01616. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, X.; Yao, Z.; Dong, X.; Zhang, D.; Hu, Y.; Zhang, S.; Lin, J.; Chen, J.; An, S.; et al. C19orf66 interrupts Zika virus replication by inducing lysosomal degradation of viral NS3. PLoS Negl. Trop. Dis. 2020, 14, e0008083. [Google Scholar] [CrossRef]
- Kinast, V.; Plociennikowska, A.; Bracht, T.; Todt, D.; Brown, R.J.P.; Boldanova, T.; Zhang, Y.; Brüggemann, Y.; Friesland, M. C19orf66 is an interferon-induced inhibitor of HCV replication that restricts formation of the viral replication organelle. J. Hepatol. 2020, 73, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.R.; Whitehead, S.S. Immune response to dengue virus and prospects for a vaccine. Annu. Rev. Immunol. 2011, 29, 587–619. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhao, Y.; Pan, J.; Yang, X.; Liang, Z.; Xie, S.; Cao, R. C19orf66 inhibits Japanese encephalitis virus replication by targeting-1 PRF and the NS3 protein. Virol. Sin. 2021, 36, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Hanners, N.W.; Mar, K.B.; Boys, I.N.; Eitson, J.L.; De La Cruz-Rivera, P.C.; Richardson, R.B.; Fan, W.; Wight-Carter, M.; Schoggins, J.W. Shiftless inhibits flavivirus replication in vitro and is neuroprotective in a mouse model of Zika virus pathogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2111266118. [Google Scholar] [CrossRef] [PubMed]
- Mangus, D.A.; Evans, M.C.; Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003, 4, 223. [Google Scholar] [CrossRef]
- Polacek, C.; Friebe, P.; Harris, E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 2009, 90, 687–692. [Google Scholar] [CrossRef]
- Burrows, C.; Latip, N.A.; Lam, S.-J.; Carpenter, L.; Sawicka, K.; Tzolovsky, G.; Gabra, H.; Bushell, M.; Glover, D.M.; Willis, A.E.; et al. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 2010, 38, 5542–5553. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.J.; Thoreen, C.C.; Dedeic, Z.; Chettle, J.; Roux, P.P.; Blagden, S.P. Controversies around the function of LARP1. RNA Biol. 2021, 18, 207–217. [Google Scholar] [CrossRef]
- Morita, E.; Suzuki, Y. Membrane-associated flavivirus replication complex—Its organization and regulation. Viruses 2021, 13, 1060. [Google Scholar] [CrossRef]
- Su, F.; Liu, X.; Jiang, Y. Roles of MOV10 in animal RNA virus infection. Front. Veter Sci. 2020, 7, 569737. [Google Scholar] [CrossRef]
- Reineke, L.C.; Lloyd, R.E. Diversion of stress granules and P-bodies during viral infection. Virology 2013, 436, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Chahar, H.S.; Chen, S.; Manjunath, N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 2013, 436, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Merret, R.; Descombin, J.; Juan, Y.-T.; Favory, J.-J.; Carpentier, M.-C.; Chaparro, C.; Charng, Y.-Y.; Deragon, J.-M.; Bousquet-Antonelli, C. XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 2013, 5, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Safaee, N.; Rosenauer, A.; Gehring, K. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the mlle domain of poly(A)-binding protein. J. Biol. Chem. 2010, 285, 13599–13606. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, S.; Maestre, A.M.; Pagni, S.; Patel, J.R.; Savage, T.; Gutman, D.; Maringer, K.; Bernal-Rubio, D.; Shabman, R.S.; Simon, V.; et al. DENV inhibits type I IFN production in infected cells by cleaving human sting. PLoS Pathog. 2012, 8, e1002934. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Chang, T.-H.; Liang, J.-J.; Chiang, R.-L.; Lee, Y.-L.; Liao, C.-L.; Lin, Y.-L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012, 8, e1002780. [Google Scholar] [CrossRef]
- Chan, Y.K.; Gack, M.U. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat. Immunol. 2016, 17, 523–530. [Google Scholar] [CrossRef]
- Daugherty, M.D.; Malik, H.S. Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 2012, 46, 677–700. [Google Scholar] [CrossRef]
- Atkins, J.F.; Loughran, G.; Bhatt, P.R.; Firth, A.E.; Baranov, P.V. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016, 44, 7007–7078. [Google Scholar] [CrossRef]
- Jacks, T.; Power, M.D.; Masiarz, F.R.; Luciw, P.A.; Barr, P.J.; Varmus, H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 1988, 331, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.E.; Atkins, J.F. A conserved predicted pseudoknot in the NS2A-encoding sequence of west Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol. J. 2009, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Melian, E.B.; Hinzman, E.; Nagasaki, T.; Firth, A.E.; Wills, N.M.; Nouwens, A.S.; Blitvich, B.J.; Leung, J.; Funk, A.; Atkins, J.F.; et al. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J. Virol. 2010, 84, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.; Lareau, C.A.; Keshishian, H.; Ganskih, S.; Schneider, C.; Hennig, T.; Melanson, R.; Werner, S.; Wei, Y.; Zimmer, M.; et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat. Microbiol. 2020, 6, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Abriola, L.; Niederer, R.O.; Pedersen, S.F.; Alfajaro, M.M.; Silva Monteiro, V.; Wilen, C.B.; Ho, Y.-C.; Gilbert, W.V.; Surovtseva, Y.V.; et al. Restriction of SARS-CoV-2 replication by targeting programmed-1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 2021, 118, e2023051118. [Google Scholar] [CrossRef]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef]
- Hahn, S.; Roberts, S. The zinc ribbon domains of the general transcription factors TFIIB and Brf: Conserved functional surfaces but different roles in transcription initiation. Genes Dev. 2000, 14, 719–730. [Google Scholar] [CrossRef]
- Xiong, W.; Contreras, D.; Irudayam, J.I.; Ali, A.; Yang, O.O.; Arumugaswami, V. C19ORF66 is an interferon-stimulated gene (ISG) which inhibits human immunodeficiency virus-1. bioRxiv 2016, 050310. [Google Scholar] [CrossRef]
- Rodriguez, W.; Srivastav, K.; Muller, M. C19ORF66 broadly escapes virus-induced endonuclease cleavage and restricts kaposi’s sarcoma-associated herpesvirus. J. Virol. 2019, 93, e00373-19. [Google Scholar] [CrossRef]
- Napthine, S.; Hill, C.H.; Nugent, H.C.M.; Brierley, I. Modulation of viral programmed ribosomal frameshifting and stop codon readthrough by the host restriction factor shiftless. Viruses 2021, 13, 1230. [Google Scholar] [CrossRef]
- Jäger, N.; Ayyub, S.A.; Korniy, N.; Peske, F.; Hoffmann, M.; Rodnina, M.V.; Pöhlmann, S. Mutagenic analysis of the HIV restriction factor shiftless. Viruses 2022, 14, 1454. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, W.; Mehrmann, T.; Muller, M. Shiftless restricts viral gene expression and influences RNA granule formation during KSHV lytic replication. bioRxiv 2022, 480778. [Google Scholar] [CrossRef]
- Wang, H.; Kong, N.; Jiao, Y.; Dong, S.; Sun, D.; Chen, X.; Zheng, H.; Tong, W.; Yu, H.; Yu, L.; et al. EGR1 suppresses porcine epidemic diarrhea virus replication by regulating IRAV to degrade viral nucleocapsid protein. J. Virol. 2021, 95, JVI0064521. [Google Scholar] [CrossRef] [PubMed]
| Flaviviruses | Proposed Mechanisms-of-Inhibition | Notes | References |
|---|---|---|---|
| DENV, WNV |
|
| [15] |
| DENV |
|
| [22] |
| ZIKV |
|
| [23] |
| JEV |
|
| [26] |
| ZIKV, YFV, WNV, DENV |
|
| [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, Y.; Murakawa, T. Restriction of Flaviviruses by an Interferon-Stimulated Gene SHFL/C19orf66. Int. J. Mol. Sci. 2022, 23, 12619. https://doi.org/10.3390/ijms232012619
Suzuki Y, Murakawa T. Restriction of Flaviviruses by an Interferon-Stimulated Gene SHFL/C19orf66. International Journal of Molecular Sciences. 2022; 23(20):12619. https://doi.org/10.3390/ijms232012619
Chicago/Turabian StyleSuzuki, Youichi, and Takeshi Murakawa. 2022. "Restriction of Flaviviruses by an Interferon-Stimulated Gene SHFL/C19orf66" International Journal of Molecular Sciences 23, no. 20: 12619. https://doi.org/10.3390/ijms232012619
APA StyleSuzuki, Y., & Murakawa, T. (2022). Restriction of Flaviviruses by an Interferon-Stimulated Gene SHFL/C19orf66. International Journal of Molecular Sciences, 23(20), 12619. https://doi.org/10.3390/ijms232012619






