Advanced Genetic Studies on Powdery Mildew Resistance in TGR-1551
Abstract
1. Introduction
2. Results
2.1. Selection of BC3, BC3S1, and BC4 Generations
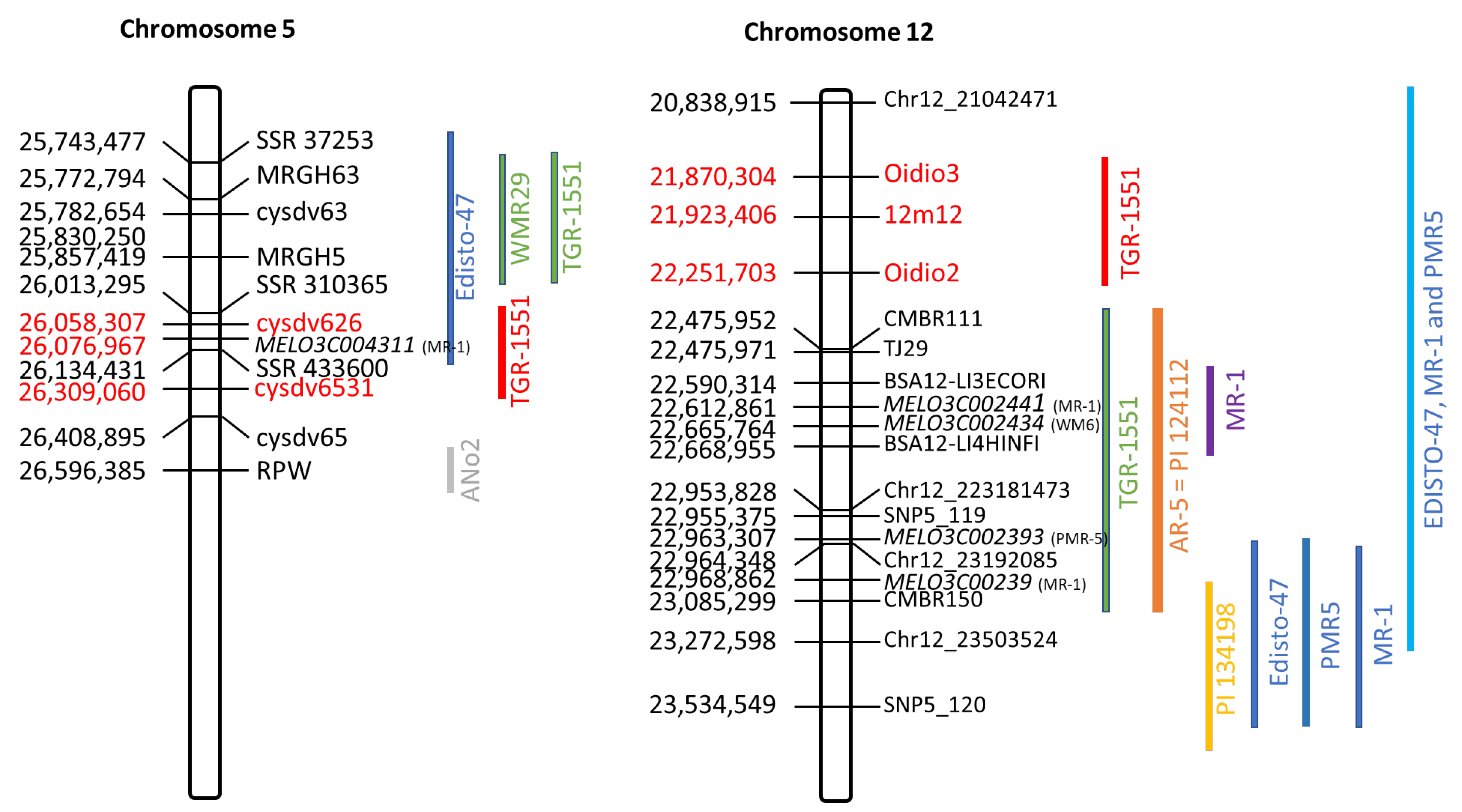
2.2. Narrowing of the Candidate Region on Chromosome 5
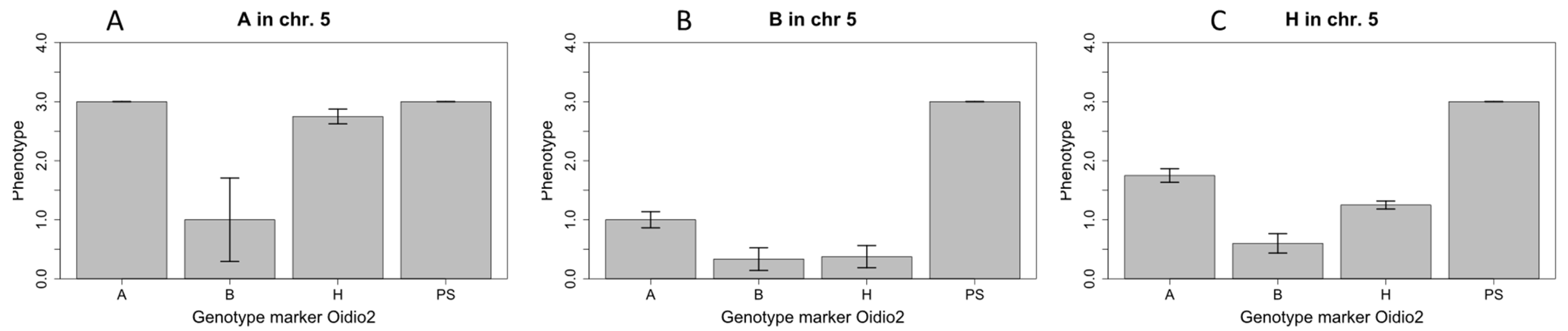
2.3. Narrowing of the Candidae Region on Chromosome 12
2.4. Changes in Resistance Levels Due to Low Temperatures
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Artificial Inoculations
4.3. Molecular Markers and Genotyping Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. FAOstats, Organización de las Naciones Unidas para la Alimentación y la Agricultura. Available online: https://www.fao.org/faostat/es/#home (accessed on 1 June 2022).
- Cohen, R.; Burger, Y.; Katzir, N. Monitoring Physiological Races of Podosphaera Xanthii (Syn. Sphaerothecafuliginea), the Causal Agent of Powdery Mildew in Cucurbits: Factors Affecting Race Identification and the Importance for Research and Commerce. Phytoparasitica 2004, 32, 174–183. [Google Scholar] [CrossRef]
- Lebeda, A.; Sedlákova, B.; Krístkova, E. Temporal Changes in Pathogenicity Structure of Cucurbit Powdery Mildew Populations. Acta Hortic. 2007, 731, 381–388. [Google Scholar] [CrossRef]
- Lebeda, A.; Sedláková, B.; Křístková, E.; Vysoudil, M. Long-Lasting Changes in the Species Spectrum of Cucurbit Powdery Mildew in the Czech Republic-Influence of Air Temperature Changes or Random Effect? Plant Prot. Sci. 2009, 45, S41–S47. [Google Scholar] [CrossRef]
- Braun, U. Taxonomic Manual of Erysiphales (Powdery Mildews). CBS Biodivers. 2012, 11, 1–707. [Google Scholar]
- Křístková, E.; Lebeda, A.; Sedláková, B. Species Spectra, Distribution and Host Range of Cucurbit Powdery Mildews in the Czech Republic, and in Some Other European and Middle Eastern Countries. Phytoparasitica 2009, 34, 337–350. [Google Scholar] [CrossRef]
- Lebeda, A.; Sedláková, B.; Křístková, E.; Widrlechner, M.P.; Kosman, E. Understanding Pathogen Population Structure and Virulence Variation for Efficient Resistance Breeding to Control Cucurbit Powdery Mildews. Plant Pathol. 2021, 70, 1364–1377. [Google Scholar] [CrossRef]
- Bardin, M.; Dogimont, C.; Nicot, P.C.; Pitrat, M. Genetic Analysis of Resistance of Melon Line PI 124112 to Sphaerotheca Fuliginea and Erysiphe Cichoracearum Studied in Recombinant Inbred Lines. In Proceedings of the International Symposium on Cucurbits, Ankara, Turkey, 1 May 1999; pp. 163–168. [Google Scholar]
- Hong, Y.J.; Hossain, M.R.; Kim, H.T.; Park, J.I.; Nou, I.S. Identification of Two New Races of Podosphaera Xanthii Causing Powdery Mildew in Melon in South Korea. Plant Pathol. J. 2018, 34, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Lebeda, A.; Křístková, E.; Sedláková, B.; McCreight, J.D.; Coffey, M.D. New Concept for Determination and Denomination of Pathotypes and Races of Cucurbit Powdery Mildew. In Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Avignon, France, 21–24 May 2008; pp. 125–134. [Google Scholar]
- Lebeda, A.; Sedláková, B. Identification and Survey of Cucurbit Powdery Mildew Races in Czech Populations. In Proceedings of the Cucurbitaceae 2006, Asheville, NC, USA, 17–21 September 2006; pp. 444–452. [Google Scholar]
- De Miccolis Angelini, R.M.; Pollastro, S.; Rotondo, P.R.; Laguardia, C.; Abate, D.; Rotolo, C.; Faretra, F. Transcriptome Sequence Resource for the Cucurbit Powdery Mildew Pathogen Podosphaera Xanthii. Sci. Data 2019, 6, 1–7. [Google Scholar] [CrossRef]
- McCreight, J.D.; Coffey, M.D.; Ando, K.; Kousik, C.S. Cucurbit Powdery Mildew Races on Melon: Current Status in the US. In Proceedings of the American Society of Horticulture Science Meeting, Orlando, FL, USA, 31 July–4 August 2018. [Google Scholar]
- Périn, C.; Hagen, L.; de Conto, V.; Katzir, N.; Danin-Poleg, Y.; Portnoy, V.; Baudracco-Arnas, S.; Chadoeuf, J.; Dogimont, C.; Pitrat, M. A Reference Map of Cucumis Melo Based on Two Recombinant Inbred Line Populations. Theor. Appl. Genet. 2002, 104, 1017–1034. [Google Scholar] [CrossRef]
- Fazza, A.C.; Dallagnol, L.J.; Fazza, A.C.; Monteiro, C.C.; de Lima, B.M.; Wassano, D.T.; Aranha Camargo, L.E. Mapping of Resistance Genes to Races 1, 3 and 5 of Podosphaera Xanthii in Melon PI 414723. Crop Breed. Appl. Biotechnol. 2013, 13, 349–355. [Google Scholar] [CrossRef]
- Zhang, C.; Ren, Y.; Guo, S.; Zhang, H.; Gong, G.; Du, Y.; Xu, Y. Application of Comparative Genomics in Developing Markers Tightly Linked to the Pm-2F Gene for Powdery Mildew Resistance in Melon (Cucumis Melo L.). Euphytica 2013, 190, 157–168. [Google Scholar] [CrossRef]
- Fukino, N.; Ohara, T.; Monforte, A.J.; Sugiyama, M.; Sakata, Y.; Kunihisa, M.; Matsumoto, S. Identification of QTLs for Resistance to Powdery Mildew and SSR Markers Diagnostic for Powdery Mildew Resistance Genes in Melon (Cucumis Melo L.). Theor. Appl. Genet. 2008, 118, 165–175. [Google Scholar] [CrossRef]
- Ning, X.; Wang, X.; Gao, X.; Zhang, Z.; Zhang, L.; Yan, W.; Li, G. Inheritances and Location of Powdery Mildew Resistance Gene in Melon Edisto47. Euphytica 2014, 195, 345–353. [Google Scholar] [CrossRef]
- Branham, S.E.; Kousik, C.; Mandal, M.K.; Wechter, W.P. Quantitative Trait Loci Mapping of Resistance to Powdery Mildew Race 1 in a Recombinant Inbred Line Population of Melon. Plant Dis. 2021, 105, 3809–3815. [Google Scholar] [CrossRef]
- Pitrat, M. Linkage Groups in Cucumis Melo L. J. Hered. 1991, 82, 406–411. [Google Scholar] [CrossRef]
- Yuste-Lisbona, F.J.; Capel, C.; Gómez-Guillamón, M.L.; Capel, J.; López-Sesé, A.I.; Lozano, R. Codominant PCR-Based Markers and Candidate Genes for Powdery Mildew Resistance in Melon (Cucumis melo L.). Theor. Appl. Genet. 2011, 122, 747–758. [Google Scholar] [CrossRef]
- Wang, X.; Li, G.; Gao, X.; Xiong, L.; Wang, W.; Han, R. Powdery Mildew Resistance Gene (Pm-AN) Located in a Segregation Distortion Region of Melon LGV. Euphytica 2011, 180, 421–428. [Google Scholar] [CrossRef]
- Perchepied, L.; Bardin, M.; Dogimont, C.; Pitrat, M. Relationship between Loci Conferring Downy Mildew and Powdery Mildew Resistance in Melon Assessed by Quantitative Trait Loci Mapping. Phytopathology 2005, 95, 556–565. [Google Scholar] [CrossRef]
- Cui, H.; Fan, C.; Ding, Z.; Wang, X.; Tang, L.; Bi, Y.; Luan, F.; Gao, P. CmPMRl and CmPMrs Are Responsible for Resistance to Powdery Mildew Caused by Podosphaera Xanthii Race 1 in Melon. Theor. Appl. Genet. 2022, 135, 1209–1222. [Google Scholar] [CrossRef]
- Cao, Y.; Diao, Q.; Chen, Y.; Jin, H.; Zhang, Y.; Zhang, H. Development of KASP Markers and Identification of a QTL Underlying Powdery Mildew Resistance in Melon (Cucumis melo L.) by Bulked Segregant Analysis and RNA-Seq. Front. Plant Sci. 2021, 11, 1819. [Google Scholar] [CrossRef]
- Li, B.; Zhao, Y.; Zhu, Q.; Zhang, Z.; Fan, C.; Amanullah, S.; Gao, P.; Luan, F. Mapping of Powdery Mildew Resistance Genes in Melon (Cucumis melo L.) by Bulked Segregant Analysis. Sci. Hortic. 2017, 220, 160–167. [Google Scholar] [CrossRef]
- Teixeira, A.P.M.; Barreto, F.A.d.S.; Camargo, L.E.A. An AFLP Marker Linked to the Pm-1 Gene that Confers Resistance to Podosphaera Xanthii Race 1 in Cucumis melo. Genet. Mol. Biol. 2008, 31, 547–550. [Google Scholar] [CrossRef]
- Hong, C.; Weiping, K.; Junfen, L.; Jiping, L. Analysis of Powdery Mildew Resistance in Wild Melon MLO Mutants. Hortic. Plant J. 2015, 1, 165–171. [Google Scholar] [CrossRef]
- Iovieno, P.; Andolfo, G.; Schiavulli, A.; Catalano, D.; Ricciardi, L.; Frusciante, L.; Ercolano, M.R.; Pavan, S. Structure, Evolution and Functional Inference on the Mildew Locus O (MLO) Gene Family in Three Cultivated Cucurbitaceae spp. BMC Genom. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Natarajan, S.; Kim, H.T.; Thamilarasan, S.K.; Veerappan, K.; Park, J.I.; Nou, I.S. Whole Genome Re-Sequencing and Characterization of Powdery Mildew Disease-Associated Allelic Variation in Melon. PLoS ONE 2016, 11, e0157524. [Google Scholar] [CrossRef]
- Yuste-Lisbona, F.J.; López-Sesé, A.I.; Gómez-Guillamón, M.L. Inheritance of Resistance to Races 1, 2 and 5 of Powdery Mildew in the Melon TGR-1551. Plant Breed. 2010, 129, 72–75. [Google Scholar] [CrossRef]
- Yuste-Lisbona, F.J.; Capel, C.; Sarria, E.; Torreblanca, R.; Gómez-Guillamón, M.L.; Capel, J.; Lozano, R.; López-Sesé, A.I. Genetic Linkage Map of Melon (Cucumis Melo L.) and Localization of a Major QTL for Powdery Mildew Resistance. Mol. Breed. 2011, 27, 181–192. [Google Scholar] [CrossRef]
- Beraldo-Hoischen, P.; Gómez-Guillamón, M.L.; López-Sesé, A.I. QTL Associated with One Recessive Gene for Powdery Mildew Resistance in the Melon Genotype TGR-1551. In Proceedings of the Xth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Antalya, Turkey, 15–18 October 2012; pp. 508–513. [Google Scholar]
- Fukino, N.; Kunihisa, M.; Matsumoto, S. Characterization of Recombinant Inbred Lines Derived from Crosses in Melon (Cucumis melo L.), ‘PMAR No. 5’ × ‘Harukei No. 3’. Breed. Sci. 2004, 54, 141–145. [Google Scholar] [CrossRef]
- Hong, J.-E.; Hossain, M.R.; Jung, H.-J. Inheritance of Resistance to Race 5 of Powdery Mildew Fungus Podosphaera Xanthii in Melon and Development of Race 5-Specific High Resolution Melting Markers. 2022. Available online: https://assets.researchsquare.com/files/rs-1433034/v1/dea556c7-b2f4-42cb-a677-789a4e435f56.pdf?c=1659760899 (accessed on 1 June 2022).
- Howlader, J.; Hong, Y.; Natarajan, S.; Sumi, K.R.; Kim, H.T.; Park, J.I.; Nou, I.S. Development of Powdery Mildew Race 5-Specific SNP Markers in Cucumis Melo L. Using Whole-Genome Resequencing. Hortic. Environ. Biotechnol. 2020, 61, 347–357. [Google Scholar] [CrossRef]
- Jorgensen, J.H. Discovery, Characterization and Exploitation of Mlo Powdery Mildew Resistance in Barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Varshney, R.K.; Ribaut, J.-M.; Buckler, E.S.; Tuberosa, R.; Antoni Rafalski, J.; Langridge, P. Can Genomics Boost Productivity of Orphan Crops? Nat. Publ. Gr. 2012, 30, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Esteras, C.; Formisano, G.; Roig, C.; Díaz, A.; Blanca, J.; Garcia-Mas, J.; Gómez-Guillamón, M.L.; López-Sesé, A.I.; Lázaro, A.; Monforte, A.J.; et al. SNP Genotyping in Melons: Genetic Variation, Population Structure, and Linkage Disequilibrium. Theor. Appl. Genet. 2013, 126, 1285–1303. [Google Scholar] [CrossRef] [PubMed]
- Leida, C.; Moser, C.; Esteras, C.; Sulpice, R.; Lunn, J.; de Langen, F.; Monforte, A.J.; Picó, B. Variability of Candidate Genes, Genetic Structure and Association with Sugar Accumulation and Climacteric Behavior in a Broad Germplasm Collection of Melon (Cucumis melo L.). BMC Genet. 2015, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Perpiñá, G.; Esteras, C.; Gibon, Y.; Monforte, A.J.; Picó, B. A New Genomic Library of Melon Introgression Lines in a Cantaloupe Genetic Background for Dissecting Desirable Agronomical Traits. BMC Plant Biol. 2016, 16, 154. [Google Scholar] [CrossRef]
- Sáez, C.; Esteras, C.; Martínez, C.; Ferriol, M.; Dhillon, N.P.S.; López, C.; Picó, B. Resistance to Tomato Leaf Curl New Delhi Virus in Melon Is Controlled by a Major QTL Located in Chromosome 11. Plant Cell Rep. 2017, 36, 1571–1584. [Google Scholar] [CrossRef]
- Pérez-de-Castro, A.; Esteras, C.; Alfaro-Fernández, A.; Daròs, J.; Monforte, A.; Picó, B.; Gómez-Guillamón, M. Fine Mapping of Wmv1551, a Resistance Gene to Watermelon Mosaic Virus in Melon. Mol. Breed. 2019, 39, 93. [Google Scholar] [CrossRef]
- Pérez-De-castro, A.; López-Martín, M.; Esteras, C.; Garcés-Claver, A.; Palomares-Ríus, F.J.; Picó, M.B.; Gómez-Guillamón, M.L. Melon Genome Regions Associated with Tgr-1551-Derived Resistance to Cucurbit Yellow Stunting Disorder Virus. Int. J. Mol. Sci. 2020, 21, 5970. [Google Scholar] [CrossRef]
- González, V.M.; Aventín, N.; Centeno, E.; Puigdomènech, P. High Presence/Absence Gene Variability in Defense-Related Gene Clusters of Cucumis melo. BMC Genom. 2013, 14, 782. [Google Scholar] [CrossRef]
- Marathe, R.; Anandalakshmi, R.; Liu, Y.; Dinesh-Kumar, S.P. The Tobacco Mosaic Virus Resistance Gene, N. Mol. Plant Pathol. 2002, 3, 167–172. [Google Scholar] [CrossRef]
- Cui, L.; Siskos, L.; Wang, C.; Schouten, H.J.; Visser, R.G.F.; Bai, Y. Breeding Melon (Cucumis melo) with Resistance to Powdery Mildew and Downy Mildew. Hortic. Plant J. 2022, 8, 545–561. [Google Scholar] [CrossRef]
- Pedersen, C.; van Themaat, E.V.L.; McGuffin, L.J.; Abbott, J.C.; Burgis, T.A.; Barton, G.; Bindschedler, L.V.; Lu, X.; Maekawa, T.; Weßling, R.; et al. Structure and Evolution of Barley Powdery Mildew Effector Candidates. BMC Genom. 2012, 13, 1–21. [Google Scholar] [CrossRef]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Corredor-Moreno, P.; Minter, F.; Davey, P.E.; Wegel, E.; Kular, B.; Brett, P.; Lewis, C.M.; Morgan, Y.M.L.; Pérez, L.A.M.; Korolev, A.V.; et al. The Branched-Chain Amino Acid Aminotransferase TaBCAT1 Modulates Amino Acid Metabolism and Positively Regulates Wheat Rust Susceptibility. Plant Cell 2021, 33, 1728. [Google Scholar] [CrossRef]
- Von Saint Paul, V.; Zhang, W.; Kanawati, B.; Geist, B.; Faus-Keßler, T.; Schmitt-Kopplin, P.; Schäffner, A.R. The Arabidopsis Glucosyltransferase UGT76B1 Conjugates Isoleucic Acid and Modulates Plant Defense and Senescence. Plant Cell 2011, 23, 4124–4145. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant Cell Wall-Mediated Immunity: Cell Wall Changes Trigger Disease Resistance Responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The Role of the Secondary Cell Wall in Plant Resistance to Pathogens. Front. Plant Sci. 2014, 5, 1–13. [Google Scholar] [CrossRef]
- Voxeur, A.; Habrylo, O.; Guénin, S.; Miart, F.; Soulié, M.C.; Rihouey, C.; Pau-Roblot, C.; Domon, J.M.; Gutierrez, L.; Pelloux, J.; et al. Oligogalacturonide Production upon Arabidopsis Thaliana-Botrytis Cinerea Interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 19743–19752. [Google Scholar] [CrossRef]
- De Azevedo Souza, C.; Li, S.; Lin, A.Z.; Boutrot, F.; Grossmann, G.; Zipfel, C.; Somerville, S.C. Cellulose-Derived Oligomers Act as Damage-Associated Molecular Patterns and Trigger Defense-like Responses. Plant Physiol. 2017, 173, 2383–2398. [Google Scholar] [CrossRef]
- Nie, J.; Wang, Y.; He, H.; Guo, C.; Zhu, W.; Pan, J.; Li, D.; Lian, H.; Pan, J.; Cai, R. Loss-of-Function Mutations in CsMLO1 Confer Durable Powdery Mildew Resistance in Cucumber (Cucumis sativus L.). Front. Plant Sci. 2015, 6, 1155. [Google Scholar] [CrossRef]
- Vogel, J.P.; Raab, T.K.; Schiff, C.; Somerville, S.C. PMR6, a Pectate Lyase-like Gene Required for Powdery Mildew Susceptibility in Arabidopsis. Plant Cell 2002, 14, 2095–2106. [Google Scholar] [CrossRef]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S.C. Mutations in PMR5 Result in Powdery Mildew Resistance and Altered Cell Wall Composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef]
- Beraldo-Hoischen, P.; Hoefle, C.; López-Sesé, A.I. Fungal Development and Callose Deposition in Compatible and Incompatible Interactions in Melon Infected with Powdery Mildew. Pathogens 2021, 10, 873. [Google Scholar] [CrossRef]
- Polonio, Á.; Pineda, M.; Bautista, R.; Martínez-Cruz, J.; Pérez-Bueno, M.L.; Barón, M.; Pérez-García, A. RNA-Seq Analysis and Fluorescence Imaging of Melon Powdery Mildew Disease Reveal an Orchestrated Reprogramming of Host Physiology. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Sarria-Villada, E.; Garzo, E.; Lopez-Sese, A.; Fereres, A.; Gomez-Guillamon, M.L. Hypersensitive Response to Aphis Gossypii Glover in Melon Genotypes Carrying the Vat Gene. J. Exp. Bot. 2009, 60, 3269–3277. [Google Scholar] [CrossRef]
- Palomares-Rius, F.; Garcés-Claver, A.; Picó, B.; Esteras, C.; Yuste-lisbona, F.; Gómez-Guillamón, M. ‘Carmen’, a Yellow Canary Melon Breeding Line Resistant to Podosphaera xanthii, Aphis gossypii, and Cucurbit Yellow Stunting Disorder Virus. Hort Sci. 2018, 53, 1072–1075. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Y.; Zhenghong, S.; Zang, H.; Zhu, W. A Sequence-Amplified Characterized Region Marker for a Single, Dominant Gene in Melon PI 134198 that Confers Resistance to a Unique Race of Podosphaera xanthii in China. Am. Soc. Hortic. Sci. 2010, 45, 1407–1410. [Google Scholar] [CrossRef]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A Recently Evolved Hexose Transporter Variant Confers Resistance to Multiple Pathogens in Wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef]
- Hayes, M.A.; Feechan, A.; Dry, I.B. Involvement of Abscisic Acid in the Coordinated Regulation of a Stress-Inducible Hexose Transporter (VvHT5) and a Cell Wall Invertase in Grapevine in Response to Biotrophic Fungal Infection. Plant Physiol. 2010, 153, 211–221. [Google Scholar] [CrossRef]
- Banday, Z.Z.; Nandi, A.K. Interconnection between Flowering Time Control and Activation of Systemic Acquired Resistance. Front. Plant Sci. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Ramonell, K.M.; Goff, K.E. The Role and Regulation of Receptor-Like Kinases in Plant Defense. Gene Regul. Syst. Biol. 2007, 1, 167–175. [Google Scholar]
- Martin, G.B.; Brommonschenkel, S.H.; Chunwongse, J.; Frary, A.; Ganal, M.W.; Spivey, R.; Wu, T.; Earle, E.D.; Tanksley, S.D. Map-Based Cloning of a Protein Kinase Gene Conferring Disease Resistance in Tomato. Science 1993, 262, 1432–1436. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H.; et al. A Receptor Kinase-Like Protein Encoded by the Rice Disease Resistance Gene, Xa21. Science 1995, 270, 1804. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, S.; Sun, Q.; Yang, L.; Zhu, Y.; Yuan, Y.; Hua, J. A Role of Cytokinin Transporter in Arabidopsis Immunity. Mol. Plant Microbe Interact. 2017, 30, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Nowara, D.; Schweizer, P. Protein Polyubiquitination Plays a Role in Basal Host Resistance of Barley. Plant Cell 2006, 18, 3321–3331. [Google Scholar] [CrossRef]
- Deng, F.; Guo, T.; Lefebvre, M.; Scaglione, S.; Antico, C.J.; Jing, T.; Yang, X.; Shan, W.; Ramonell, K.M. Expression and Regulation of ATL9, an E3 Ubiquitin Ligase Involved in Plant Defense. PLoS ONE 2017, 12, e0188458. [Google Scholar] [CrossRef]
- Li, W.; Zhong, S.; Li, G.; Li, Q.; Mao, B.; Deng, Y.; Zhang, H.; Zeng, L.; Song, F.; He, Z. Rice RING Protein OsBBI1 with E3 Ligase Activity Confers Broad-Spectrum Resistance against Magnaporthe Oryzae by Modifying the Cell Wall Defence. Cell Res. 2011, 21, 835–848. [Google Scholar] [CrossRef]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; Tholl, D. The Major Volatile Organic Compound Emitted from Arabidopsis Thaliana Flowers, the Sesquiterpene (E)-β-Caryophyllene, Is a Defense against a Bacterial Pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef]
- Godard, K.A.; White, R.; Bohlmann, J. Monoterpene-Induced Molecular Responses in Arabidopsis Thaliana. Phytochemistry 2008, 69, 1838–1849. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, P.; Wan, Y.; Cui, H.; Fan, C.; Liu, S.; Luan, F. Comparative Transcriptome Profiling of Genes and Pathways Related to Resistance against Powdery Mildew in Two Contrasting Melon Genotypes. Sci. Hortic. 2018, 227, 169–180. [Google Scholar] [CrossRef]
- Sun, J.; Dong, Y.; Wang, C.; Xiao, S.; Jiao, Z.; Gao, C. Identification and Characterization of Melon Circular RNAs Involved in Powdery Mildew Responses through Comparative Transcriptome Analysis. PeerJ 2021, 9, e11216. [Google Scholar] [CrossRef]
- Zhou, X.; Cui, J.; Cui, H.; Jiang, N.; Hou, X.; Liu, S.; Gao, P.; Luan, Y.; Meng, J.; Luan, F. Identification of LncRNAs and Their Regulatory Relationships with Target Genes and Corresponding MiRNAs in Melon Response to Powdery Mildew Fungi. Gene 2020, 735, 144403. [Google Scholar] [CrossRef]
- Fukino, N.; Yoshioka, Y.; Sugiyama, M.; Sakata, Y.; Matsumoto, S. Identification and Validation of Powdery Mildew (Podosphaera Xanthii)-Resistant Loci in Recombinant Inbred Lines of Cucumber (Cucumis sativus L.). Mol. Breed. 2013, 32, 267–277. [Google Scholar] [CrossRef]
- Hosoya, K.; Kuzuya, M.; Murakami, T.; Kato, K.; Narisawa, K.; Ezura, H. Impact of Resistant Melon Cultivars on Sphaerotheca Fuliginea. Plant Breed. 2000, 119, 286–288. [Google Scholar] [CrossRef]
- Tores, J.A.; Gomez-Guillamon, M.L.; Canovas, I. Temperature-Conditioned Response to Sphaerotheca Fuliginea Race 1 in the Spanish Melon Cultivar ANC57. Cucurbit Genet. Coop. Rep. 1996, 19, 59–60. [Google Scholar]
- Sakata, Y.; Kubo, N.; Morishita, M.; Kitadani, E.; Sugiyama, M.; Hirai, M. QTL Analysis of Powdery Mildew Resistance in Cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2006, 112, 243–250. [Google Scholar] [CrossRef]
- Ferriere, H.; Molot, P.M. Effect of Leaf Position on the Susceptibility of Melon Plants to Artificial Infection with Powdery Mildew, Sphaerotheca Fuliginea. J. Phytopathol. 1988, 121, 250–254. [Google Scholar] [CrossRef]
- Bohn, G.W.; Whitaker, T.W. Genetics of resistance to powdery mildew race 2 in muskmelon. Phytopathology 1964, 54, 587–591. [Google Scholar]
- Boiteux, L.S.; Reifschneider, F.J.B.; Pessoa, H.B.S.V. Phenotypic Expression of Quantitative and Qualitative Components of Partial Resistance to Powdery Mildew (Sphaerotheca Fuliginea Race 1) in Melon (Cucumis melo) Germplasm. Plant Breed. 1995, 114, 185–187. [Google Scholar] [CrossRef]
- McCreight, J.D. Genes for Resistance to Powdery Mildew Races 1 and 2 U.S. in Melon PI 313970. HortScience 2003, 38, 591–594. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. Isolation of Plant DNa from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. Available online: https://arxiv.org/abs/1207.3907 (accessed on 12 October 2022).
- Cingolani, P.; Platss, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
| Number of BC3 Plant | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Markers Set | Marker | Chr a | Genomic Position (bp) | 15 | 19 | 34 | 64 | 78 | 95 | 96 | 105 | 139 | 141 | 166 | 24 | 28 | 37 | 88 | 89 | 146 | 148 | 159 | 198 |
| Background1 | CMPSNP898 | 5 | 256,610 | A | A | A | A | A | A | A | H | A | A | H | A | A | A | H | H | n.d | A | A | A |
| Background1 | CMPSNP387 | 5 | 1,260,621 | A | A | A | H | A | A | A | H | A | A | H | A | A | A | A | H | n.d | A | A | A |
| Background1 | CMPSNP437 | 5 | 1,711,810 | A | A | A | H | A | A | A | A | A | H | H | A | A | A | A | H | n.d | H | A | A |
| Background1 | CMPSNP726 | 5 | 2,373,146 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | H | n.d | H | A | A |
| Background1 | SSH9G15 | 5 | 5,781,400 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | n.d | n.d | H | A | A |
| CYSDV1 | cysdv10B | 5 | 6,298,039 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | A | A | H | H | A |
| CYSDV1 | cysdv11 | 5 | 9,494,514 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | A | A | n.d | H | A |
| Background1 | CMPSNP788 | 5 | 12,024,140 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | A | A | n.d | H | A |
| WMV1 | b5wmv2 | 5 | 15,026,076 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | A | A | H | H | A |
| CYSDV1 | cysdv14 | 5 | 17,216,441 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | A | A | n.d | H | A |
| Background1 | 60k41.243 | 5 | 19,612,771 | H | H | A | H | A | H | A | H | A | H | H | A | A | A | A | A | A | n.d | H | A |
| WMV1 | b5wmv3 | 5 | 19,968,717 | n.d | A | A | H | A | A | A | A | A | n.d | H | H | n.d | A | A | A | A | n.d | H | A |
| CYSDV1 | cysdv17 | 5 | 22,544,163 | A | A | A | H | A | A | A | A | A | A | H | H | H | A | A | A | A | n.d | H | A |
| CYSDV1 | cysdv18 | 5 | 24,125,662 | A | H | A | H | A | A | A | A | A | A | H | A | H | A | A | A | A | n.d | H | A |
| CYSDV1 | cysdv19 | 5 | 24,192,280 | A | H | A | H | A | A | A | A | A | A | H | A | H | A | A | A | A | n.d | H | A |
| CYSDV1 | cysdv21 | 5 | 24,427,738 | A | H | A | H | A | A | A | A | A | H | H | A | H | A | A | A | H | n.d | H | A |
| CYSDV1 | cysdv22 | 5 | 24,434,940 | A | H | A | H | A | A | A | A | A | H | H | A | H | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv40 | 5 | 24,474,795 | A | H | A | H | A | A | A | A | A | H | H | A | H | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv42 | 5 | 24,608,464 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv43 | 5 | 24,678,113 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv44 | 5 | 24,704,705 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv45 | 5 | 24,761,527 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv46 | 5 | 24,778,081 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv48 | 5 | 24,842,569 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv49 | 5 | 24,842,825 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| WMV2 | SNP25 | 5 | 24,898,321 | A | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | A | H | A |
| WMV2 | SNP26 | 5 | 25,045,765 | H | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv50 | 5 | 25,052,002 | H | H | A | H | A | A | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv51 | 5 | 25,132,216 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| WMV1 | b5wmv4 | 5 | 25,132,292 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| WMV1 | b5wmv4A | 5 | 25,144,133 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv53 | 5 | 25,144,085 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv54 | 5 | 25,210,212 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv55 | 5 | 25,210,573 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv56 | 5 | 25,233,221 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv57 | 5 | 25,341,873 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv58 | 5 | 25,356,079 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv59 | 5 | 25,435,244 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| WMV1 | b5wmv5 | 5 | 25,694,699 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv60 | 5 | 25,744,140 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv61 | 5 | 25,756,801 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv61 | 5 | 25,772,357 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv62 | 5 | 25,776,015 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| CYSDV2 | cysdv63 | 5 | 25,782,654 | H | H | A | H | A | H | A | A | A | H | H | A | A | A | A | A | H | n.d | H | A |
| Background1 | CMPSNP464 | 5 | 26,405,006 | H | H | A | H | A | H | A | H | A | A | H | A | A | A | A | A | n.d | A | H | H |
| CYSDV2 | cysdv65 | 5 | 26,408,895 | H | H | A | H | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV2 | cysdv69 | 5 | 26,467,357 | H | H | A | H | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv24 | 5 | 26,547,204 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV1 | b5wmv7 | 5 | 26,592,732 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV1 | b5wmv7B | 5 | 26,592,732 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv23 | 5 | 26,752,698 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv25 | 5 | 26,769,546 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv26 | 5 | 26,940,168 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv27 | 5 | 26,957,159 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV1 | b5wmv8 | 5 | 26,963,176 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv28 | 5 | 27,118,062 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV1 | b2wmv9 | 5 | 27,273,256 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV1 | b2wmv10 | 5 | 27,464,146 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV2 | SNP29 | 5 | 27,538,308 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| WMV1 | b5wmv11 | 5 | 27,570,154 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| CYSDV1 | cysdv30B | 5 | 27,570,488 | H | H | A | A | A | H | A | H | A | A | A | A | A | A | A | A | A | n.d | H | H |
| Background1 | AI_13-H12 | 5 | 28,039,739 | H | H | A | A | H | A | A | H | A | A | A | A | A | A | A | A | A | A | H | H |
| Background1 | CMPSNP385 | 12 | 344,819 | A | A | A | A | A | H | A | A | A | A | A | A | A | A | A | H | n.d | A | A | A |
| Background1 | CMPSNP310 | 12 | 5,032,799 | A | A | A | A | A | H | A | A | A | A | A | A | A | A | A | A | n.d | A | A | H |
| Background1 | AI_35-A08 | 12 | 12,750,025 | A | A | A | A | H | A | A | A | A | A | A | A | A | A | A | A | n.d | A | A | H |
| Background1 | ai09g07 | 12 | 16,532,245 | A | A | A | A | H | A | A | A | A | A | A | A | A | A | A | A | n.d | A | A | H |
| Background1 | CMPSNP285 | 12 | 20,421,309 | A | A | H | A | H | H | A | A | H | H | A | A | A | A | H | A | n.d | A | A | H |
| Background1/2 | CMPSNP361 | 12 | 23,000,406 | A | A | H | A | A | H | H | H | H | H | A | A | A | A | H | H | A | H | A | A |
| Background1 | CMPSNP5 | 12 | 24,246,762 | A | A | H | A | A | H | H | A | A | H | A | H | A | A | A | H | n.d | H | A | A |
| Background1 | fr14f22 | 12 | 25,050,570 | H | A | A | A | A | H | H | A | A | H | A | H | A | A | A | H | n.d | A | A | A |
| Background1 | P02.03 | 12 | 25,661,792 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | n.d | A | A | A |
| SE | SE | SE | SE | SE | SE | SE | SE | SE | SE | SE | SU | SU | SU | SU | SU | SU | SU | SE | SU | ||||
| Marker Name | Chr a | Genomic Position (bp) v.4.0 | BC4(159)4xPS-7S1-13 | BC4(159)4xPS-7S1-2 | BC5(159)4-14S1-5 |
|---|---|---|---|---|---|
| Cysdv63 | 5 | 25,782,654 | H | H | H |
| Cysdv626 | 5 | 26,058,307 | H | H | H |
| Cysdv6531 | 5 | 26,309,060 | A | H | A |
| Cysdv65 | 5 | 26,408,895 | A | A | A |
| Phenotype | Resistant | Resistant | Susceptible |
| BC4(95)10xPS Plants | ||||
|---|---|---|---|---|
| Marker | Chr a | Genomic Position v.4.0 (bp) | 3 | 5 |
| S5_23380459 | 5 | 24,192,281 | A | A |
| cysdv63 | 5 | 25,782,654 | A | A |
| cysdv65 | 5 | 26,408,896 | A | A |
| S5_25653869 | 5 | 26,419,648 | A | A |
| S12_18721450 | 12 | 18,530,394 | A | A |
| Oidio3 | 12 | 21,870,304 | A | A |
| S12_22130778 | 12 | 21,923,406 | A | A |
| Oidio 2 | 12 | 22,251,703 | H | H |
| Oidio1 | 12 | 22,309,972 | H | H |
| CMPSNP361 | 12 | 23,000,406 | H | H |
| Phenotype | SU | SU | ||
| Number of BC3 Plant | ||||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Chr a | Genomic Position (bp) | 34 | 96 | 139 | 88 | 89 | 148 |
| CMPSNP285 | 12 | 20,421,309 | H | A | H | H | A | A |
| Oidio8 | 12 | 20,920,054 | H | A | A | n.d 1 | A | A |
| Oidio5 | 12 | 21,470,961 | H | H | A | n.d | A | A |
| Oidio4 | 12 | 21,866,445 | H | H | A | n.d | A | A |
| Oidio3 | 12 | 21,870,304 | H | H | A | A | A | A |
| Oidio2 | 12 | 22,251,703 | H | H | H | A | A | A |
| Oidio1 | 12 | 22,309,972 | H | H | H | A | A | A |
| OidioC | 12 | 22,875,711 | H | H | H | H | A | H |
| OidoB | 12 | 22,913,643 | H | H | H | H | H | H |
| OidioA | 12 | 22,915,510 | H | H | H | H | H | H |
| CMPSNP361 | 12 | 23,000,406 | H | H | H | H | H | H |
| Phenotype | SE | SE | SE | SU | SU | SU | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Martín, M.; Pérez-de-Castro, A.; Picó, B.; Gómez-Guillamón, M.L. Advanced Genetic Studies on Powdery Mildew Resistance in TGR-1551. Int. J. Mol. Sci. 2022, 23, 12553. https://doi.org/10.3390/ijms232012553
López-Martín M, Pérez-de-Castro A, Picó B, Gómez-Guillamón ML. Advanced Genetic Studies on Powdery Mildew Resistance in TGR-1551. International Journal of Molecular Sciences. 2022; 23(20):12553. https://doi.org/10.3390/ijms232012553
Chicago/Turabian StyleLópez-Martín, María, Ana Pérez-de-Castro, Belén Picó, and María Luisa Gómez-Guillamón. 2022. "Advanced Genetic Studies on Powdery Mildew Resistance in TGR-1551" International Journal of Molecular Sciences 23, no. 20: 12553. https://doi.org/10.3390/ijms232012553
APA StyleLópez-Martín, M., Pérez-de-Castro, A., Picó, B., & Gómez-Guillamón, M. L. (2022). Advanced Genetic Studies on Powdery Mildew Resistance in TGR-1551. International Journal of Molecular Sciences, 23(20), 12553. https://doi.org/10.3390/ijms232012553








