BOA/DHB/Na: An Efficient UV-MALDI Matrix for High-Sensitivity and Auto-Tagging Glycomics
Abstract
1. Introduction
2. Results and Discussion
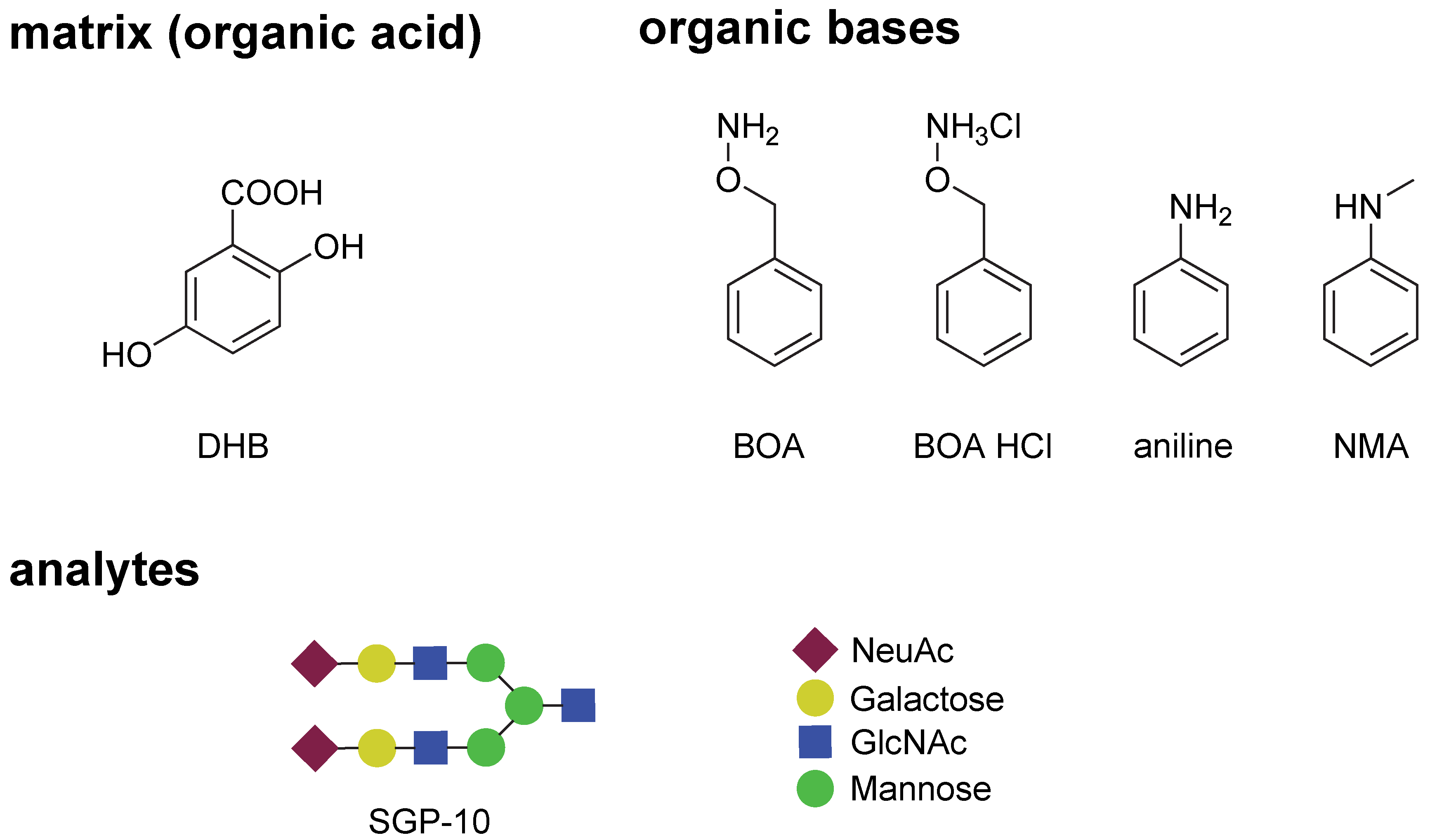
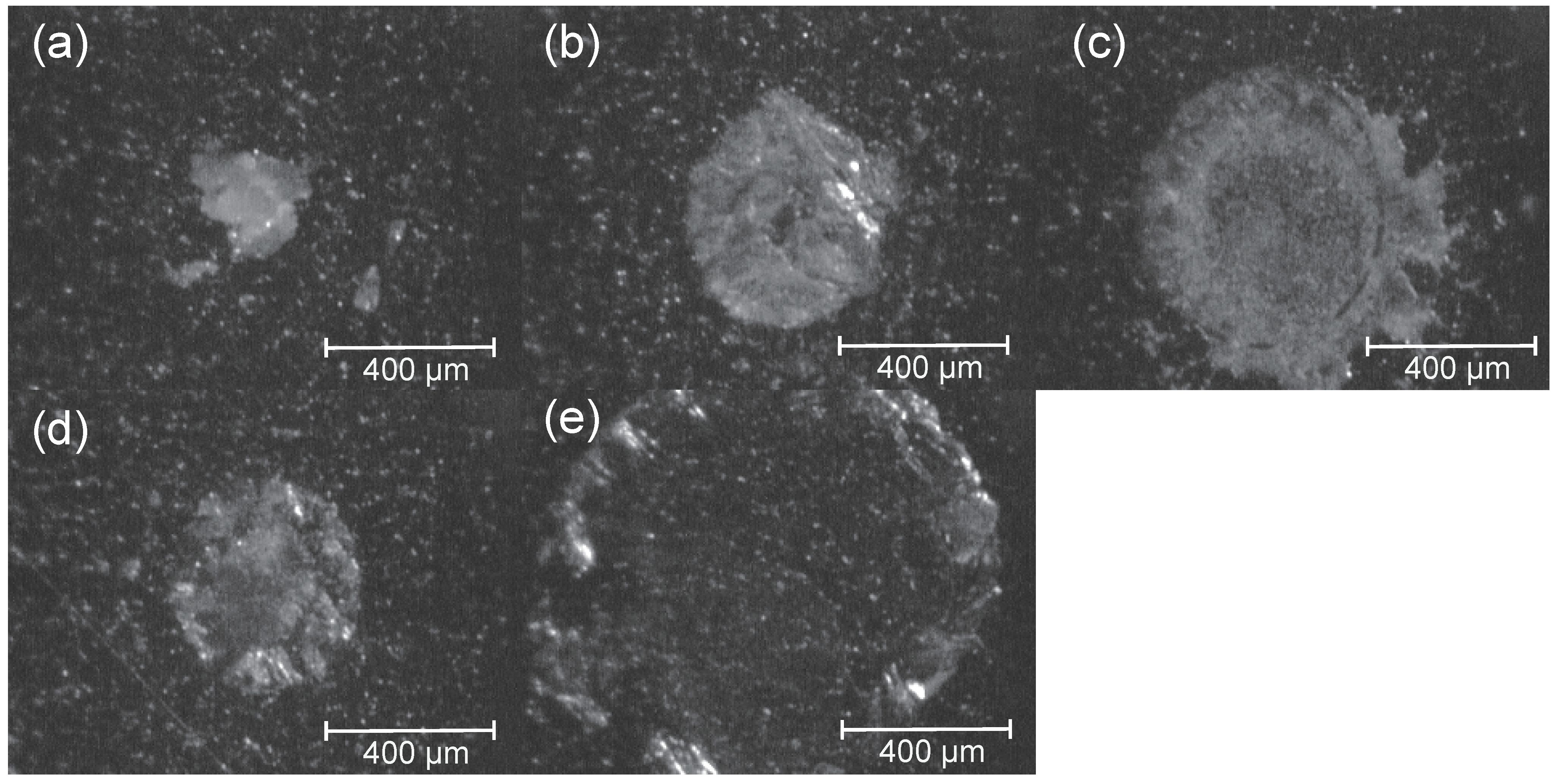
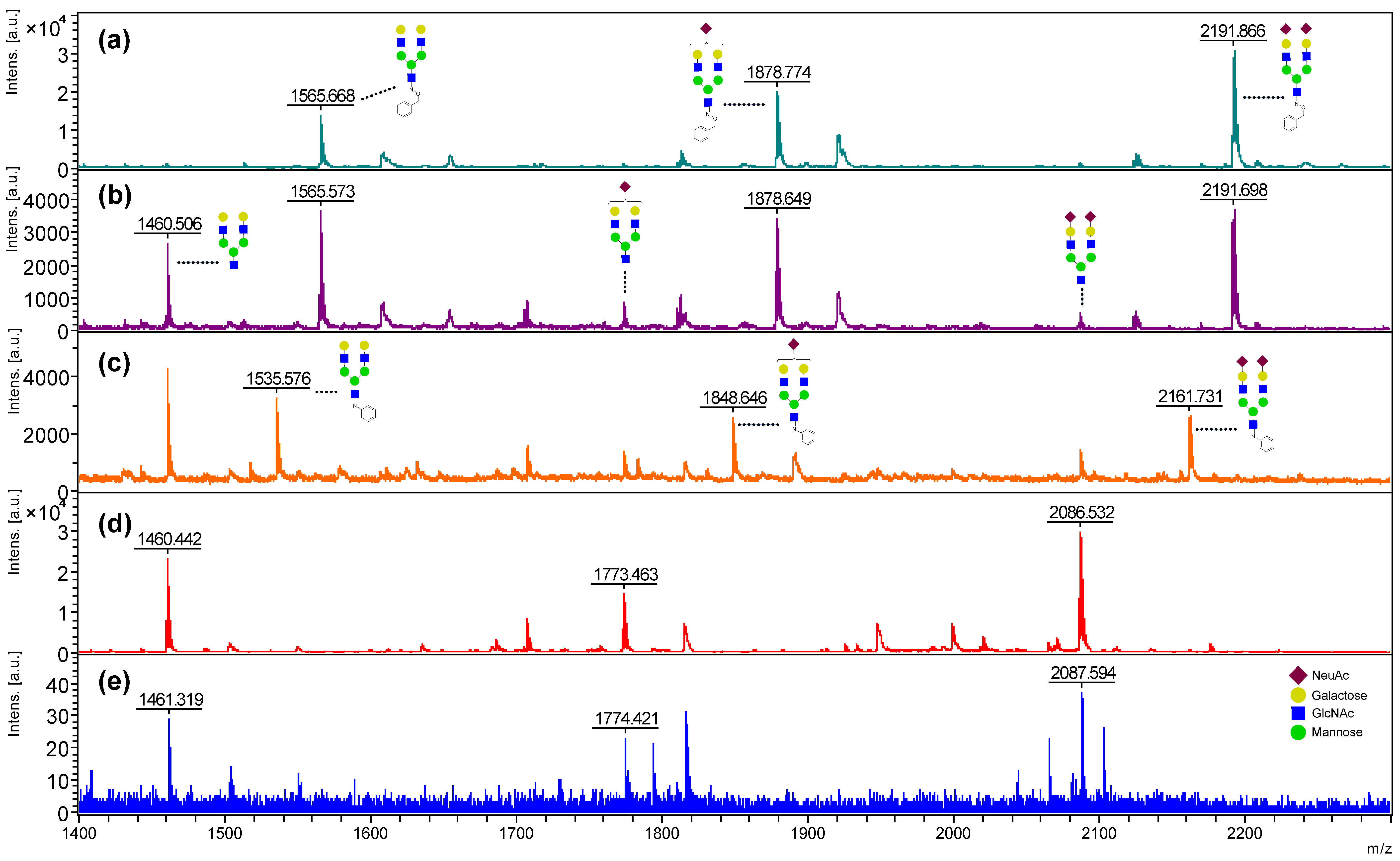
2.1. Evaluation of the BOA Matrix
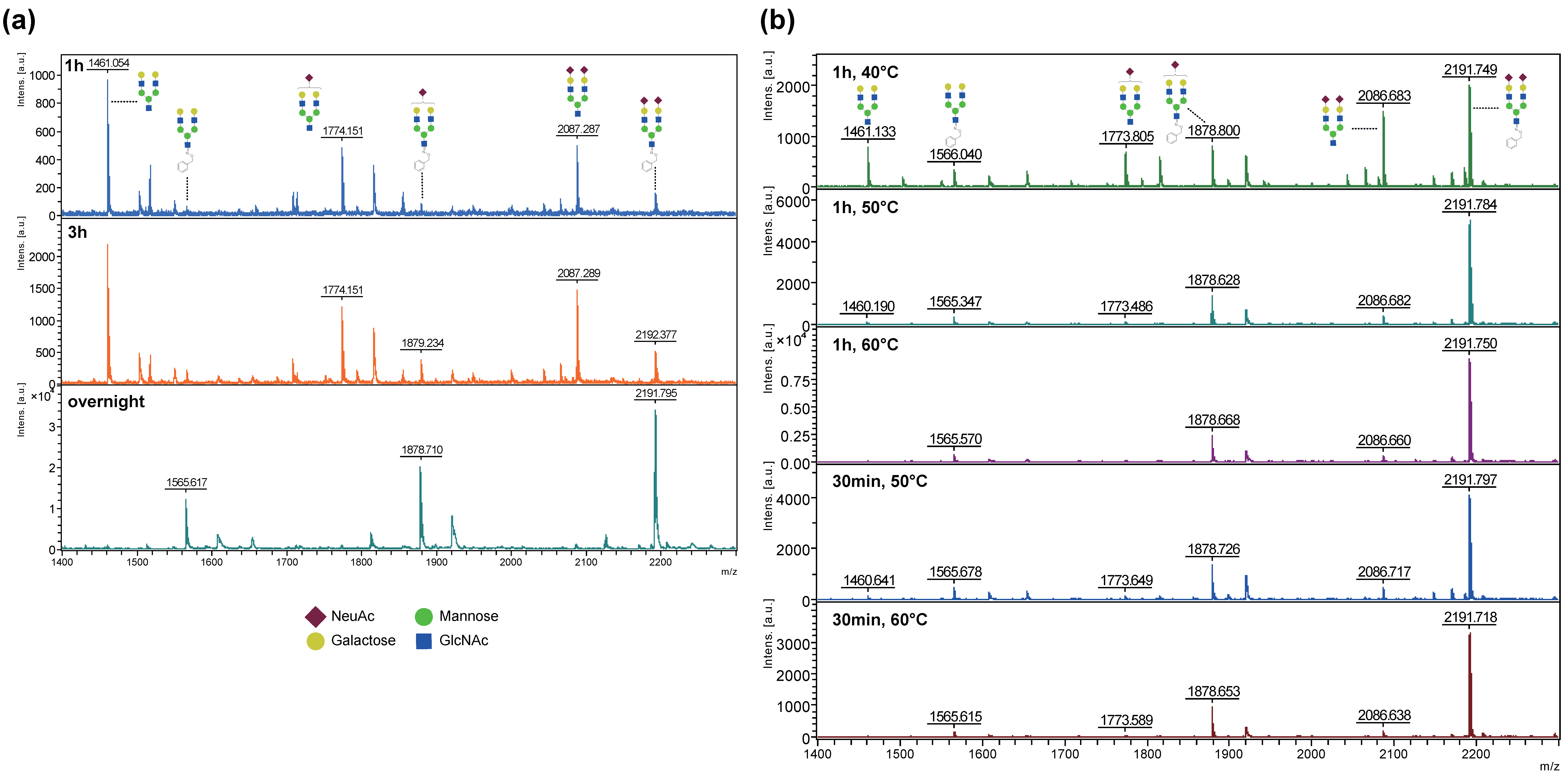
2.2. Optimization of the BOA-Tagging and Solidification Time by the BOA/DHB/Na Matrix with Analytes
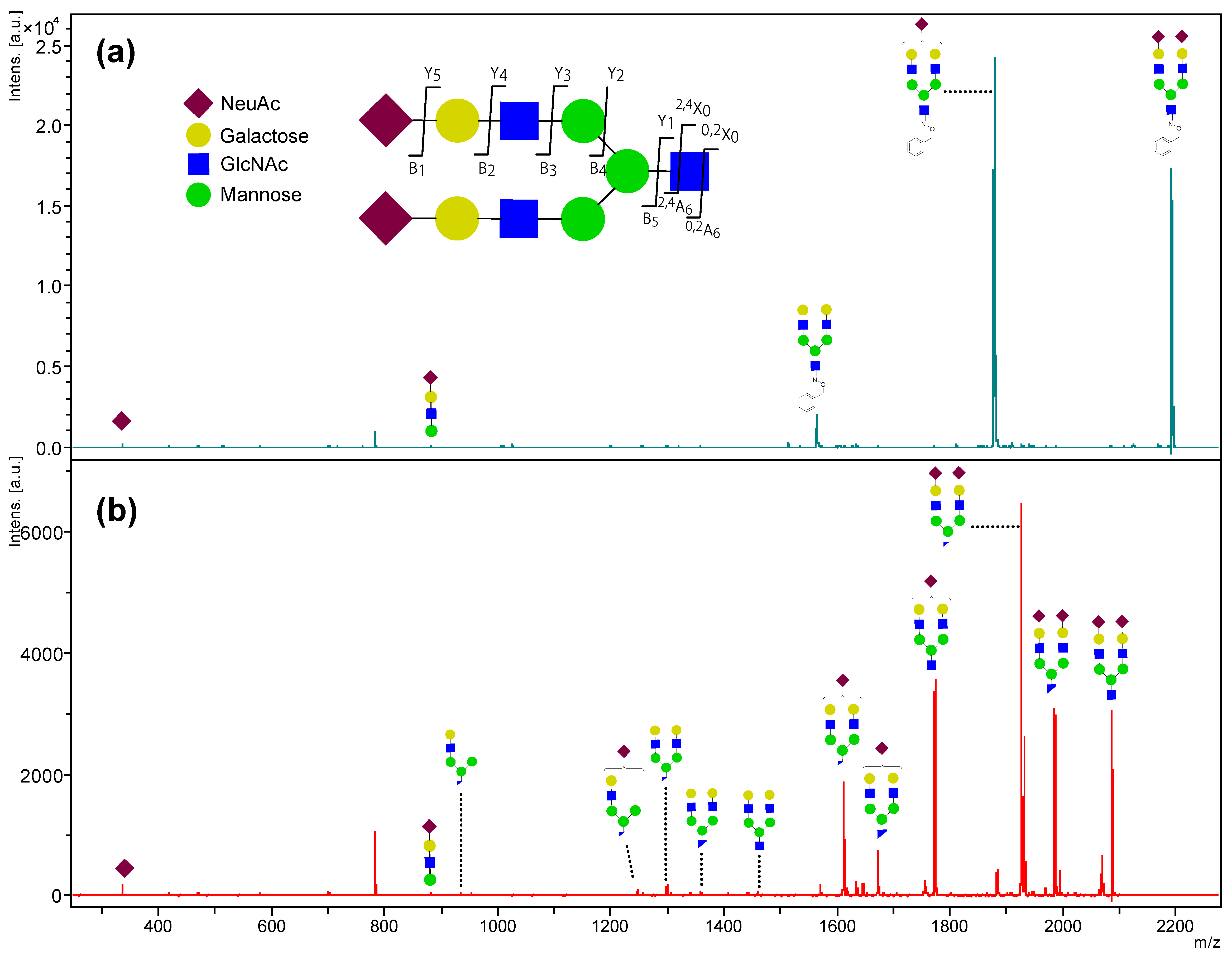
2.3. Stabilizing Effect of the Reducing End of Oligosaccharides by BOA Modification from In-Source and Post-Source Decay in MALDI
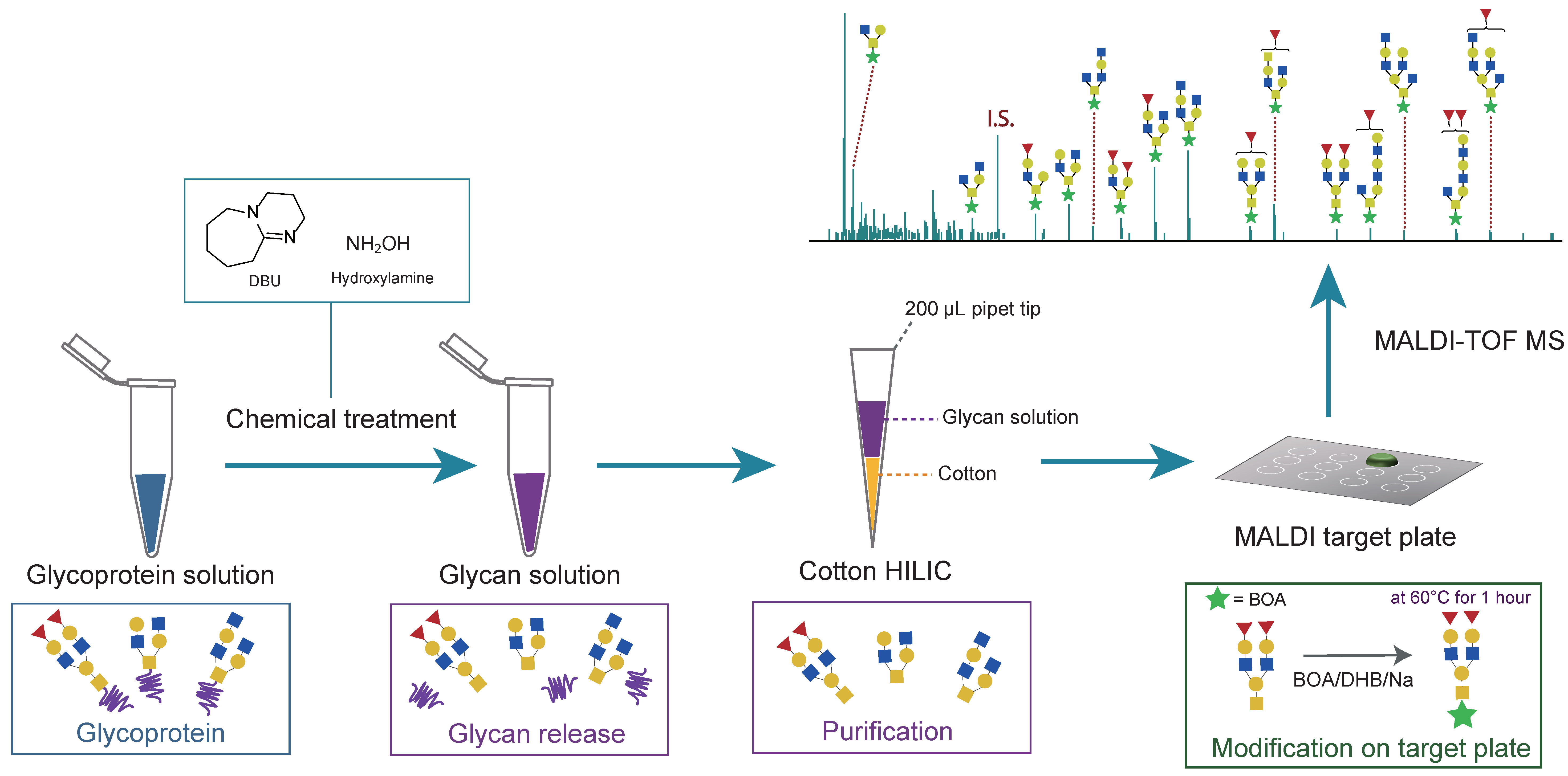
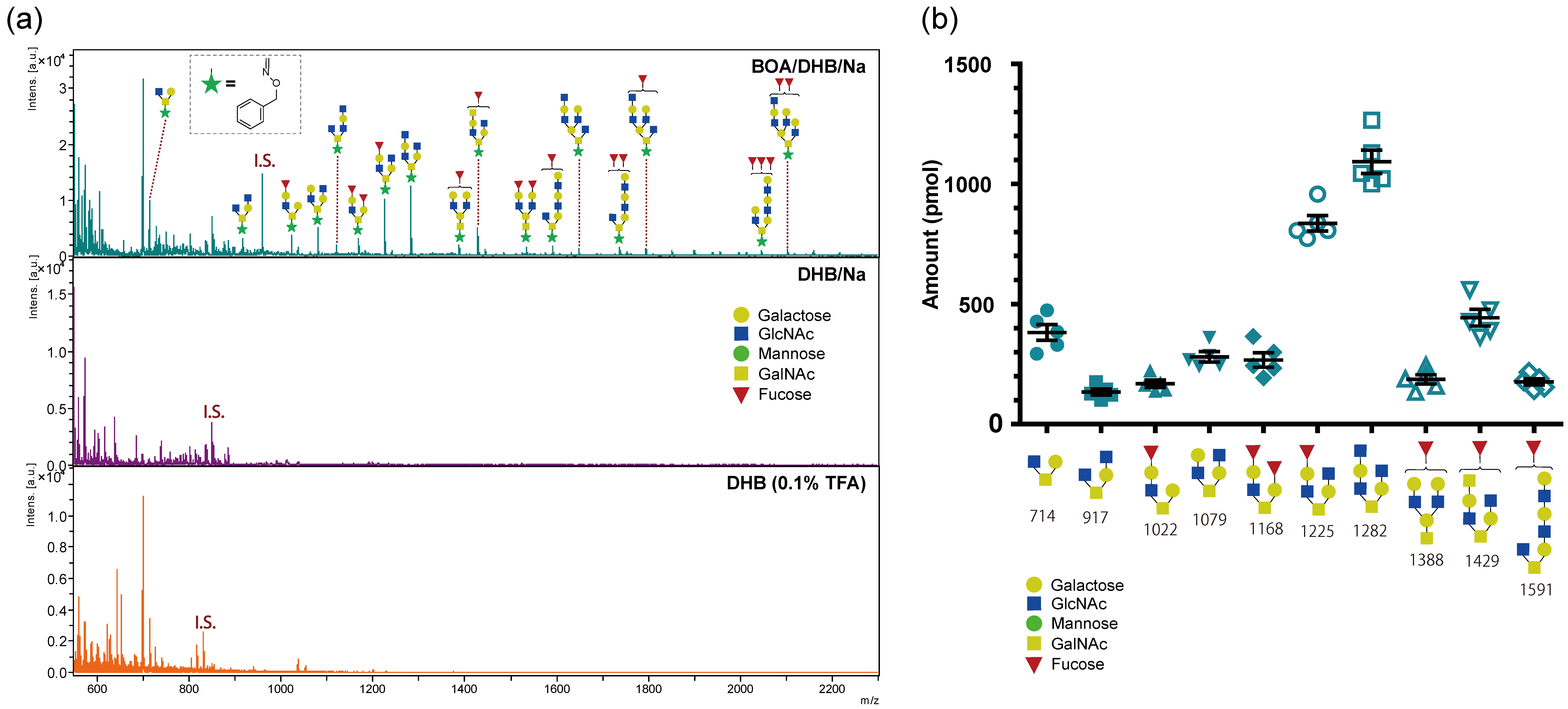
2.4. Profiling of O-Glycans from Mucin Using the BOA/DHB/Na Matrix
3. Materials and Methods
3.1. Materials and Reagents
3.2. Matrices
3.3. Preparation of Pre-Spotted MALDI Target Plates and Analytes
3.4. MALDI–TOFMS
3.5. Activation of Cotton HILIC Microtips
3.6. O-Linked Glycan Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Dobie, C.; Skropeta, D. Insights into the Role of Sialylation in Cancer Progression and Metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Harveys, D.J.; Kuster, B.K.; Naven, T.J.P. Perspectives in the Glycosciences-Matrix-Assisted Laser Desorption/Ionization (MALDI) Mass Spectrometry of Carbohydrates. Glycoconj. J. 1998, 15, 333–338. [Google Scholar] [CrossRef]
- Strupat, K.; Karas, M.; Hlllenkamp, F. 2,5-Dlhydroxybenzoic Acid: A New Matrix for Laser Desorption-Iomzation Mass Spectrometry. Int. J. Mass Spectrom. 1991, 111, 89–102. [Google Scholar] [CrossRef]
- Nie, H.; Li, Y.; Sun, X.L. Recent Advances in Sialic Acid-Focused Glycomics. J. Proteom. 2012, 75, 3098–3112. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Shinohara, Y.; Furukawa, J.I.; Nagahori, N.; Nishimura, S.I. Rapid and Simple Solid-Phase Esterification of Sialic Acid Residues for Quantitative Glycomics by Mass Spectrometry. Chem. Eur. J. 2007, 13, 4797–4804. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Wada, Y.; Tanaka, K. Derivatization for Stabilizing Sialic Acids in MALDI-MS. Anal. Chem. 2005, 77, 4962–4968. [Google Scholar] [CrossRef]
- Kobylis, P.; Stepnowski, P.; Caban, M. Review of the applicability of ionic liquid matrices for the quantification of small molecules by MALDI MS. Microchem. J. 2021, 164, 105983. [Google Scholar] [CrossRef]
- Snovida, S.I.; Chen, V.C.; Perreault, H. Use of a 2,5-Dihydroxybenzoic Acid/Aniline MALDI Matrix for Improved Detection and on-Target Derivatization of Glycans: A Preliminary Report. Anal. Chem. 2006, 78, 8561–8568. [Google Scholar] [CrossRef]
- Hinou, H. Aniline Derivative/DHB/Alkali Metal Matrices for Reflectron Mode MALDI-TOF and TOF/TOF MS Analysis of Unmodified Sialylated Oligosaccharides and Glycopeptides. Int. J. Mass Spectrom. 2019, 443, 109–115. [Google Scholar] [CrossRef]
- Urakami, S.; Hinou, H. Glycan-Selective MALDI In-souce Decay Analysis of Intact Glycoprotein. Anal. Sens. 2022, 2, e202100040. [Google Scholar]
- Saggiomo, V.; Lüning, U. On the Formation of Imines in Water-a Comparison. Tetrahedron Lett. 2009, 50, 4663–4665. [Google Scholar] [CrossRef]
- Kölmel, D.K.; Kool, E.T. Oximes and Hydrazones in Bioconjugation: Mechanism and Catalysis. Chem. Rev. 2017, 117, 10358–10376. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, S.; Boturyn, D.; Marra, A.; Renaudet, O.; Dumy, P. Oxime Ligation: A Chemoselective Click-Type Reaction for Accessing Multifunctional Biomolecular Constructs. Chem. Eur. J. 2014, 20, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Gebrehiwot, A.G.; Melka, D.S.; Kassaye, Y.M.; Rehan, F.; Rangappa, S.; Hinou, H.; Kamiyama, T.; Nishimura, S.I. Healthy Human Serum N-Glycan Profiling Reveals the Influence of Ethnic Variation on the Identified Cancer-Relevant Glycan Biomarkers. PLoS ONE 2018, 13, e0209515. [Google Scholar] [CrossRef] [PubMed]
- Kudina, O.; Eral, B.; Mugele, F. e-MALDI: An An Electrowetting-Enhanced Drop Drying Method for MALDI Mass Spectrometry. Anal. Chem. 2016, 88, 4669–4675. [Google Scholar] [CrossRef]
- Du, Q.; Jia, W.; Dai, J.; Du, C.; Shao, A.; Zhang, R.; Ji, Y. Effect of Dissociation Constant (PKa) of Natural Organic Matter on Photo-Generation of Reactive Oxygen Species (ROS). J. Photochem. Photobiol. A 2020, 391, 112345. [Google Scholar] [CrossRef]
- Kurogochi, M.; Nishimura, S.I. Structural Characterization of N-Glycopeptides by Matrix-Dependent Selective Fragmentation of MALDI-TOF/TOF Tandem Mass Spectrometry. Anal. Chem. 2004, 76, 6097–6101. [Google Scholar] [CrossRef]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A Novel MALDI LIFT-TOF/TOF Mass Spectrometer for Proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef]
- Cooper, C.A.; Gasteiger, E.; Packer, N.H. GlycoMod-A software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 2001, 1, 340–349. [Google Scholar] [CrossRef]
- Miura, Y.; Kato, K.; Takegawa, Y.; Kurogochi, M.; Furukawa, J.I.; Shinohara, Y.; Nagahori, N.; Amano, M.; Hinou, H.; Nishimura, S.I. Glycoblotting-Assisted O-Glycomics: Ammonium Carbamate Allows for Highly Efficient O-Glycan Release from Glycoproteins. Anal. Chem. 2010, 82, 10021–10029. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, A.; Thet Tin, W.W.; Toyoda, M.; Sakaguchi, M. A Practical Method of Liberating O-Linked Glycans from Glycoproteins Using Hydroxylamine and an Organic Superbase. Biochem. Biophys. Res. Commun. 2019, 513, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Selman, M.H.J.; Hemayatkar, M.; Deelder, A.M.; Wuhrer, M. Cotton HILIC SPE Microtips for Microscale Purification and Enrichment of Glycans and Glycopeptides. Anal. Chem. 2011, 83, 2492–2499. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, J.I.; Fujitani, N.; Araki, K.; Takegawa, Y.; Odama, K.; Shinohara, Y. A Versatile Method for Analysis of Serine/Threonine Posttranslational Modifications by β-Elimination in the Presence of Pyrazolone Analogues. Anal. Chem. 2011, 83, 9060–9067. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Hirabayashi, J.; Kakei, K. Analysis of O-Glycans as 9-Fluorenylmethyl Derivatives and Its Application to the Studies on Glycan Array. Anal. Chem. 2013, 85, 3325–3333. [Google Scholar] [CrossRef]
- Goso, Y.; Sugaya, T.; Ishihara, K.; Kurihara, M. Comparison of Methods to Release Mucin-Type O-Glycans for Glycomic Analysis. Anal. Chem. 2017, 89, 8870–8876. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barada, E.; Hinou, H. BOA/DHB/Na: An Efficient UV-MALDI Matrix for High-Sensitivity and Auto-Tagging Glycomics. Int. J. Mol. Sci. 2022, 23, 12510. https://doi.org/10.3390/ijms232012510
Barada E, Hinou H. BOA/DHB/Na: An Efficient UV-MALDI Matrix for High-Sensitivity and Auto-Tagging Glycomics. International Journal of Molecular Sciences. 2022; 23(20):12510. https://doi.org/10.3390/ijms232012510
Chicago/Turabian StyleBarada, Erina, and Hiroshi Hinou. 2022. "BOA/DHB/Na: An Efficient UV-MALDI Matrix for High-Sensitivity and Auto-Tagging Glycomics" International Journal of Molecular Sciences 23, no. 20: 12510. https://doi.org/10.3390/ijms232012510
APA StyleBarada, E., & Hinou, H. (2022). BOA/DHB/Na: An Efficient UV-MALDI Matrix for High-Sensitivity and Auto-Tagging Glycomics. International Journal of Molecular Sciences, 23(20), 12510. https://doi.org/10.3390/ijms232012510




