Different RNA Elements Control Viral Protein Synthesis in Polerovirus Isolates Evolved in Separate Geographical Regions
Abstract
1. Introduction
2. Results
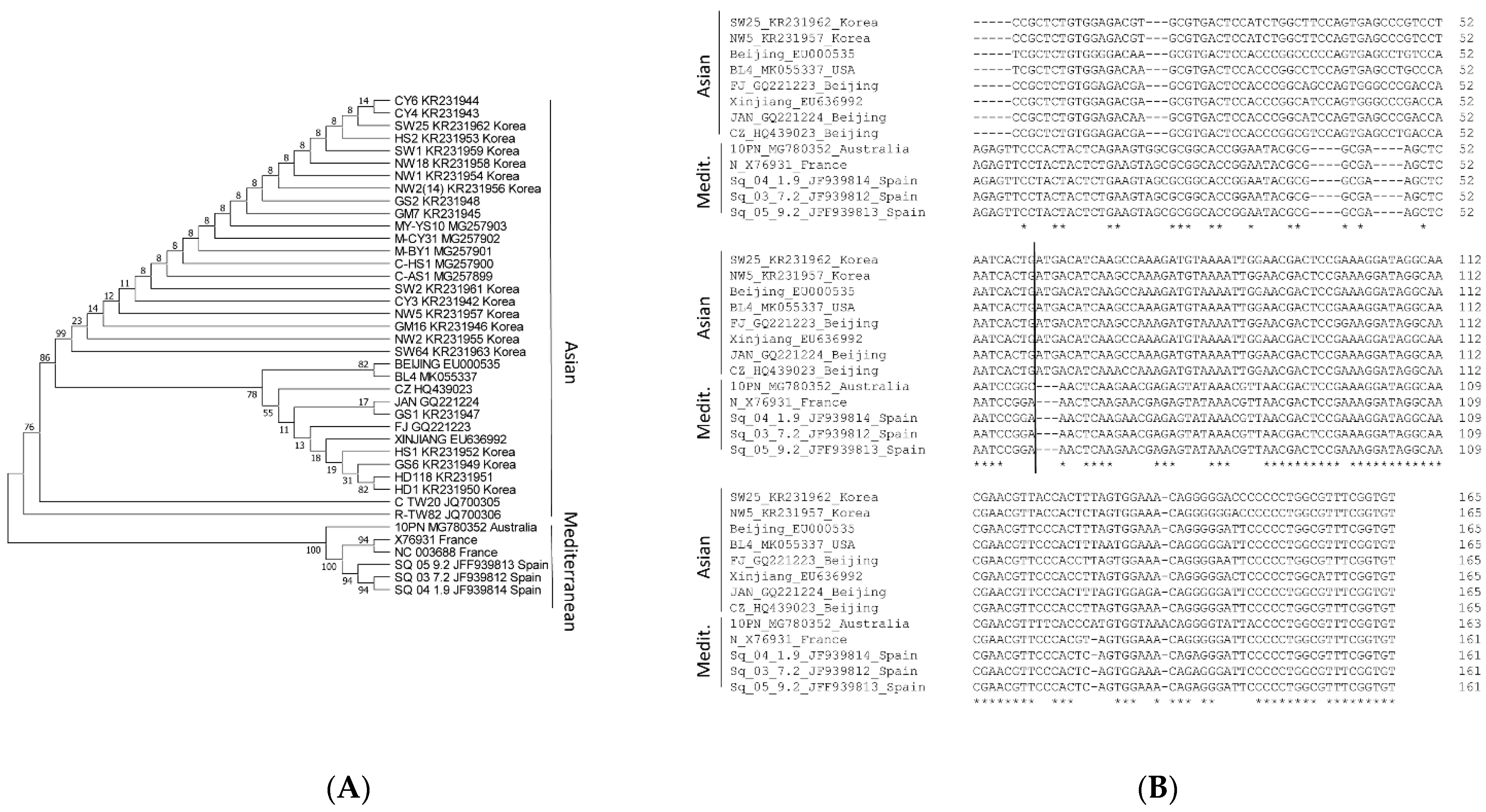
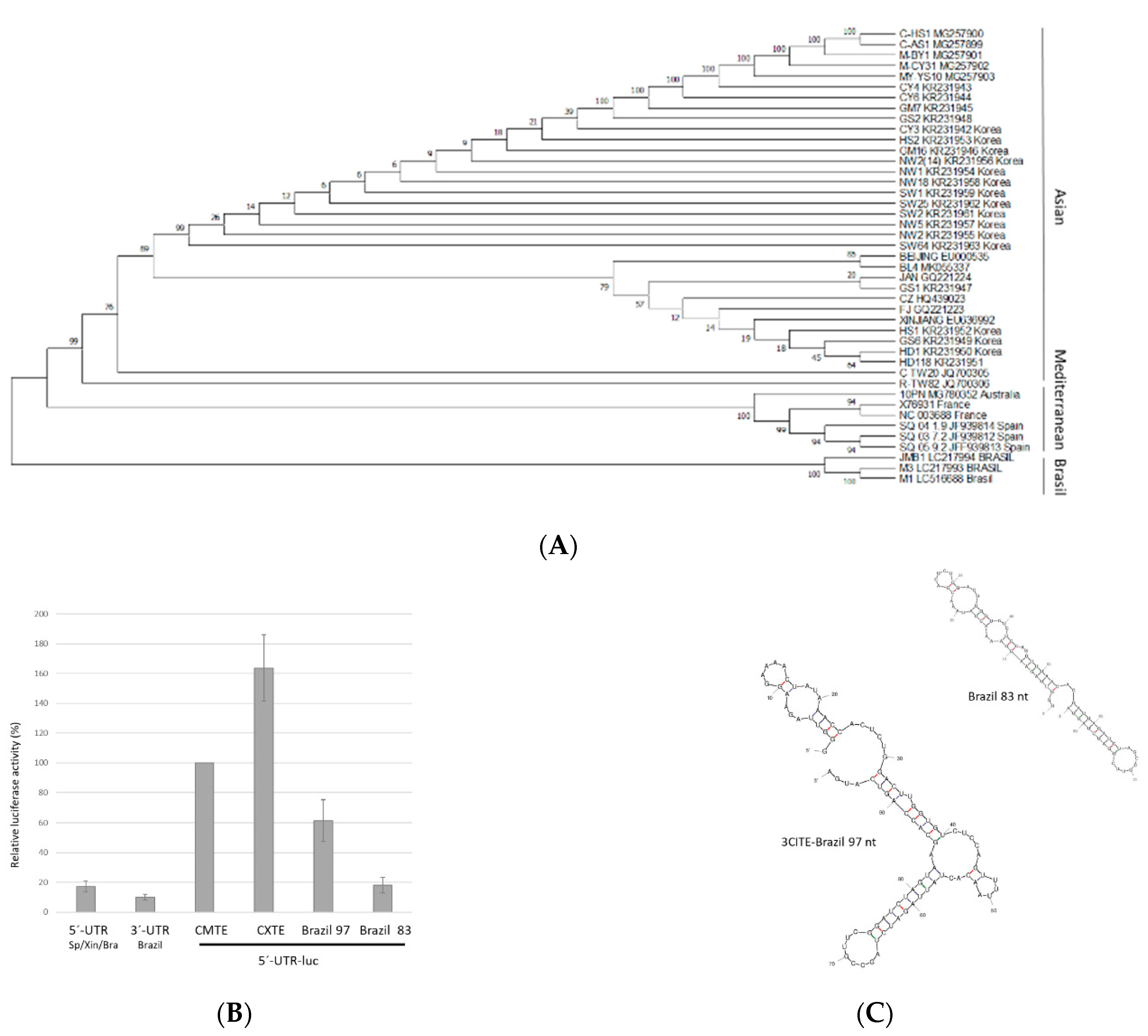
2.1. Conservation of the 3′-UTR Sequences of CABYV Isolates
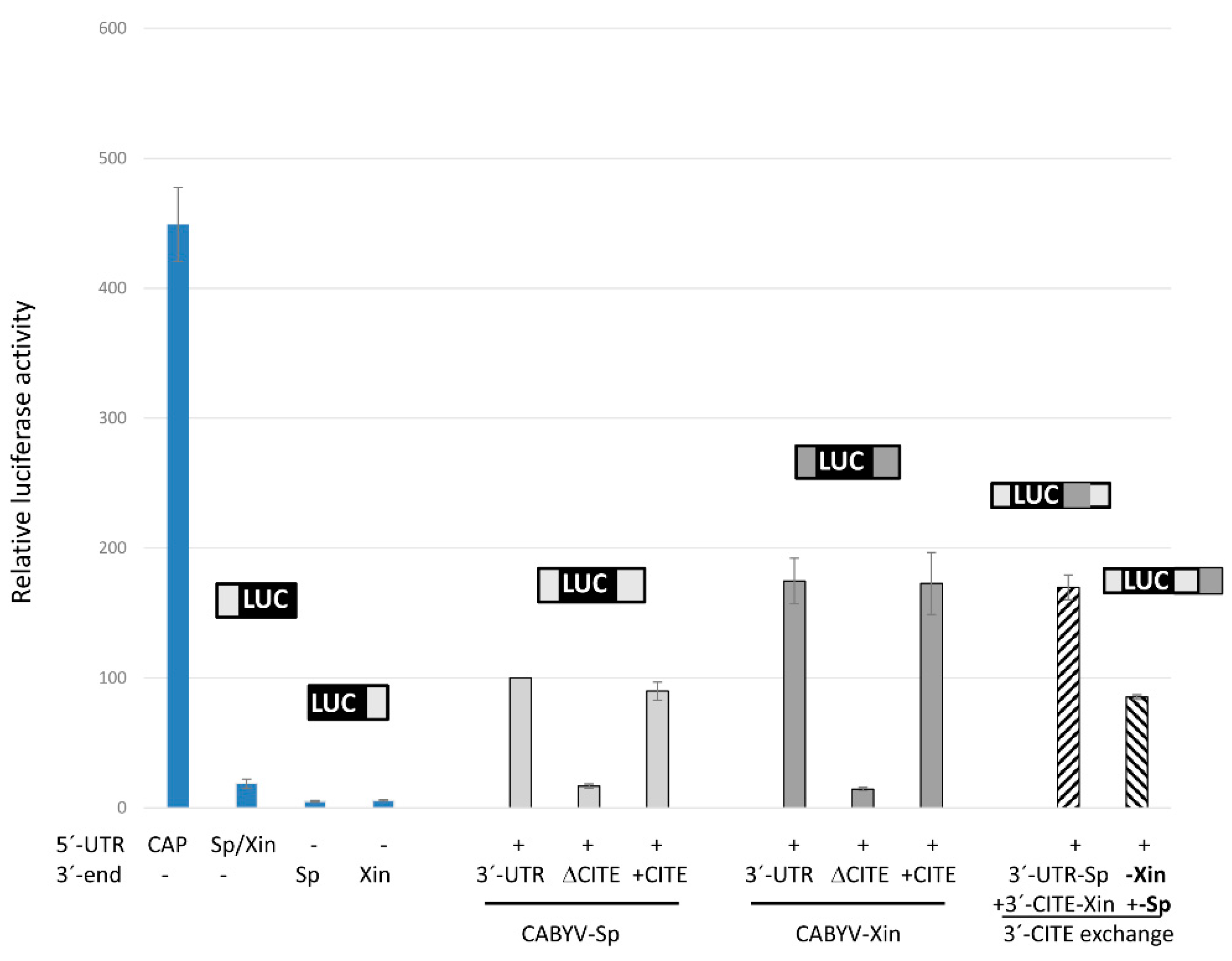
2.2. Identification of 3′-CITEs in CABYV 3′-UTRs
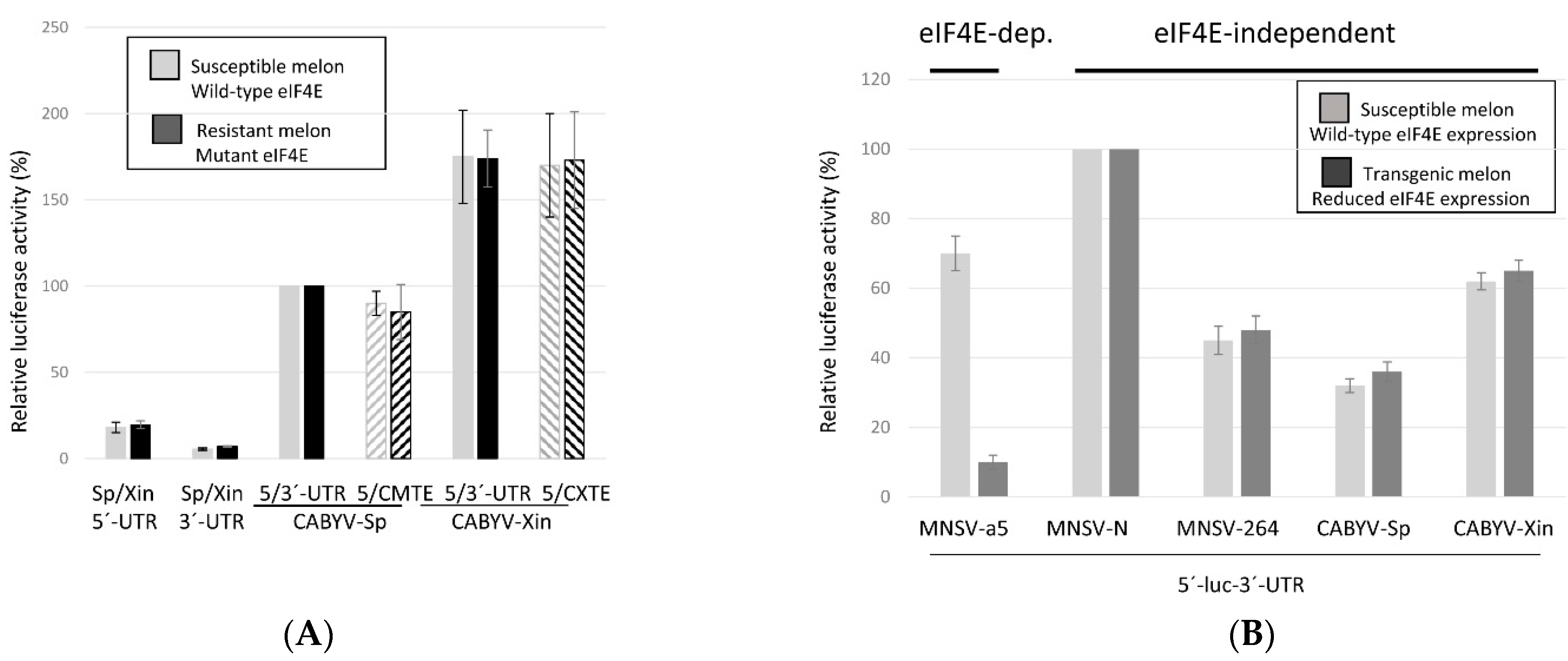
2.3. Translation Mediated by the CABYV 3′-CITEs Is eIF4E-Independent
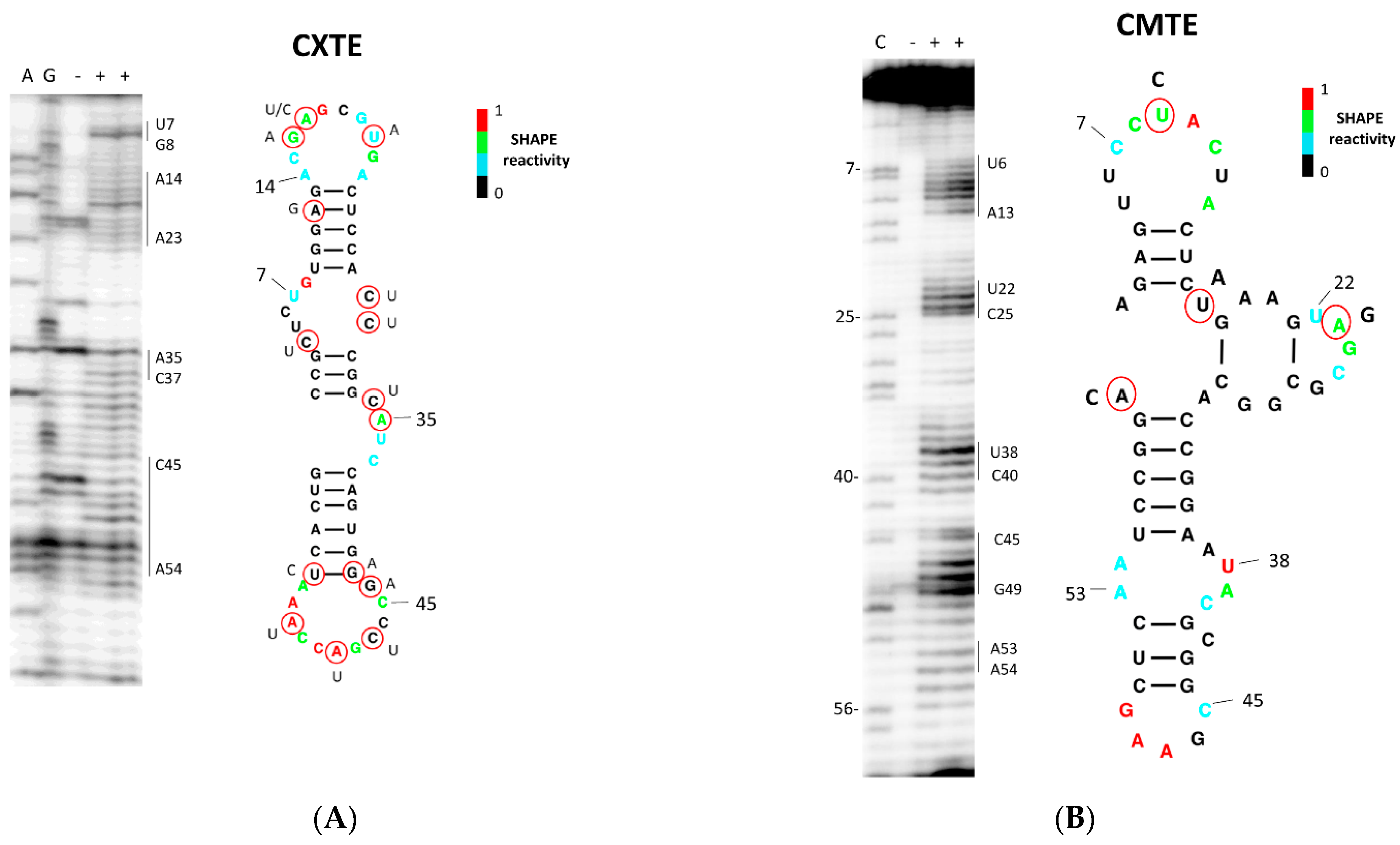
2.4. CABYV 3′-CITEs Vary in Sequence and Structure
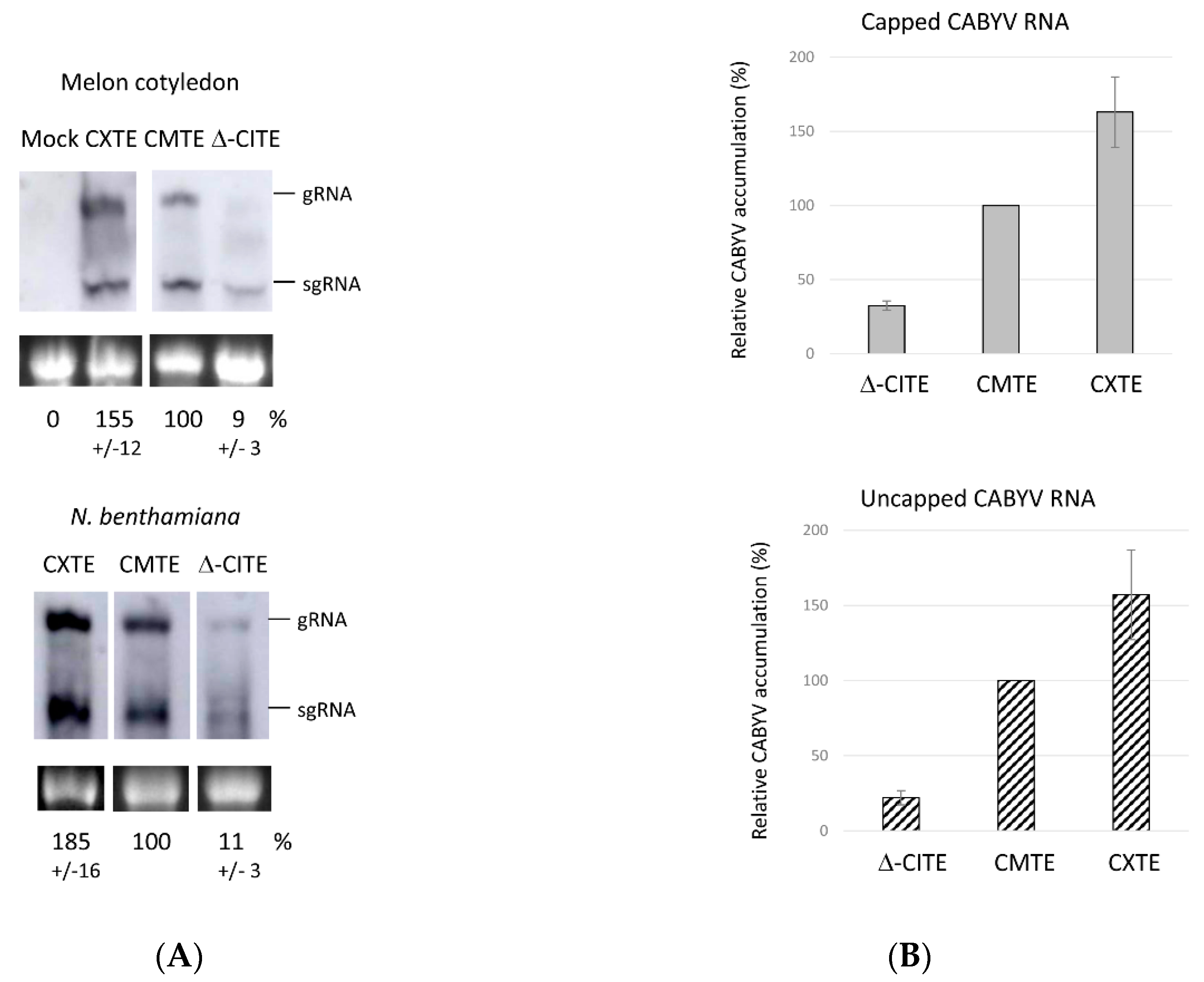
2.5. Role of 3′-CITEs on Virus Multiplication Activity of CABYV
2.6. Possible 3′-CITE of New Brazilian CABYV Isolates
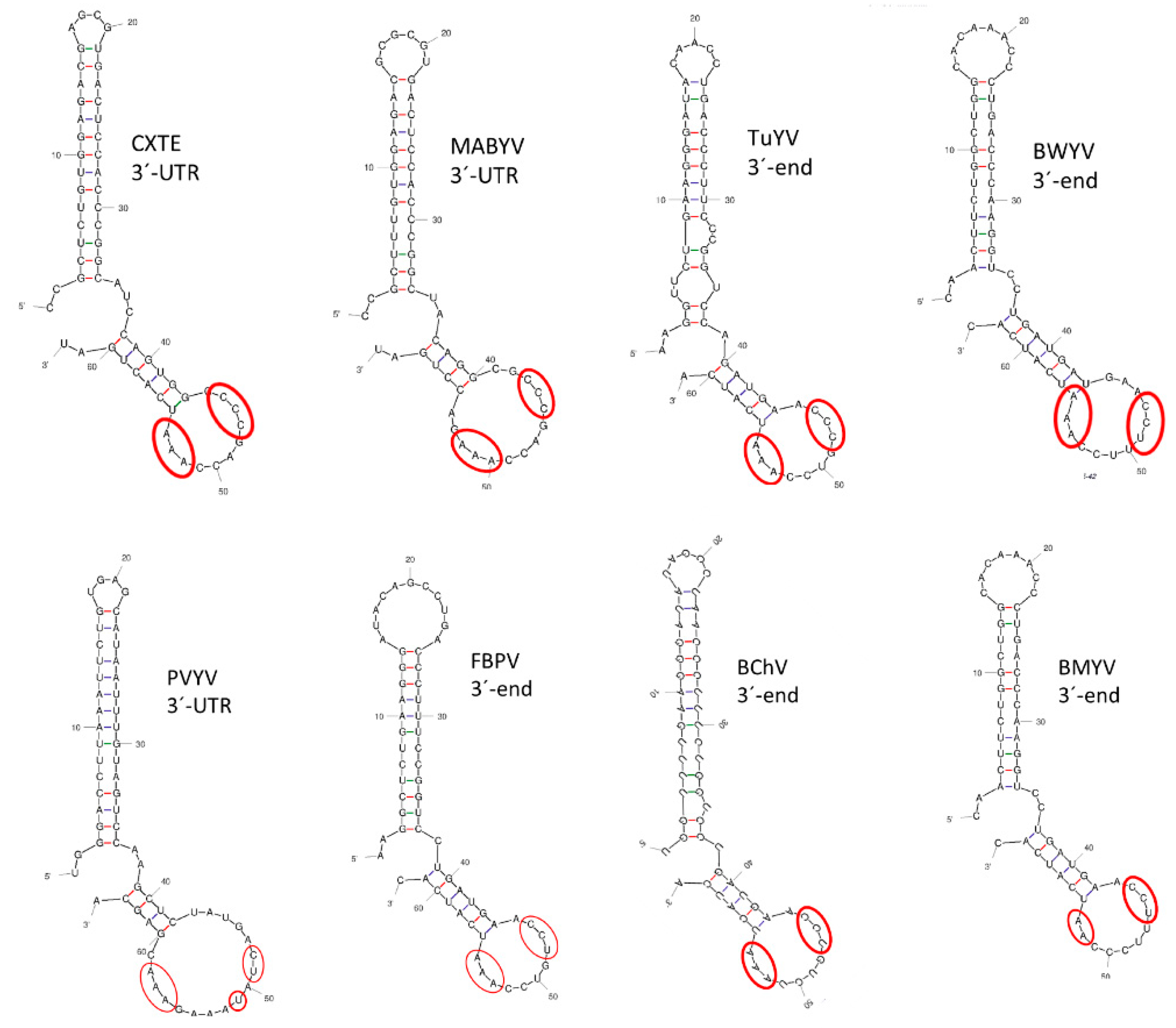
2.7. Searching for Possible 3′-CITEs in Polerovirus 3′-Ends
3. Discussion
4. Materials and Methods
4.1. Luc-Constructs
4.2. In Vivo Translation in Melon Protoplasts
4.3. RNA Structure Analysis
4.4. Construction and Multiplication Analysis of Mutant Viruses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lecoq, H.; Bourdin, D.; Wipf-Scheibel, C.; Bon, M.; Lot, H.; Lemaire, O.; Herrbach, E. A New Yellowing Disease of Cucurbits Caused by a Luteovirus, Cucurbit Aphid-Borne Yellows Virus. Plant Pathol. 1992, 41, 749–761. [Google Scholar] [CrossRef]
- Kassem, M.A.; Sempere, R.N.; Juarez, M.; Aranda, M.A.; Truniger, V. Cucurbit Aphid-Borne Yellows Virus Is Prevalent in Field-Grown Cucurbit Crops of Southeastern Spain. Plant Dis. 2007, 91, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Mnari Hattab, M.; Kummert, J.; Roussel, S.; Ezzaier, K.; Zouba, A.; Jijakli, M.H. First Report of Cucurbit Aphid-Borne Yellows Virus in Tunisia Causing Yellows on Five Cucurbitacious Species. Plant Dis. 2005, 89, 776. [Google Scholar] [CrossRef] [PubMed]
- Knierim, D.; Deng, T.C.; Tsai, W.S.; Green, S.K.; Kenyon, L. Molecular Identification of Three Distinct Polerovirus Species and a Recombinant Cucurbit Aphid-Borne Yellows Virus Strain Infecting Cucurbit Crops in Taiwan. Plant Pathol. 2010, 59, 991–1002. [Google Scholar] [CrossRef]
- Boubourakas, I.N.; Avgelis, A.D.; Kyriakopoulou, P.E.; Katis, N.I. Occurrence of Yellowing Viruses (Beet Pseudo-Yellows Virus, Cucurbit Yellow Stunting Disorder Virus and Cucurbit Aphid-Borne Yellows Virus) Affecting Cucurbits in Greece. Plant Pathol. 2006, 55, 276–283. [Google Scholar] [CrossRef]
- Xiang, H.Y.; Shang, Q.X.; Han, C.G.; Li, D.W.; Yu, J.L. Complete Sequence Analysis Reveals Two Distinct Poleroviruses Infecting Cucurbits in China. Arch. Virol. 2008, 153, 1155–1160. [Google Scholar] [CrossRef]
- Orfanidou, C.; Maliogka, V.I.; Katis, N.I. First Report of Cucurbit Chlorotic Yellows Virus in Cucumber, Melon, and Watermelon in Greece. Plant Dis. 2014, 98, 1446. [Google Scholar] [CrossRef]
- Bananej, K.; Desbiez, C.; Wipf-Scheibel, C.; Vahdat, I.; Kheyr-Pour, A.; Ahoonmanesh, A.; Lecoq, H. First Report of Cucurbit Aphid-Borne Yellows Virus in Iran Causing Yellows on Four Cucurbit Crops. Plant Dis. 2006, 90, 526. [Google Scholar] [CrossRef]
- Maina, S.; Barbetti, M.J.; Edwards, O.R.; Minemba, D.; Areke, M.W.; Jones, R.A.C. First Complete Genome Sequence of Cucurbit Aphid-Borne Yellows Virus from Papua New Guinea. Genome Announc. 2018, 6, 11. [Google Scholar] [CrossRef]
- Costa, T.M.; Blawid, R.; Aranda, M.A.; Freitas, D.M.S.; Andrade, G.P.; Inoue-Nagata, A.K.; Nagata, T. Cucurbit Aphid-Borne Yellows Virus from Melon Plants in Brazil Is an Interspecific Recombinant. Arch. Virol. 2019, 164, 249–254. [Google Scholar] [CrossRef]
- Menzel, W.; Maeritz, U.; Seigner, L. First Report of Cucurbit Aphid-Borne Yellows Virus Infecting Cucurbits in Germany. New Dis. Rep. 2020, 41, 1. [Google Scholar] [CrossRef]
- Listihani, L.; Damayanti, T.A.; Hidayat, S.H.; Wiyono, S. First Report of Cucurbit Aphid-Borne Yellows Virus on Cucumber in Java, Indonesia. J. Gen. Plant Pathol. 2020, 86, 219–223. [Google Scholar] [CrossRef]
- Mnari-Hattab, M.; Gauthier, N.; Zouba, A. Biological and Molecular Characterization of the Cucurbit Aphid-Borne Yellows Virus Affecting Cucurbits in Tunisia. Plant Dis. 2009, 93, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.M.; Inoue-Nagata, A.K.; Vidal, A.H.; da G. Ribeiro, S.; Nagata, T. The Recombinant Isolate of Cucurbit Aphid-Borne Yellows Virus from Brazil Is a Polerovirus Transmitted by Whiteflies. Plant Pathol. 2020, 69, 1042–1050. [Google Scholar] [CrossRef]
- Sõmera, M.; Fargette, D.; Hébrard, E.; Sarmiento, C. ICTV Virus Taxonomy Profile: Solemoviridae 2021. J. Gen. Virol. 2021, 102, 1–2. [Google Scholar] [CrossRef]
- Prufer, D.; Wipf-Scheibel, C.A.T.H.; Richards, K.; Guilley, H.; Lecoq, H.; Jonard, G.; Pru, D. Synthesis of a Full-Length Infectious CDNA Clone of Cucurbit Aphid-Borne Yellows Virus and Its Use in Gene Exchange Experiments with Structural Proteins from Other Luteoviruses. Virology 1995, 214, 150–158. [Google Scholar] [CrossRef][Green Version]
- Pe, S.; Reinbold, C.; Erdinger, M.; Herrbach, E.; Richards, K.; Brault, V.; Perigon, S.; Reinbold, C.; Erdinger, M.; Scheidecker, D.; et al. The Polerovirus Minor Capsid Protein Determines Vector Specificity and Intestinal Tropism in the Aphid. J. Virol. 2005, 79, 9685–9693. [Google Scholar] [CrossRef]
- Delfosse, V.C.; Barrios Barón, M.P.; Distéfano, A.J. What We Know about Poleroviruses: Advances in Understanding the Functions of Polerovirus Proteins. Plant Pathol. 2021, 70, 1047–1061. [Google Scholar] [CrossRef]
- Smirnova, E.; Firth, A.E.; Miller, W.A.; Scheidecker, D.; Brault, V.; Reinbold, C.; Rakotondrafara, A.M.; Chung, B.Y.W.; Ziegler-Graff, V. Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement. PLoS Pathog. 2015, 11, 1–31. [Google Scholar] [CrossRef]
- Taliansky, M.; Mayo, M.A.; Barker, H. Potato Leafroll Virus: A Classic Pathogen Shows Some New Tricks. Mol. Plant Pathol. 2003, 4, 81–89. [Google Scholar] [CrossRef]
- Reinbold, C.; Lacombe, S.; Ziegler-Graff, V.; Scheidecker, D.; Wiss, L.; Beuve, M.; Caranta, C.; Brault, V. Closely Related Poleroviruses Depend on Distinct Translation Initiation Factors to Infect Arabidopsis Thaliana. Mol. Plant-Microbe Interact. 2013, 26, 257. [Google Scholar] [CrossRef] [PubMed]
- Kassem, M.A.; Juarez, M.; Gómez, P.; Mengual, C.M.; Sempere, R.N.; Plaza, M.; Elena, S.F.; Moreno, A.; Fereres, A.; Aranda, M.A. Genetic Diversity and Potential Vectors and Reservoirs of Cucurbit Aphid-Borne Yellows Virus in Southeastern Spain. Phytopathology 2013, 103, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Maachi, A.; Donaire, L.; Hernando, Y.; Aranda, M.A. Genetic Differentiation and Migration Fluxes of Viruses from Melon Crops and Crop Edge Weeds. J. Virol. 2022, 96, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Xiang, H.; Han, C.; Li, D.; Yu, J. Distribution and Molecular Diversity of Three Cucurbit-Infecting Poleroviruses in China. Virus Res. 2009, 145, 341–346. [Google Scholar] [CrossRef]
- Kwak, H.R.; Lee, H.J.; Kim, E.A.; Seo, J.K.; Kim, C.S.; Lee, S.G.; Kim, J.S.; Choi, H.S.; Kim, M. Complete Genome Sequences and Evolutionary Analysis of Cucurbit Aphid-Borne Yellows Virus Isolates from Melon in Korea. Plant Pathol. J. 2018, 34, 532–543. [Google Scholar] [CrossRef]
- van Regenmortel, M.H.; Fauquet, C.M.; Bishop, D.H.L.; Carstens, E.B.; Estes, M.K.; Lemon, S.M.; Maniliff, J.; Mayo, M.A.; McGeoch, D.J.; Pringle, C.R.; et al. Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- Dreher, T.W.; Miller, W.A. Translational Control in Positive Strand RNA Plant Viruses. Virology 2006, 344, 185–197. [Google Scholar] [CrossRef]
- Kneller, E.L.P.; Rakotondrafara, A.M.; Miller, W.A. Cap-Independent Translation of Plant Viral RNAs. Virus Res. 2006, 119, 63–75. [Google Scholar] [CrossRef]
- Miras, M.; Miller, W.A.; Truniger, V.; Aranda, M.A. Non-Canonical Translation in Plant RNA Viruses. Front. Plant Sci. 2017, 8, 494. [Google Scholar] [CrossRef]
- Miller, W.A.; White, K.A. Long-Distance RNA-RNA Interactions in Plant Virus Gene Expression and Replication. Annu. Rev. Phytopathol. 2006, 44, 447–467. [Google Scholar] [CrossRef]
- Simon, A.E.; Miller, W.A. 3′ Cap-Independent Translation Enhancers of Plant Viruses. Annu. Rev. Microbiol. 2013, 67, 21–42. [Google Scholar] [CrossRef]
- Truniger, V.; Miras, M.; Aranda, M.A. Structural and Functional Diversity of Plant Virus 3′-Cap-Independent Translation Enhancers (3′-CITEs). Front. Plant Sci. 2017, 8, 2047. [Google Scholar] [CrossRef] [PubMed]
- Miras, M.; Sempere, R.N.; Kraft, J.J.; Miller, W.A.; Aranda, M.A.; Truniger, V. Interfamilial Recombination between Viruses Led to Acquisition of a Novel Translation-Enhancing RNA Element That Allows Resistance Breaking. New Phytol. 2014, 202, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Nieto, C.; Gonzalez-Ibeas, D.; Aranda, M. Mechanism of Plant EIF4E-Mediated Resistance against a Carmovirus (Tombusviridae): Cap-Independent Translation of a Viral RNA Controlled in Cis by an (a)Virulence Determinant. Plant J. 2008, 56, 716–727. [Google Scholar] [CrossRef]
- Miras, M.; Truniger, V.; Querol-Audi, J.; Aranda, M.A. Analysis of the Interacting Partners EIF4F and 3′-CITE Required for Melon Necrotic Spot Virus Cap-Independent Translation. Mol. Plant Pathol. 2017, 18, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Nieto, C.; Rodríguez-Moreno, L.; Rodríguez-Hernández, A.M.; Aranda, M.A.; Truniger, V. Nicotiana Benthamiana Resistance to Non-Adapted Melon Necrotic Spot Virus Results from an Incompatible Interaction between Virus RNA and Translation Initiation Factor 4E. Plant J. 2011, 66, 492–501. [Google Scholar] [CrossRef]
- Martin, D.P.; Lemey, P.; Lott, M.; Moulton, V.; Posada, D.; Lefeuvre, P. RDP3: A Flexible and Fast Computer Program for Analyzing Recombination. Bioinformatics 2010, 26, 2462–2463. [Google Scholar] [CrossRef]
- Nieto, C.; Morales, M.; Orjeda, G.; Clepet, C.; Monfort, A.; Sturbois, B.; Puigdomenech, P.; Pitrat, M.; Caboche, M.; Dogimont, C.; et al. An EIF4E Allele Confers Resistance to an Uncapped and Non-Polyadenylated RNA Virus in Melon. Plant J. 2006, 48, 452–462. [Google Scholar] [CrossRef]
- Rodríguez-Hernández, A.M.; Gosalvez, B.; Sempere, R.N.; Burgos, L.; Aranda, M.A.; Truniger, V. Melon RNA Interference (RNAi) Lines Silenced for Cm-EIF4E Show Broad Virus Resistance. Mol. Plant Pathol. 2012, 13, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Diaz, J.A.; Nieto, C.; Moriones, E.; Truniger, V.; Aranda, M.A. Molecular Characterization of a Melon Necrotic Spot Virus Strain That Overcomes the Resistance in Melon and Nonhost Plants. Mol. Plant-Microbe Interact. 2004, 17, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension (SHAPE): Quantitative RNA Structure Analysis at Single Nucleotide Resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, S.A.; Weeks, K.M. Time-Resolved RNA SHAPE Chemistry. J. Am. Chem. Soc. 2008, 130, 16178–16180. [Google Scholar] [CrossRef] [PubMed]
- Rabadán, M.P.; Juárez, M.; De Moya-Ruiz, C.; Gómez, P. Aphid-Borne Viruses Infecting Cultivated Watermelon and Squash in Spain: Characterization of a Variant of Cucurbit Aphid-Borne Yellows Virus (CABYV). Plant Pathol. 2021, 70, 1476–1485. [Google Scholar] [CrossRef]
- McPherson, J.C.; Nester, E.W.; Gordon, M.P. Proteins Encoded by Agrobacterium Tumefaciens Ti Plasmid DNA (T-DNA) in Crown Gall Tumors. Proc. Natl. Acad. Sci. USA 1980, 77, 2666–2670. [Google Scholar] [CrossRef]
- Van Der Wilk, F.; Verbeek, M.; Dullemans, A.M.; Van Den Heuvel, J.F.J.M. The Genome-Linked Protein of Potato Leafroll Virus Is Located Downstream of the Putative Protease Domain of the ORF1 Product. Virology 1997, 234, 300–303. [Google Scholar] [CrossRef]
- Jiang, J.; Laliberté, J.F. The Genome-Linked Protein VPg of Plant Viruses—A Protein with Many Partners. Curr. Opin. Virol. 2011, 1, 347–354. [Google Scholar] [CrossRef]
- Miller, W.A.; Lozier, Z. Yellow Dwarf Viruses of Cereals: Taxonomy and Molecular Mechanisms. Annu. Rev. Phytopathol. 2022, 60, 121–141. [Google Scholar] [CrossRef]
- Shen, R.; WA, M.; Miller, W.A. The 3′ Untranslated Region of Tobacco Necrosis Virus RNA Contains a Barley Yellow Dwarf Virus-like Cap-Independent Translation Element. J. Virol. 2004, 78, 4655–4664. [Google Scholar] [CrossRef]
- Allen, E.; Wang, S.; Miller, W.A. Barley Yellow Dwarf Virus RNA Requires a Cap-Independent Translation Sequence Because It Lacks a 5′ Cap. Virology 1999, 253, 139–144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, F.; Kasprzak, W.K.; Szarko, C.; Shapiro, B.A.; Simon, A.E. The 3′ Untranslated Region of Pea Enation Mosaic Virus Contains Two T-Shaped, Ribosome-Binding, Cap-Independent Translation Enhancers. J. Virol. 2014, 88, 11696–11712. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Mäkinen, K. Insights into the Functions of EIF4E-Binding Motif of VPG in Potato Virus A Infection. Viruses 2020, 12, 197. [Google Scholar] [CrossRef]
- Mayo, M.A.; Robinson, D.J.; Jolly, C.A.; Hyman, L. Nucleotide Sequence of Potato Leafroll Luteovirus RNA. J. Gen. Virol. 1989, 70 Pt 5, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L.; White, K.A. 3′ Cap-Independent Translation Enhancers of Positive-Strand RNA Plant Viruses. Curr. Opin. Virol. 2011, 1, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Moonan, F.; Molina, J.; Mirkov, T.E. Sugarcane Yellow Leaf Virus: An Emerging Virus That Has Evolved by Recombination between Luteoviral and Poleroviral Ancestors. Virology 2000, 269, 156–171. [Google Scholar] [CrossRef]
- Domier, L.L.; McCoppin, N.K.; Larsen, R.C.; D’Arcy, C.J. Nucleotide Sequence Shows That Bean Leafroll Virus Has a Luteovirus-like Genome Organization. J. Gen. Virol. 2002, 83, 1791–1798. [Google Scholar] [CrossRef][Green Version]
- Gibbs, M.J.; Cooper, J.I. A Recombinational Event in the History of Luteoviruses Probably Induced by Base-Pairing between the Genomes of Two Distinct Viruses. Virology 1995, 206, 1129–1132. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Lab Press: New York, NY, USA, 2001. [Google Scholar]
- Miras, M.; Sempere, R.N.; Kraft, J.J.; Miller, W.A.; Aranda, M.A.; Truniger, V. Determination of the Secondary Structure of an RNA Fragment in Solution: Selective 2‘-Hydroxyl Acylation Analyzed by Primer Extension Assay (SHAPE). Bio-Protocol 2015, 5, e1386. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.; Kraft, J.J.; Hui, A.Y.; Miller, W.A. Structural Plasticity of Barley Yellow Dwarf Virus-like Cap-Independent Translation Elements in Four Genera of Plant Viral RNAs. Virology 2010, 402, 177. [Google Scholar] [CrossRef]
- Kraft, J.J.; Treder, K.; Peterson, M.S.; Miller, W.A. Cation-Dependent Folding of 3′ Cap-Independent Translation Elements Facilitates Interaction of a 17-Nucleotide Conserved Sequence with EIF4G. Nucleic. Acids Res. 2013, 41, 3398–3413. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Laederach, A.; Pearlman, S.M.; Herschlag, D.; Altman, R.B. SAFA: Semi-Automated Footprinting Analysis Software for High-Throughput Quantification of Nucleic Acid Footprinting Experiments. RNA 2005, 11, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Parisien, M.; Major, F. The MC-Fold and MC-Sym Pipeline Infers RNA Structure from Sequence Data. Nature 2008, 452, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Kassem, M.A.; Gosalvez, B.; Garzo, E.; Fereres, A.; Gómez-Guillamón, M.L.; Aranda, M.A. Resistance to Cucurbit Aphid-Borne Yellows Virus in Melon Accession TGR-1551. Phytopathology 2015, 105, 1389–1396. [Google Scholar] [CrossRef][Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miras, M.; Aranda, M.A.; Truniger, V. Different RNA Elements Control Viral Protein Synthesis in Polerovirus Isolates Evolved in Separate Geographical Regions. Int. J. Mol. Sci. 2022, 23, 12503. https://doi.org/10.3390/ijms232012503
Miras M, Aranda MA, Truniger V. Different RNA Elements Control Viral Protein Synthesis in Polerovirus Isolates Evolved in Separate Geographical Regions. International Journal of Molecular Sciences. 2022; 23(20):12503. https://doi.org/10.3390/ijms232012503
Chicago/Turabian StyleMiras, Manuel, Miguel A. Aranda, and Verónica Truniger. 2022. "Different RNA Elements Control Viral Protein Synthesis in Polerovirus Isolates Evolved in Separate Geographical Regions" International Journal of Molecular Sciences 23, no. 20: 12503. https://doi.org/10.3390/ijms232012503
APA StyleMiras, M., Aranda, M. A., & Truniger, V. (2022). Different RNA Elements Control Viral Protein Synthesis in Polerovirus Isolates Evolved in Separate Geographical Regions. International Journal of Molecular Sciences, 23(20), 12503. https://doi.org/10.3390/ijms232012503






