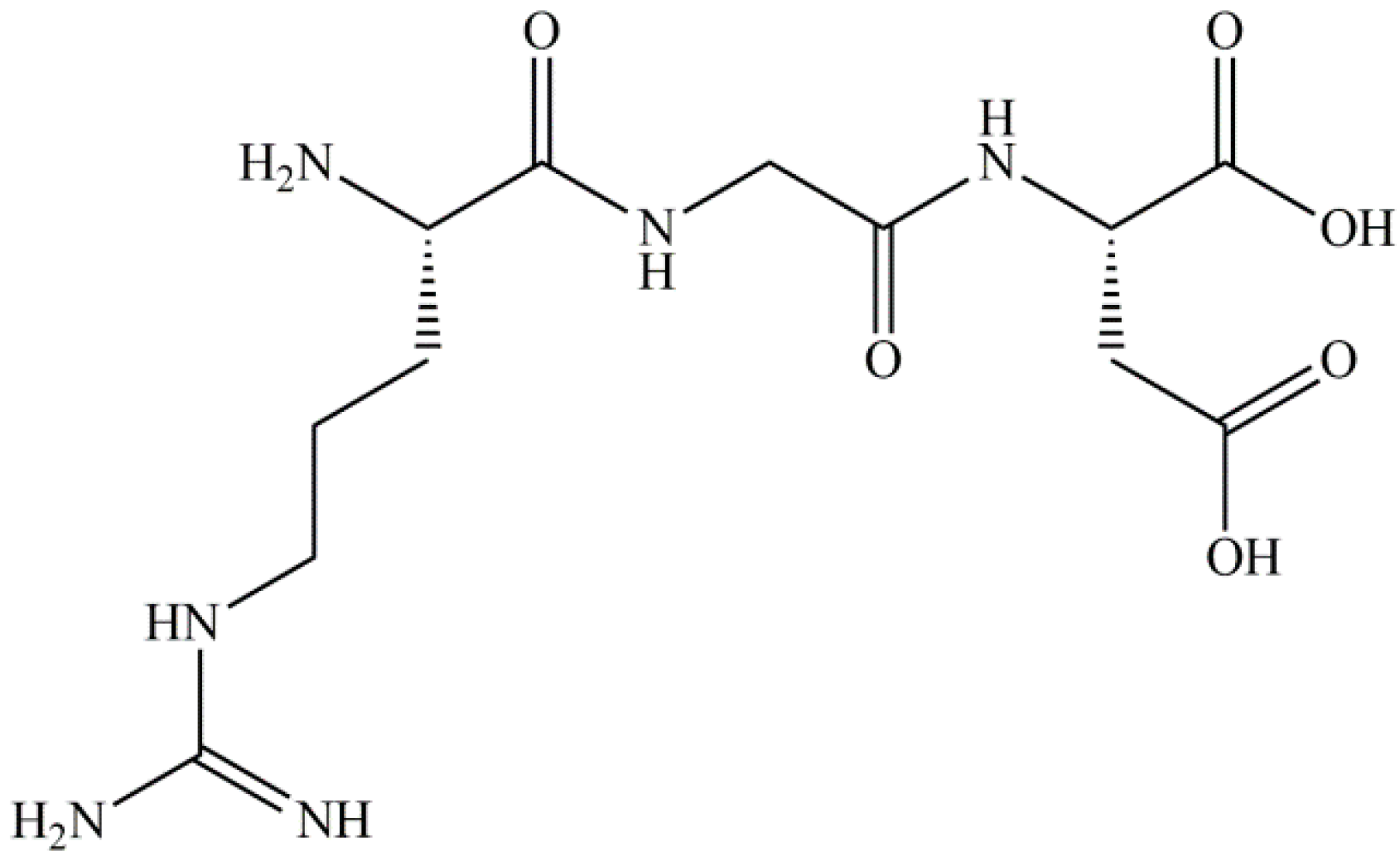
Comparative Assessment of the Inhibitory Potential of the Herbicide Glyphosate and Its Structural Analogs on RGD-Specific Integrins Using Enzyme-Linked Immunosorbent Assays
Abstract
1. Introduction
2. Results and Discussion
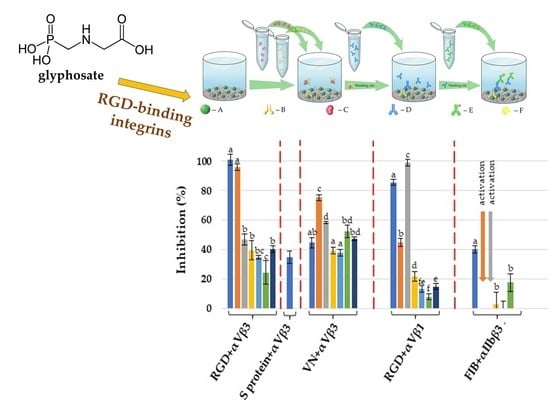
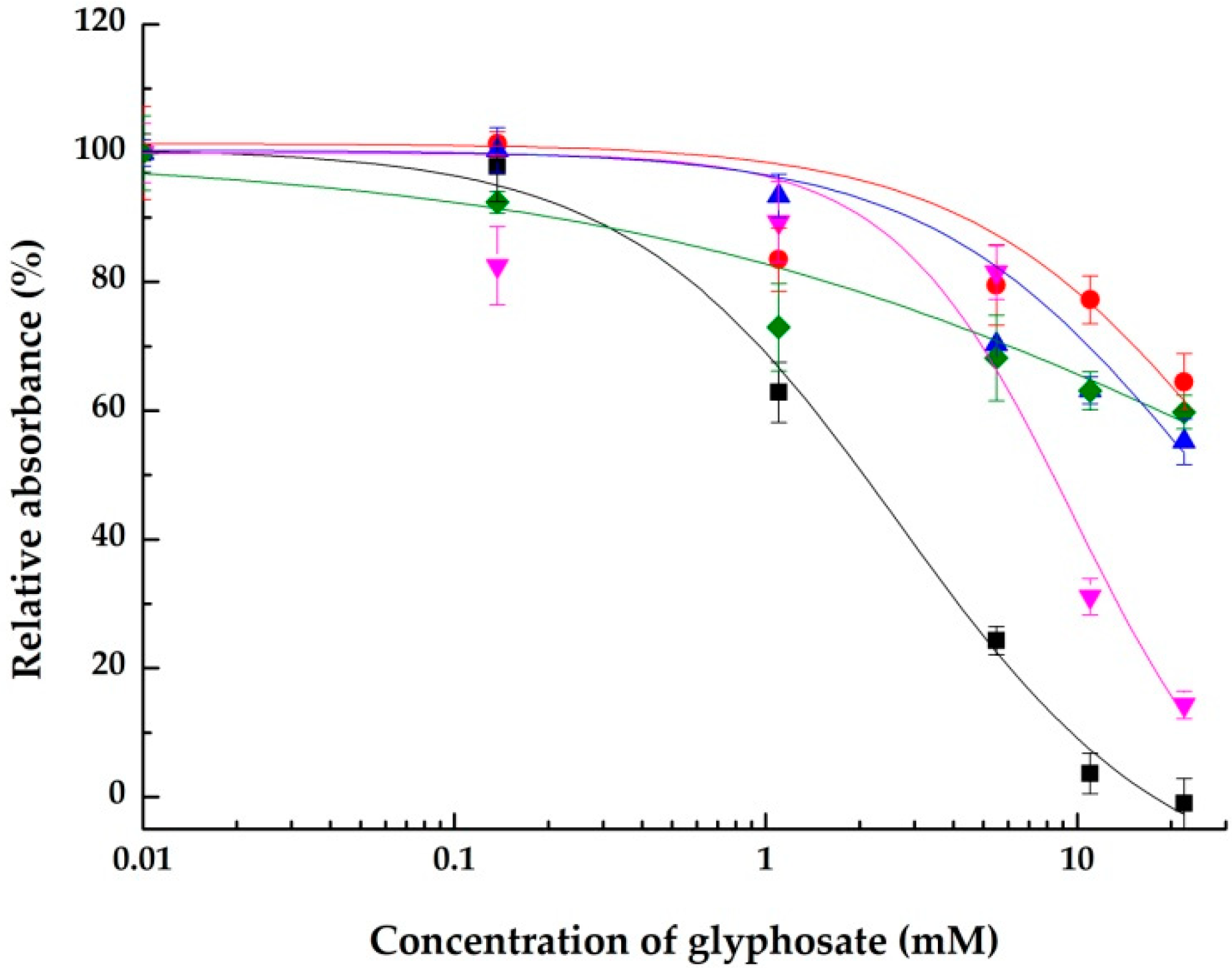
2.1. Inhibitory Effects on the Integrin αVβ3
2.1.1. Using a Synthetic Polymer Containing the RGD Motif as A Surface Affinity Binding Agent
2.1.2. Using Vitronectin as a Surface Affinity Binding Agent
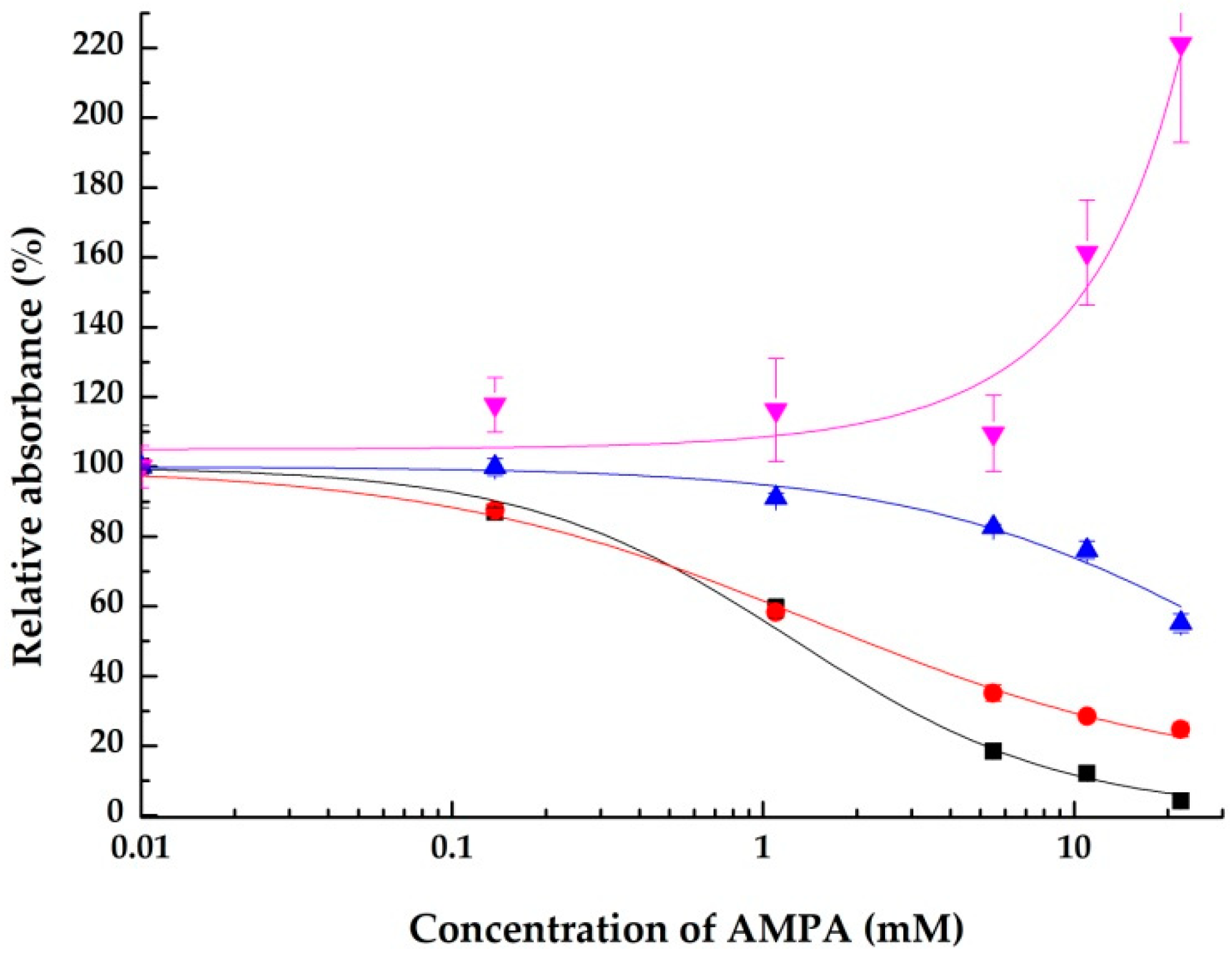
2.1.3. Using S-Protein, the SARS-CoV-2 Spike Protein Receptor Binding Domain, as a Surface Affinity Binding Agent
2.2. Inhibitory Effects on the Integrin α5β1
2.3. Inhibitory Effects on the Integrin αllbβ3
2.4. Biochemical and Possible Medical Relevance
3. Materials and Methods
3.1. Chemicals
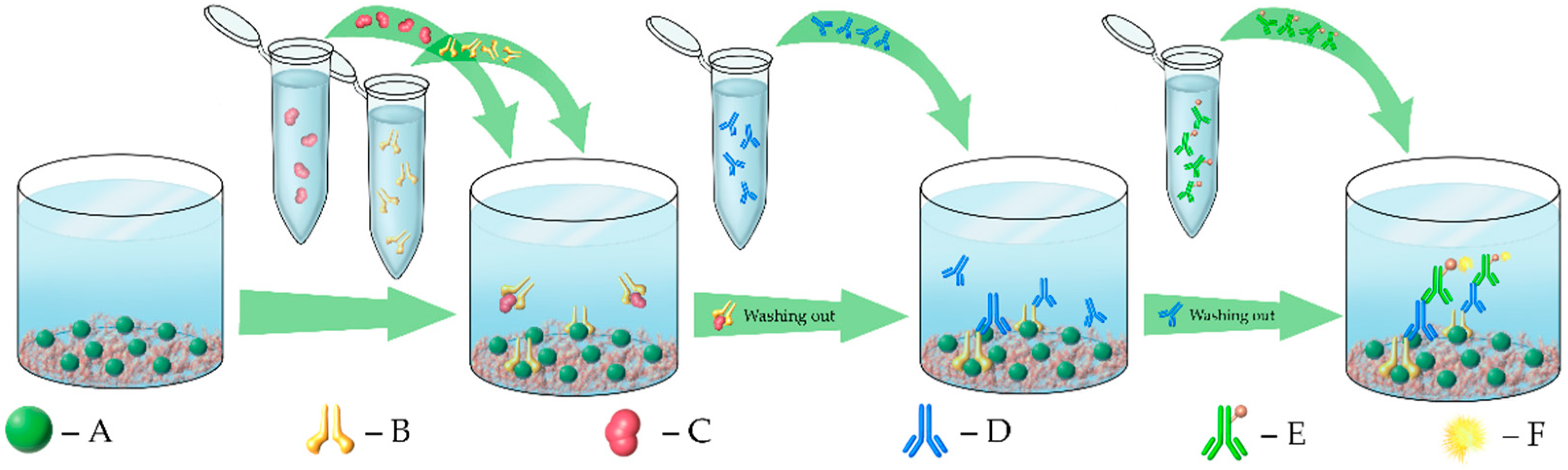
3.2. Enzyme-Linked Immunosorbent Assays (ELISAs)
3.3. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef]
- Morse, E.M.; Brahme, N.N.; Calderwood, D.A. Integrin cytoplasmic tail interactions. Biochemistry 2014, 53, 810–820. [Google Scholar] [CrossRef]
- Haas, T.A.; Plow, E.F. Integrin-ligarid interactions: A year in review. Curr. Opin. Cell Biol. 1994, 6, 656–662. [Google Scholar] [CrossRef]
- Miranti, C.K.; Brugge, J.S. Sensing the environment: A historical perspective on integrin signal transduction. Nat. Cell Biol. 2002, 4, E83–E90. [Google Scholar] [CrossRef]
- Takada, Y.; Ye, X.; Simon, S. The integrins. Genome Biol. 2007, 8, 215. [Google Scholar] [CrossRef]
- Mezu-Ndubuisi, O.J.; Maheshwari, A. The role of integrins in inflammation and angiogenesis. Pediatr. Res. 2021, 89, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.H.; Carman, C.V.; Springer, T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Imunol. 2007, 25, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Takagi, J. Structural basis for ligand recognition by RGD (Arg-Gly-Asp)-dependent integrins. Biochem. Soc. Trans. 2004, 32, 403–406. [Google Scholar] [CrossRef]
- Wilkinson, A.L.; John, A.E.; Barrett, J.W.; Gower, E.; Morrison, V.S.; Man, Y.; TaoPun, K.; Roper, J.A.; Luckett, J.C.; Borthwick, L.A.; et al. Pharmacological characterisation of GSK3335103, an oral αvβ6 integrin small molecule RGD-mimetic inhibitor for the treatment of fibrotic disease. Eur. J. Pharmacol. 2021, 913, 174618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Z.; Li, T.; Chen, L.; Lyu, J.; Li, C.; Lin, Y.; Hao, N.; Zhou, M.; Zhong, Z. Enhanced therapeutic effect of RGD-modified polymeric micelles loaded with low-dose methotrexate and nimesulide on rheumatoid arthritis. Theranostics 2019, 9, 708–720. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Z.; Ye, X.; Chen, Z.; Chen, Z.S. Mechanisms of thrombosis and research progress on targeted antithrombotic drugs. Drug Discov. Today 2021, 26, 2282–2302. [Google Scholar] [CrossRef]
- Cheng, Y.; Ji, Y. RGD-modified polymer and liposome nanovehicles: Recent research progress for drug delivery in cancer therapeutics. Eur. J. Pharm. Sci. 2019, 128, 8–17. [Google Scholar] [CrossRef]
- Claes, J.; Liesenborghs, L.; Peetermans, M.; Veloso, T.R.; Missiakas, D.; Schneewind, O.; Mancini, S.; Entenza, J.M.; Hoylaerts, M.F.; Heying, R.; et al. Clumping factor A, von Willebrand factor-binding protein and von Willebrand factor anchor Staphylococcus aureus to the vessel wall. J. Thromb. Haemost. 2017, 15, 1009–1019. [Google Scholar] [CrossRef]
- Voss, S.; Hallström, T.; Saleh, M.; Burchhardt, G.; Pribyl, T.; Singh, B.; Riesbeck, K.; Zipfel, P.F.; Hammerschmidt, S. The choline-binding protein PspC of Streptococcus pneumoniae interacts with the C-terminal heparin-binding domain of vitronectin. J. Biol. Chem. 2013, 288, 15614–15627. [Google Scholar] [CrossRef]
- Nader, D.; Curley, G.F.; Kerrigan, S.W. A new perspective in sepsis treatment: Could RGD-dependent integrins be novel targets? Drug Discov. Today 2020, 25, 2317–2325. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Sigrist, C.J.; Bridge, A.; Le Mercier, P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020, 177, 104759. [Google Scholar] [CrossRef]
- Omiecinski, C.J.; Vanden Heuvel, J.P.; Perdew, G.H.; Peters, J.M. Xenobiotic metabolism, disposition, and regulation by receptors: From biochemical phenomenon to predictors of major toxicities. Toxicol. Sci. 2011, 120 (Suppl. 1), 49–75. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef]
- Heckmann, D.; Kessler, H. Design and synthesis of integrin ligands. Methods Enzymol. 2007, 426, 463–503. [Google Scholar] [CrossRef] [PubMed]
- Stadel, J.M.; Nichols, A.J.; Bertolini, D.R.; Samanen, J.M. Therapeutic Potential of Integrin Antagonists. In Integrins: Molecular and Biological Responses to the Extracellular Matrix.; Cheresh, D.A., Mecham, R.M., Eds.; Academic Press: Cambridge, MA, USA, 1994; pp. 237–271. ISBN 978-0-08-091729-0. [Google Scholar] [CrossRef]
- Hartman, G.D.; Egbertson, M.S.; Halczenko, W.; Laswell, W.L.; Duggan, M.E.; Smith, R.L.; Naylor, A.M.; Manno, P.D.; Lynch, R.J.; Zhang, G.; et al. Non-peptide fibrinogen receptor antagonists. 1. Discovery and design of exosite inhibitors. J. Med. Chem. 1992, 35, 4640–4642. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.H.; Keenan, R.M.; Willette, R.N.; Lark, M.W. Identification and in vivo efficacy of small-molecule antagonists of integrin αvβ3 (the vitronectin receptor). Drug Discov. Today 2000, 5, 397–408. [Google Scholar] [CrossRef]
- Shimaoka, M.; Salas, A.; Yang, W.; Weitz-Schmidt, G.; Springer, T.A. Small Molecule Integrin Antagonists that Bind to the β2 Subunit I-like Domain and Activate Signals in One Direction and Block Them in the Other. Immunity 2003, 19, 391–402. [Google Scholar] [CrossRef]
- Smalheer, J.M.; Weigelt, C.A.; Woerner, F.J.; Wells, J.S.; Daneker, W.F.; Mousa, S.A.; Wexler, R.R.; Jadhav, P.K. Synthesis and biological evaluation of nonpeptide integrin antagonists containing spirocyclic scaffolds. Bioorg. Med. Chem. Lett. 2004, 14, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.R.; Meyer, J.M.; Miyashiro, J.M.; Engleman, V.W.; Freeman, S.K.; Griggs, D.W.; Klover, J.A.; Nickols, G.A. Discovery of Diphenylmethanepropionic and Dihydrostilbeneacetic Acids as Antagonists of the Integrin αvβ3. Chem. Biol. Drug. Des. 2006, 67, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Székács, I.; Farkas, E.; Gémes, B.L.; Takács, E.; Székács, A.; Horváth, R. Integrin targeting of glyphosate and its cell adhesion modulation effects on osteoblastic MC3T3-E1 cells revealed by label-free optical biosensing. Sci. Rep. 2018, 8, 17401. [Google Scholar] [CrossRef] [PubMed]
- Székács, A.; Darvas, B. Forty years with glyphosate. In Herbicides—Properties, Synthesis and Control of Weeds; Hasaneen, M.N., Ed.; InTech: Rijeka, Croatia, 2012; pp. 247–284. ISBN 978-953-307-803-8. [Google Scholar] [CrossRef]
- Székács, A.; Darvas, B. Re-registration challenges of glyphosate in the European Union. Front. Environ. Sci. 2018, 6, 78. [Google Scholar] [CrossRef]
- Soares, D.; Silva, L.; Duarte, S.; Pena, A.; Pereira, A. Glyphosate Use, Toxicity and Occurrence in Food. Foods 2021, 10, 2785. [Google Scholar] [CrossRef] [PubMed]
- Dill, G.M. Glyphosate-resistant crops: History, status and future. Pest Manag. Sci. 2005, 61, 219–224. [Google Scholar] [CrossRef]
- Baylis, A.D. Why glyphosate is a global herbicide: Strengths, weaknesses and prospects. Pest Manag. Sci. 2000, 56, 299–308. [Google Scholar] [CrossRef]
- Battaglin, W.A.; Meyer, M.T.; Kuivila, K.M.; Dietze, J.E. Glyphosate and Its Degradation Product AMPA Occur Frequently and Widely in U.S. Soils, Surface Water, Groundwater, and Precipitation. J. Am. Water Resour. Assoc. 2014, 50, 275–290. [Google Scholar] [CrossRef]
- Carles, L.; Gardon, H.; Joseph, L.; Sanchís, J.; Farré, M.; Artigas, J. Meta-analysis of glyphosate contamination in surface waters and dissipation by biofilms. Environ. Int. 2019, 124, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Peillex, C.; Pelletier, M. The impact and toxicity of glyphosate and glyphosate-based herbicides on health and immunity. J. Immunotoxicol. 2020, 17, 163–174. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). Some organophosphate insecticides and herbicides. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 2017; Volume 112, pp. 1–452. Available online: http://monographs.iarc.fr/ENG/Monographs/vol112/mono112.pdf (accessed on 11 October 2022).
- Portier, C.J. A comprehensive analysis of the animal carcinogenicity data for glyphosate from chronic exposure rodent carcinogenicity studies. Environ. Health 2020, 19, 18. [Google Scholar] [CrossRef]
- European Food Safety Authority. Evaluation of the impact of glyphosate and its residues in feed on animal health. EFSA J. 2018, 16, 5283. [Google Scholar] [CrossRef]
- European Chemicals Agency (ECHA). Committee for Risk Assessment (RAC) Opinion Proposing Harmonised Classification and Labeling at EU level of Glyphosate (ISO); N-(phosphonomethyl)glycine; ECHA: Hesinki, Finland, 2021. Available online: https://echa.europa.eu/documents/10162/882a2dc7-9e6f-b0ac-491a-ed3526b4018a (accessed on 11 October 2022).
- Mesnage, R.; Antoniou, M. Facts and fallacies in the debate on glyphosate toxicity. Front. Public Health 2017, 5, 316. [Google Scholar] [CrossRef] [PubMed]
- Morvillo, M. Glyphosate effect: Has the glyphosate controversy affected the EU’s regulatory epistemology? Eur. J. Risk Regul. 2020, 11, 422–435. [Google Scholar] [CrossRef]
- Grandcoin, A.; Piel, S.; Baurès, E. AminoMethylPhosphonic acid (AMPA) in natural waters: Its sources, behavior and environmental fate. Water Res. 2017, 117, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Siehl, D.L.; Castle, L.A.; Gorton, R.; Keenan, R.J. The molecular basis of glyphosate resistance by an optimized microbial acetyltransferase. J. Biol. Chem. 2007, 282, 11446–11455. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 11 October 2022).
- International Union of Pure and Applied Chemistry: Glyphosate-Isopropylamine (Ref: MON 0139). Available online: http://sitem.herts.ac.uk/aeru/iupac/Reports/2395.htm (accessed on 11 October 2022).
- Kapp, T.G.; Rechenmacher, F.; Neubauer, S.; Maltsev, O.V.; Cavalcanti-Adam, E.A.; Zarka, R.; Reuning, U.; Notni, J.; Wester, H.J.; Mas-Moruno, C.; et al. A Comprehensive Evaluation of the Activity and Selectivity Profile of Ligands for RGD-binding Integrins. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Yan, B.; Smith, J.W. Mechanism of integrin activation by disulfide bond reduction. Biochemistry 2001, 40, 8861–8867. [Google Scholar] [CrossRef]
- Kim, C.; Ye, F.; Ginsberg, M.H. Regulation of integrin activation. Ann. Rev. Cell Dev. Biol. 2011, 27, 321–345. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, P.; Dard, M.M. Structure and function of RGD peptides involved in bone biology. Cell. Mol. Life Sci. 2003, 60, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Xu, M.; Zhang, L.; Qu, L.; Liu, X. Role of αVβ3 in prostate cancer: Metastasis initiator and important therapeutic target. Onco Targets Ther. 2020, 13, 7411–7422. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, F.; Chen, X. Integrin αVβ3 targeted cancer therapy. Drug Dev. Res. 2008, 69, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Nader, D.; Kerrigan, S. Vascular dysregulation following SARS-CoV-2 infection involves integrin signalling through a VE-Cadherin mediated pathway. bioRxiv 2022. [Google Scholar] [CrossRef]
- Antonov, A.S.; Antonova, G.N.; Munn, D.H.; Mivechi, N.; Lucas, R.; Catravas, J.D.; Verin, A.D. αVβ3 integrin regulates macrophage inflammatory responses via PI3 kinase/Akt-dependent NF-κB activation. J Cell Physiol. 2011, 226, 469–476. [Google Scholar] [CrossRef]
- Francis, S.E.; Goh, K.L.; Hodivala-Dilke, K.; Bader, B.L.; Stark, M.; Davidson, D.; Hynes, R.O. Central roles of α5β1 integrin and fibronectin in vascular development in mouse embryos and embryoid bodies. Arteroscler. Thromb. Vasc. Biol. 2002, 22, 927–933. [Google Scholar] [CrossRef]
- Li, R.; Maminishkis, A.; Zahn, G.; Vossmeyer, D.; Miller, S.S. Integrin α5β1 mediates attachment, migration, and proliferation in human retinal pigment epithelium: Relevance for proliferative retinal disease. Investig. Ophthalmol. Vis. Sci. 2009, 50, 5988–5996. [Google Scholar] [CrossRef] [PubMed]
- Pimton, P.; Sarkar, S.; Sheth, N.; Perets, A.; Marcinkiewicz, C.; Lazarovici, P.; Lelkes, P.I. Fibronectin-mediated upregulation of α5β1 integrin and cell adhesion during differentiation of mouse embryonic stem cells. Cell Adhes. Migr. 2011, 5, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, X.; Shi, X.; Zhu, M.; Wang, J.; Huang, S.; Huang, X.; Wang, H.; Li, L.; Deng, H.; et al. Platelet integrin αIIbβ3: Signal transduction, regulation, and its therapeutic targeting. J. Hematol. Oncol. 2019, 12, 26. [Google Scholar] [CrossRef]
- Blue, R.; Kowalska, A.M.; Hirsch, J.; Murcia, M.; Janczak, C.A.; Harrington, A.; Jirouskova, M.; Li, J.; Fuentes, R.; Thornton, M.A.; et al. Structural and therapeutics insights from the species specificity and in vivo antithrombotic activity of a novel alpha IIb-specific alpha IIb beta 3 anatonist. Blood 2009, 114, 195. [Google Scholar] [CrossRef]
- Gan, Z.R.; Gould, R.J.; Jacobs, J.W.; Friedman, P.A.; Polokoff, M.A. Echistatin—A potent platelet aggregation inhibitor from the venom of the viper, Echis carinatus. J. Biol. Chem. 1988, 263, 19827–19832. [Google Scholar] [CrossRef]
- Pfaff, M.; McLane, M.A.; Beviglia, L.; Niewiarowski, S.; Timpl, R. Comparison of Disintegrins with limited variation in the RGD Loop in their binding to purified integrins aIIbb3, avb3 and a5b1 and in cell adhesion inhibition. Cell Adhes. Commun. 1994, 2, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, R.M.; Naughton, M.A.; Teng, W.; Rose, J.W.; Phillips, D.R.; Nannizzi, L.; Arfsten, A.; Campbell, A.M.; Charo, I.F. Design of potent and specific integrin antagonists. Peptide antagonists with high specificity for glycoprotein IIb-IIIa. J. Biol. Chem. 1993, 268, 1066–1073. [Google Scholar] [CrossRef]
- Rodbard, D. Mathematics and statistics of ligand assays: An illustrated guide. In Ligand Assay: Analysis of International Developments on Isotopic and Nonisotopic Immunoassay; Langan, J., Clapp, J.J., Eds.; Masson Publishing: New York, NY, USA, 1981; pp. 45–99. ISBN 0893520942. [Google Scholar]


| Compound | CAS No. 1 | Molecular Mass | Water Solubility (g/L) | Clog Po/w 2 | Comment |
|---|---|---|---|---|---|
| Glyphosate Glyphosate isopropylammonium salt 3 | 1071-83-6 386411-94-0 | 169.07 228.19 | 157 (pH 7.0) 1050 | −4.6 * −4.16 | Active ingredient of herbicides |
| Aminomethylphosphonic acid (AMPA) | 1066-51-9 | 111.04 | 56 (20 °C) | −4.7 * | Metabolite |
| N-acetylglyphosate | 129660-96-4 | 211.11 | slight | −2.4 * | Metabolite |
| Glycine | 56-40-6 | 75.07 | 250 (25 °C) | −3.2 * | Amino acid, metabolite |
| Acetylglycine | 543-24-8 | 117.10 | 26.3 (15 °C) | −1.2 * | Acylamino acid |
| Sarcosine | 107-97-1 | 89.09 | 89.1 (20 °C) | −3.2 −2.8 * | Metabolite |
| Iminodiacetic acid | 142-73-4 | 133.10 | 24.3 (5 °C) | −3.3 * | Minor impurity |
| ELISA | IC50 Value (mM) 1 (Maximal Inhibition Achieved) | |||
|---|---|---|---|---|
| Integrin | αVβ3 | αVβ3 | α5β1 | αllbβ3 |
| Immobilized affinity reagent | RGD | Vitronectin | RGD | Fibrinogen |
| Compounds | ||||
| Glyphosate | 2.7 ± 0.5 (100.0 ± 3.9%) | >22 (44.8 ± 3.6%) | 9.7 ± 3.2 (85.7 ± 2.1%) | >22 (40.2 ± 2.6%) |
| Aminomethylphosphonic acid (AMPA) | 1.3 ± 0.2 (95.9 ± 2.2%) | 1.5 ± 0.4 (75.3 ± 2.1%) | >22 (44.8 ± 2.6%) | >22 (activation) 2 |
| N-acetylglyphosate | >22 (40.6 ± 2.2%) | >22 (47.5 ± 1.2%) | >22 (15.0 ± 2.0%) | >22 (n. d. 3) |
| Glycine | >22 (39.5 ± 6.6%) | >22 (39.5 ± 2.4%) | >22 (22.1 ± 3.3%) | >22 (n. d.) |
| Acetylglycine | >22 (46.8 ± 3.7%) | 9.2 ± 1.0 (58.5 ± 0.8%) | 5.8 ± 0.9 (98.9 ± 2.3%) | >22 (activation) |
| Sarcosine | >22 (35.0 ± 1.2%) | >22 (37.9 ± 2.3%) | >22 (13.4 ± 2.1%) | >22 (n. d.) |
| Iminodiacetic acid | >22 (24.6 ± 7.9%) | 11.5 ± 1.5 (52.4 ± 4.2%) | >22 (8.0 ± 2.0%) | >22 (n. d.) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gémes, B.; Takács, E.; Székács, I.; Horvath, R.; Székács, A. Comparative Assessment of the Inhibitory Potential of the Herbicide Glyphosate and Its Structural Analogs on RGD-Specific Integrins Using Enzyme-Linked Immunosorbent Assays. Int. J. Mol. Sci. 2022, 23, 12425. https://doi.org/10.3390/ijms232012425
Gémes B, Takács E, Székács I, Horvath R, Székács A. Comparative Assessment of the Inhibitory Potential of the Herbicide Glyphosate and Its Structural Analogs on RGD-Specific Integrins Using Enzyme-Linked Immunosorbent Assays. International Journal of Molecular Sciences. 2022; 23(20):12425. https://doi.org/10.3390/ijms232012425
Chicago/Turabian StyleGémes, Borbála, Eszter Takács, Inna Székács, Robert Horvath, and András Székács. 2022. "Comparative Assessment of the Inhibitory Potential of the Herbicide Glyphosate and Its Structural Analogs on RGD-Specific Integrins Using Enzyme-Linked Immunosorbent Assays" International Journal of Molecular Sciences 23, no. 20: 12425. https://doi.org/10.3390/ijms232012425
APA StyleGémes, B., Takács, E., Székács, I., Horvath, R., & Székács, A. (2022). Comparative Assessment of the Inhibitory Potential of the Herbicide Glyphosate and Its Structural Analogs on RGD-Specific Integrins Using Enzyme-Linked Immunosorbent Assays. International Journal of Molecular Sciences, 23(20), 12425. https://doi.org/10.3390/ijms232012425