Unraveling the Potential Role of NEDD4-like E3 Ligases in Cancer
Abstract
1. Introduction
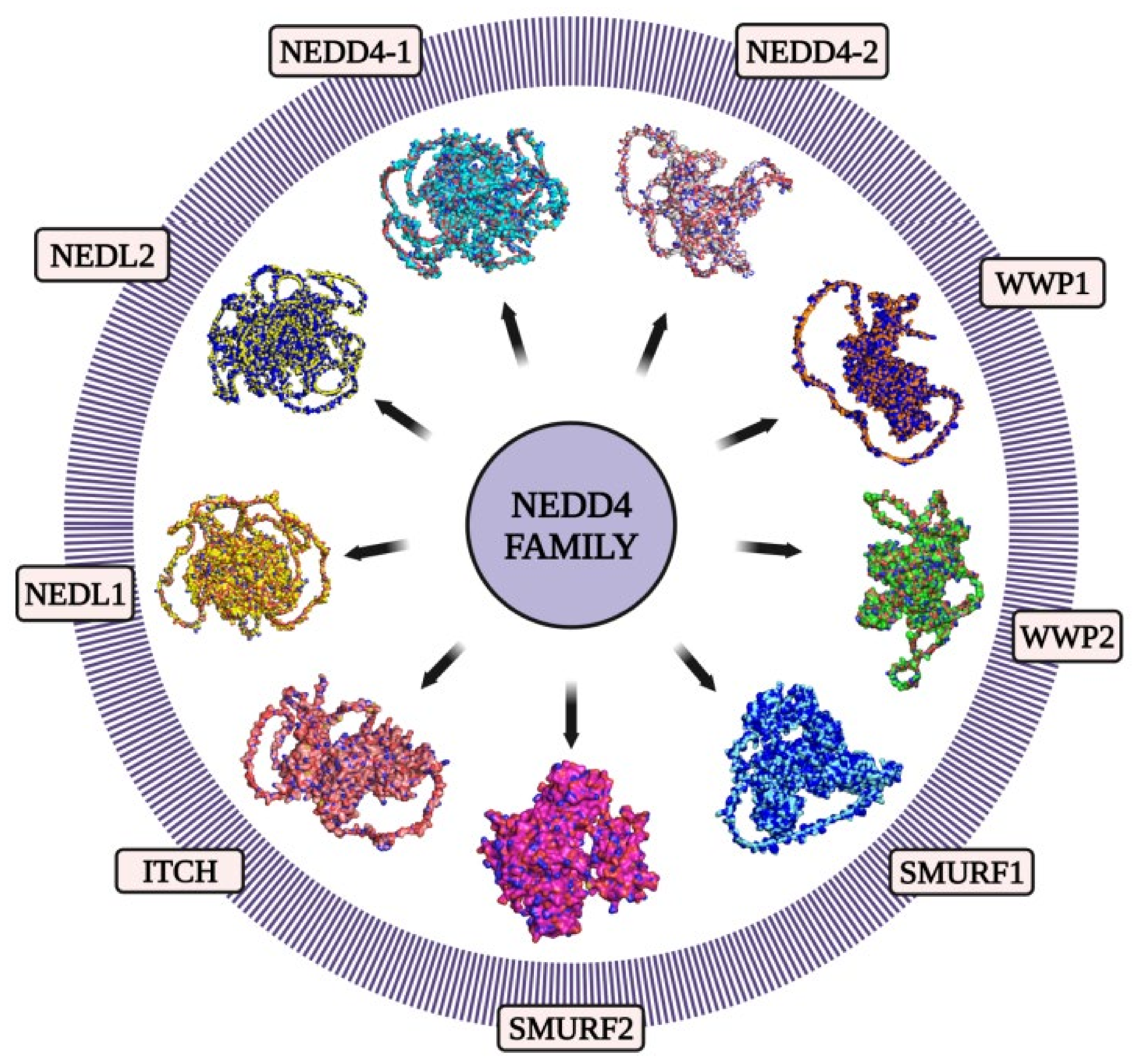
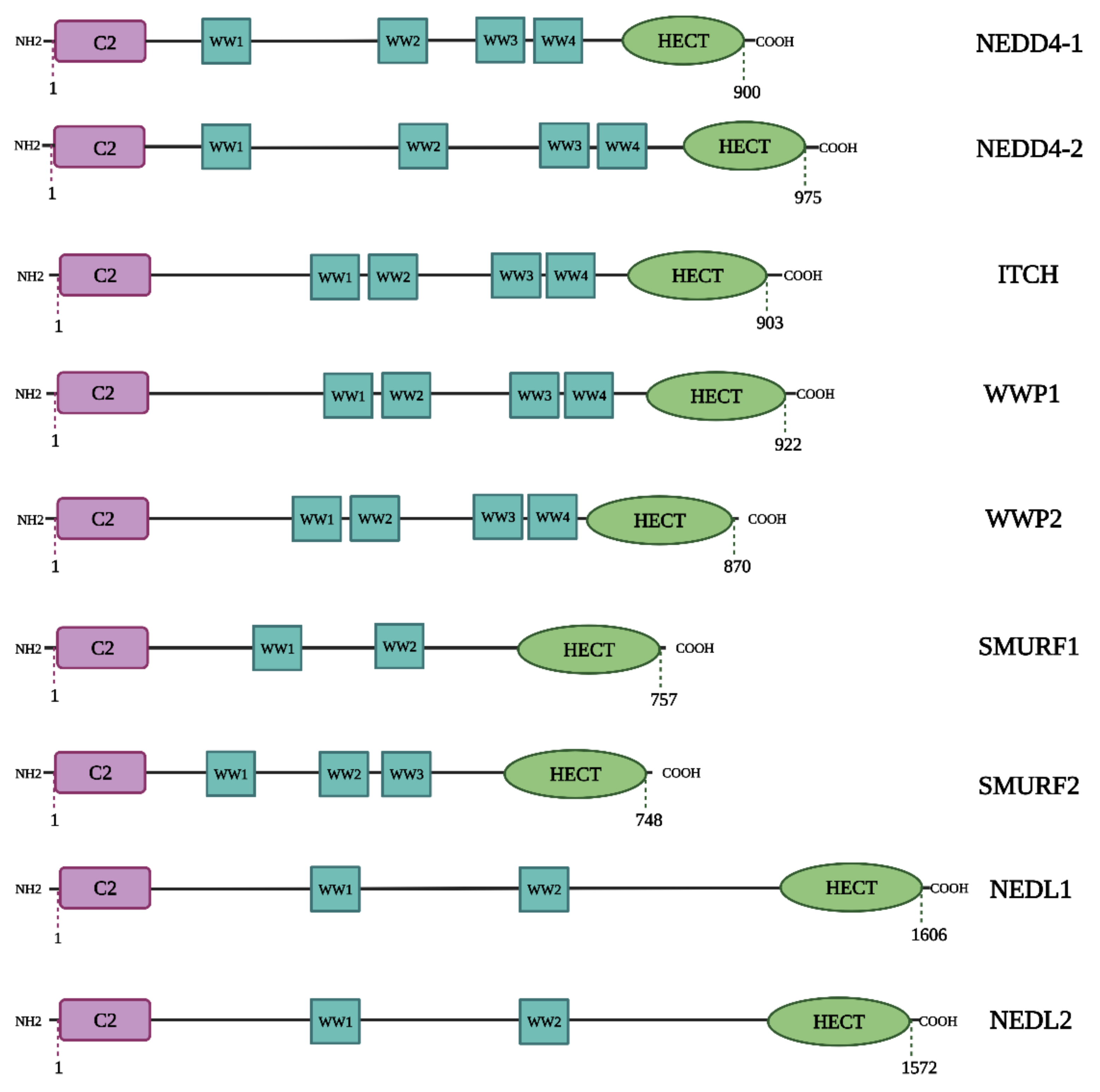
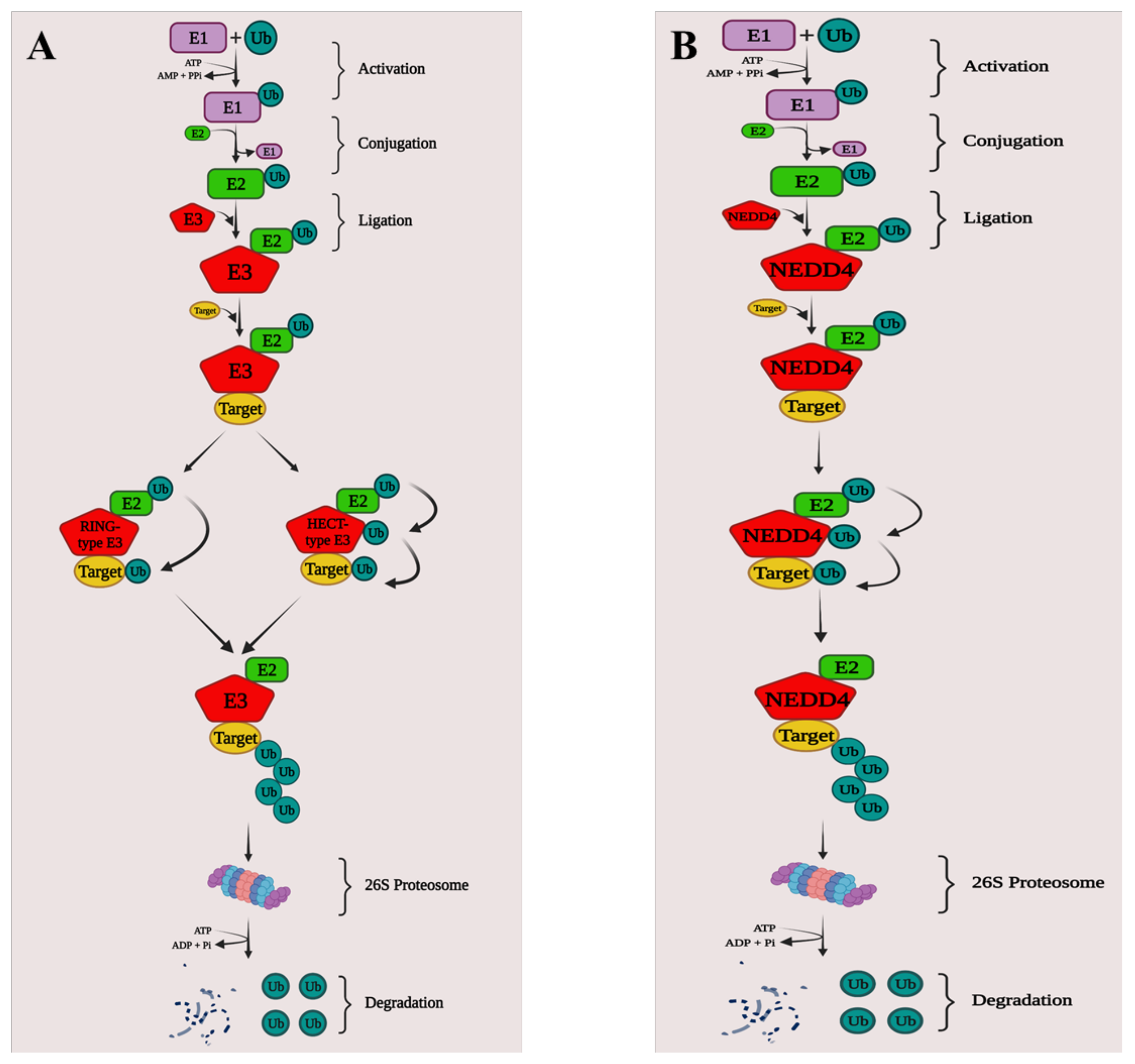
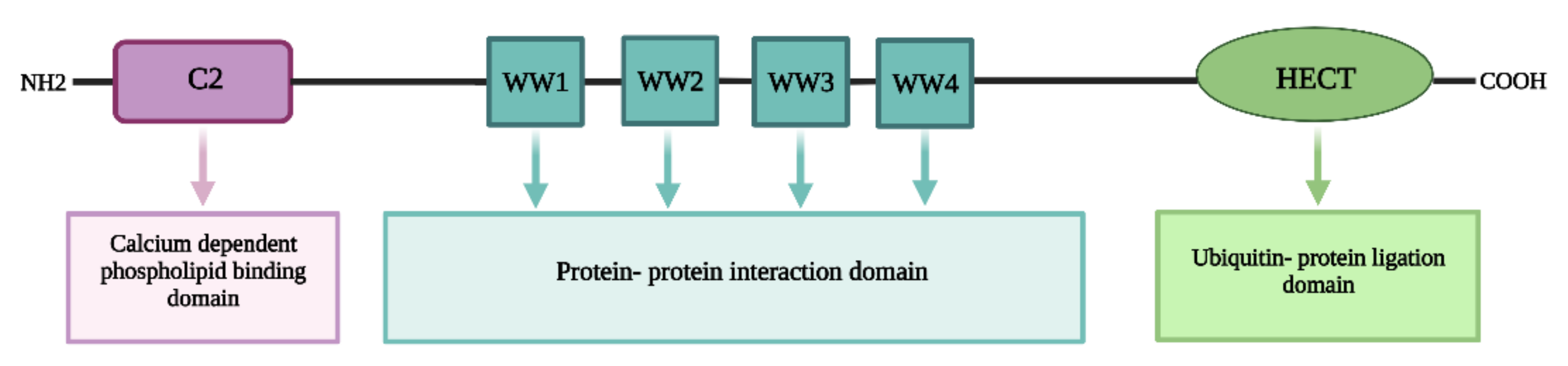
2. NEDD4 and Cancer
3. NEDD4 Functions in Human Cancers
3.1. NEDD4 and Cell Survival
3.2. NEDD4 and Cell Proliferation
3.3. NEDD4 and Autophagy
3.4. NEDD4 and Cell Migration and Invasion
3.5. NEDD4 and Metastasis
3.6. NEDD4 and EMT
3.7. NEDD4 and Chemoresistance
3.8. NEDD4 and Multiple Signaling Pathways
4. Effect of NEED4 on Various Types of Cancer
4.1. Bladder Cancer (BCa)
4.2. Breast Cancer (BC)
4.3. Cervical Cancer
4.4. Colorectal Cancer (CRC)
4.5. Endometrial Cancer (EC)
4.6. Gastric Cancer (GC)
4.7. Glioma/Glioblastoma
4.8. Liver Cancer
4.9. Lung Cancer
4.10. Melanoma
4.11. Nasopharyngeal Carcinoma (NPC)
4.12. Neuroblastoma (NB)
4.13. Ovarian Cancer
4.14. Pancreatic Cancer (PCa)
4.15. Prostate Cancer
4.16. Other Cancers
5. Oncogenic Role of NEDD4 in Cancer
6. Tumor-Suppressive Role of NEDD4
7. NEDD4 Knockout Mice
8. NEDD4 as a Therapeutic Target
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zaimy, M.; Saffarzadeh, N.; Mohammadi, A.; Pourghadamyari, H.; Izadi, P.; Sarli, A.; Moghaddam, L.; Paschepari, S.; Azizi, H.; Torkamandi, S. New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther. 2017, 24, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Girisa, S.; Henamayee, S.; Parama, D.; Rana, V.; Dutta, U.; Kunnumakkara, A.B. Targeting Farnesoid X receptor (FXR) for developing novel therapeutics against cancer. Mol. Biomed. 2021, 2, 1–23. [Google Scholar] [CrossRef]
- Hausman, D.M. What is cancer? Perspect. Biol. Med. 2019, 62, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Ranaware, A.M.; Banik, K.; Deshpande, V.; Padmavathi, G.; Roy, N.K.; Sethi, G.; Fan, L.; Kumar, A.P.; Kunnumakkara, A.B. Magnolol: A neolignan from the magnolia family for the prevention and treatment of cancer. Int. J. Mol. Sci. 2018, 19, 2362. [Google Scholar] [CrossRef]
- Banik, K.; Harsha, C.; Bordoloi, D.; Sailo, B.L.; Sethi, G.; Leong, H.C.; Arfuso, F.; Mishra, S.; Wang, L.; Kumar, A.P. Therapeutic potential of gambogic acid, a caged xanthone, to target cancer. Cancer Lett. 2018, 416, 75–86. [Google Scholar] [CrossRef]
- Padmavathi, G.; Rathnakaram, S.R.; Monisha, J.; Bordoloi, D.; Roy, N.K.; Kunnumakkara, A.B. Potential of butein, a tetrahydroxychalcone to obliterate cancer. Phytomedicine 2015, 22, 1163–1171. [Google Scholar] [CrossRef]
- Jia, L.Y.; Shanmugam, M.K.; Sethi, G.; Bishayee, A. Potential role of targeted therapies in the treatment of triple-negative breast cancer. Anticancer Drugs 2016, 27, 147–155. [Google Scholar] [CrossRef]
- Hassanpour, S.H.; Dehghani, M. Review of cancer from perspective of molecular. J. Cancer Res. Pract. 2017, 4, 127–129. [Google Scholar] [CrossRef]
- Banik, K.; Ranaware, A.M.; Harsha, C.; Nitesh, T.; Girisa, S.; Deshpande, V.; Fan, L.; Nalawade, S.P.; Sethi, G.; Kunnumakkara, A.B. Piceatannol: A natural stilbene for the prevention and treatment of cancer. Pharmacol. Res. 2020, 153, 104635. [Google Scholar] [CrossRef]
- Shabnam, B.; Padmavathi, G.; Banik, K.; Girisa, S.; Monisha, J.; Sethi, G.; Fan, L.; Wang, L.; Mao, X.; Kunnumakkara, A.B. Sorcin a potential molecular target for cancer therapy. Transl. Oncol. 2018, 11, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Banik, K.; Ranaware, A.M.; Deshpande, V.; Nalawade, S.P.; Padmavathi, G.; Bordoloi, D.; Sailo, B.L.; Shanmugam, M.K.; Fan, L.; Arfuso, F. Honokiol for cancer therapeutics: A traditional medicine that can modulate multiple oncogenic targets. Pharmacol. Res. 2019, 144, 192–209. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Parama, D.; Daimari, E.; Girisa, S.; Banik, K.; Harsha, C.; Dutta, U.; Kunnumakkara, A.B. Rationalizing the therapeutic potential of apigenin against cancer. Life Sci. 2021, 267, 118814. [Google Scholar] [CrossRef]
- Kunnumakkara, A.B.; Bordoloi, D.; Sailo, B.L.; Roy, N.K.; Thakur, K.K.; Banik, K.; Shakibaei, M.; Gupta, S.C.; Aggarwal, B.B. Cancer drug development: The missing links. Exp. Biol. Med. 2019, 244, 663–689. [Google Scholar] [CrossRef] [PubMed]
- Khwairakpam, A.D.; Banik, K.; Girisa, S.; Shabnam, B.; Shakibaei, M.; Fan, L.; Arfuso, F.; Monisha, J.; Wang, H.; Mao, X. The vital role of ATP citrate lyase in chronic diseases. J. Mol. Med. 2020, 98, 71–95. [Google Scholar] [CrossRef] [PubMed]
- Girisa, S.; Saikia, Q.; Bordoloi, D.; Banik, K.; Monisha, J.; Daimary, U.D.; Verma, E.; Ahn, K.S.; Kunnumakkara, A.B. Xanthohumol from Hop: Hope for cancer prevention and treatment. IUBMB Life 2021, 73, 1016–1044. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, E.; Banik, K.; Harsha, C.; Sailo, B.L.; Thakur, K.K.; Khwairakpam, A.D.; Vikkurthi, R.; Devi, T.B.; Gupta, S.C.; Kunnumakkara, A.B. Phytochemicals in cancer cell chemosensitization: Current knowledge and future perspectives. Semin. Cancer Biol. 2020, 80, 306–339. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Vuković, P.; Lugović-Mihić, L.; Ćesić, D.; Novak-Bilić, G.; Šitum, M.; Spoljar, S. Melanoma development: Current knowledge on melanoma pathogenesis. Acta Dermatovenerol. Croat. 2020, 28, 163–168. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Costa-Pinheiro, P.; Montezuma, D.; Henrique, R.; Jerónimo, C. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics 2015, 7, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Aronson, J.K.; Ferner, R.E. Biomarkers—A general review. Curr. Protoc. Pharmacol. 2017, 76, 9.23.1–9.23.17. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, E.P. Cancer biomarkers: Can we turn recent failures into success? J. Natl. Cancer Inst. 2010, 102, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Schwesinger, C. The ubiquitin-proteasome system in kidney physiology and disease. Nat. Rev. Nephrol. 2019, 15, 393–411. [Google Scholar] [CrossRef]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. 2020, 5, 11. [Google Scholar] [CrossRef]
- Rieser, E.; Cordier, S.M.; Walczak, H. Linear ubiquitination: A newly discovered regulator of cell signalling. Trends Biochem. Sci. 2013, 38, 94–102. [Google Scholar] [CrossRef]
- Lin, C.H.; MacGurn, J.A.; Chu, T.; Stefan, C.J.; Emr, S.D. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 2008, 135, 714–725. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Haas, A.L.; Warms, J.V.; Rose, I.A. Ubiquitin adenylate: Structure and role in ubiquitin activation. Biochemistry 1983, 22, 4388–4394. [Google Scholar] [CrossRef] [PubMed]
- Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005, 6, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Gilberto, S.; Peter, M. Dynamic ubiquitin signaling in cell cycle regulation. J. Cell Biol. 2017, 216, 2259–2271. [Google Scholar] [CrossRef]
- Shmueli, A.; Oren, M. Life, death, and ubiquitin: Taming the mule. Cell 2005, 121, 963–965. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Hunter, T. The regulatory crosstalk between kinases and proteases in cancer. Nat. Rev. Cancer 2010, 10, 278–292. [Google Scholar] [CrossRef]
- Sun, T.; Liu, Z.; Yang, Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 2020, 19, 146. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Ikeda, F.; Dikic, I. Atypical ubiquitin chains: New molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008, 9, 536–542. [Google Scholar] [CrossRef]
- Mansour, M.A. Ubiquitination: Friend and foe in cancer. Int. J. Biochem. Cell Biol. 2018, 101, 80–93. [Google Scholar] [CrossRef]
- Hodson, C.; Purkiss, A.; Miles, J.A.; Walden, H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection. Structure 2014, 22, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.; Kim, K.B.; Crews, C.M. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 2001, 21, 245–273. [Google Scholar] [CrossRef] [PubMed]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Xia, L.; Song, Y.; Liu, H.; Wang, Z.-w.; Zhu, X. Insights into the biological role of NEDD4L E3 ubiquitin ligase in human cancers. Front. Oncol. 2021, 11, 774648. [Google Scholar] [CrossRef]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef]
- Fajner, V.; Maspero, E.; Polo, S. Targeting HECT-type E3 ligases—Insights from catalysis, regulation and inhibitors. FEBS Lett. 2017, 591, 2636–2647. [Google Scholar] [CrossRef]
- Sluimer, J.; Distel, B. Regulating the human HECT E3 ligases. Cell Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Hoeller, D.; Hecker, C.M.; Dikic, I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat. Rev. Cancer 2006, 6, 776–788. [Google Scholar] [CrossRef]
- Weissman, A.M.; Shabek, N.; Ciechanover, A. The predator becomes the prey: Regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 2011, 12, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J. 1998, 17, 7151–7160. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.K.; Lin, H.-K.; Sun, S.-C.; Zhang, S. Targeting ubiquitination for cancer therapies. Future Med. Chem. 2015, 7, 2333–2350. [Google Scholar] [CrossRef]
- Wan, L.; Liu, T.; Hong, Z.; Pan, Y.; Sizemore, S.T.; Zhang, J.; Ma, Z. NEDD4 expression is associated with breast cancer progression and is predictive of a poor prognosis. Breast Cancer Res. 2019, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Xu, Y.J.; Jiang, S.Y.; Xu, Z.; Cao, B.Y.; Sethi, G.; Zeng, Y.Y.; Kong, Y.; Mao, X.L. Suppression of the USP10/CCND1 axis induces glioblastoma cell apoptosis. Acta Pharm. Sin. 2021, 42, 1338–1346. [Google Scholar] [CrossRef]
- Ren, Y.; Xu, X.; Mao, C.Y.; Han, K.K.; Xu, Y.J.; Cao, B.Y.; Zhang, Z.B.; Sethi, G.; Tang, X.W.; Mao, X.L. RNF6 promotes myeloma cell proliferation and survival by inducing glucocorticoid receptor polyubiquitination. Acta Pharm. Sin. 2020, 41, 394–403. [Google Scholar] [CrossRef]
- Sailo, B.L.; Banik, K.; Girisa, S.; Bordoloi, D.; Fan, L.; Halim, C.E.; Wang, H.; Kumar, A.P.; Zheng, D.; Mao, X. FBXW7 in cancer: What has been unraveled thus far? Cancers 2019, 11, 246. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, X.; Lin, P.; He, Y.; Zhang, Y.; Cao, B.; Zhang, Z.; Sethi, G.; Liu, J.; Zhou, X. Inhibition of the deubiquitinase USP9x induces pre-B cell homeobox 1 (PBX1) degradation and thereby stimulates prostate cancer cell apoptosis. J. Biol. Chem. 2019, 294, 4572–4582. [Google Scholar] [CrossRef]
- Mao, X.; Sethi, G.; Zhang, Z.; Wang, Q. The emerging roles of the HERC ubiquitin ligases in cancer. Curr. Pharm. Des. 2018, 24, 1676–1681. [Google Scholar] [CrossRef]
- Huang, X.; Chen, J.; Cao, W.; Yang, L.; Chen, Q.; He, J.; Yi, Q.; Huang, H.; Zhang, E.; Cai, Z. The many substrates and functions of NEDD4-1. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Goel, P.; Manning, J.A.; Kumar, S. NEDD4-2 (NEDD4L): The ubiquitin ligase for multiple membrane proteins. Gene 2015, 557, 1–10. [Google Scholar] [CrossRef]
- Ye, X.; Wang, L.; Shang, B.; Wang, Z.; Wei, W. NEDD4: A promising target for cancer therapy. Curr. Cancer Drug Targets 2014, 14, 549–556. [Google Scholar] [CrossRef]
- Wang, Z.-w.; Hu, X.; Ye, M.; Lin, M.; Chu, M.; Shen, X. NEDD4 E3 ligase: Functions and mechanism in human cancer. Semin. Cancer Biol. 2020, 67, 92–101. [Google Scholar] [CrossRef]
- Hatstat, A.K.; Pupi, M.D.; McCafferty, D.G. Predicting PY motif-mediated protein-protein interactions in the Nedd4 family of ubiquitin ligases. PLoS ONE 2021, 16, e0258315. [Google Scholar] [CrossRef]
- Boase, N.A.; Kumar, S. NEDD4: The founding member of a family of ubiquitin-protein ligases. Gene 2015, 557, 113–122. [Google Scholar] [CrossRef]
- Spagnol, G.; Kieken, F.; Kopanic, J.L.; Li, H.; Zach, S.; Stauch, K.L.; Grosely, R.; Sorgen, P.L. Structural studies of the Nedd4 WW domains and their selectivity for the Connexin43 (Cx43) carboxyl terminus. J. Biol. Chem. 2016, 291, 7637–7650. [Google Scholar] [CrossRef]
- Pohl, P.; Joshi, R.; Petrvalska, O.; Obsil, T.; Obsilova, V. 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains. Commun. Biol. 2021, 4, 1–15. [Google Scholar] [CrossRef]
- Yao, W.; Shan, Z.; Gu, A.; Fu, M.; Shi, Z.; Wen, W. WW domain-mediated regulation and activation of E3 ubiquitin ligase Suppressor of Deltex. J. Biol. Chem. 2018, 293, 16697–16708. [Google Scholar] [CrossRef]
- Zhu, K.; Shan, Z.; Chen, X.; Cai, Y.; Cui, L.; Yao, W.; Wang, Z.; Shi, P.; Tian, C.; Lou, J.; et al. Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase Itch. EMBO Rep. 2017, 18, 1618–1630. [Google Scholar] [CrossRef]
- Haouari, S.; Vourc’h, P.; Jeanne, M.; Marouillat, S.; Veyrat-Durebex, C.; Lanznaster, D.; Laumonnier, F.; Corcia, P.; Blasco, H.; Andres, C.R. The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 3882. [Google Scholar] [CrossRef]
- Henshall, T.L.; Manning, J.A.; Alfassy, O.S.; Goel, P.; Boase, N.A.; Kawabe, H.; Kumar, S. Deletion of Nedd4-2 results in progressive kidney disease in mice. Cell Death Differ. 2017, 24, 2150–2160. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Kawabe, H.; Rotin, D. The Ubiquitin Ligase Nedd4L Regulates the Na/K/2Cl Co-transporter NKCC1/SLC12A2 in the Colon. J. Biol. Chem. 2017, 292, 3137–3145. [Google Scholar] [CrossRef]
- Leitz, D.H.W.; Duerr, J.; Mulugeta, S.; Seyhan Agircan, A.; Zimmermann, S.; Kawabe, H.; Dalpke, A.H.; Beers, M.F.; Mall, M.A. Congenital Deletion of Nedd4-2 in Lung Epithelial Cells Causes Progressive Alveolitis and Pulmonary Fibrosis in Neonatal Mice. Int. J. Mol. Sci. 2021, 22, 6146. [Google Scholar] [CrossRef]
- Gao, S.; Alarcón, C.; Sapkota, G.; Rahman, S.; Chen, P.-Y.; Goerner, N.; Macias, M.J.; Erdjument-Bromage, H.; Tempst, P.; Massagué, J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 2009, 36, 457–468. [Google Scholar] [CrossRef]
- Lee, D.-E.; Yoo, J.E.; Kim, J.; Kim, S.; Kim, S.; Lee, H.; Cheong, H. NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. Cell Death Dis. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, J.P.; Chen, X.; Coffey, R.J. NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. PLoS ONE 2013, 8, e81514. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; De Simone, M.; Pollice, A.; Santoro, R.; La Mantia, G.; Guerrini, L.; Calabro, V. Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle 2006, 5, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Kathania, M.; Khare, P.; Zeng, M.; Cantarel, B.; Zhang, H.; Ueno, H.; Venuprasad, K. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-gammat ubiquitination. Nat. Immunol. 2016, 17, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Chen, C. WWP1: A versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol. Life Sci. 2012, 69, 1425–1434. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, X.; Luo, Z. WWP2: A multifunctional ubiquitin ligase gene. Pathol. Oncol. Res. 2014, 20, 799–803. [Google Scholar] [CrossRef]
- Kavsak, P.; Rasmussen, R.K.; Causing, C.G.; Bonni, S.; Zhu, H.; Thomsen, G.H.; Wrana, J.L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 2000, 6, 1365–1375. [Google Scholar] [CrossRef]
- Zhu, H.; Kavsak, P.; Abdollah, S.; Wrana, J.L.; Thomsen, G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999, 400, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Koganti, P.; Levy-Cohen, G.; Blank, M. Smurfs in Protein Homeostasis, Signaling, and Cancer. Front. Oncol. 2018, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dong, K.; Wang, Y.; Peng, G.; Song, X.; Yu, Y.; Shen, P.; Cui, X. Integrating HECW1 expression into the clinical indicators exhibits high accuracy in assessing the prognosis of patients with clear cell renal cell carcinoma. BMC Cancer 2021, 21, 890. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Fujita, T.; Ozaki, T.; Kato, C.; Kurose, Y.; Sakamoto, M.; Kato, S.; Goto, T.; Itoyama, Y.; Aoki, M.; et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 2004, 279, 11327–11335. [Google Scholar] [CrossRef]
- Lu, L.; Hu, S.; Wei, R.; Qiu, X.; Lu, K.; Fu, Y.; Li, H.; Xing, G.; Li, D.; Peng, R.; et al. The HECT type ubiquitin ligase NEDL2 is degraded by anaphase-promoting complex/cyclosome (APC/C)-Cdh1, and its tight regulation maintains the metaphase to anaphase transition. J. Biol. Chem. 2013, 288, 35637–35650. [Google Scholar] [CrossRef]
- Qiu, X.; Wei, R.; Li, Y.; Zhu, Q.; Xiong, C.; Chen, Y.; Zhang, Y.; Lu, K.; He, F.; Zhang, L. NEDL2 regulates enteric nervous system and kidney development in its Nedd8 ligase activity-dependent manner. Oncotarget 2016, 7, 31440–31453. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Mooers, B.H. Shortcuts for faster image creation in PyMOL. Protein Sci. 2020, 29, 268–276. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Levy-Cohen, G.; Blank, M. Molecular functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2015, 1856, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Totland, M.Z.; Bergsland, C.H.; Fykerud, T.A.; Knudsen, L.M.; Rasmussen, N.L.; Eide, P.W.; Yohannes, Z.; Sørensen, V.; Brech, A.; Lothe, R.A. The E3 ubiquitin ligase NEDD4 induces endocytosis and lysosomal sorting of connexin 43 to promote loss of gap junctions. J. Cell Sci. 2017, 130, 2867–2882. [Google Scholar] [PubMed]
- Wasserman, S.S.; Shteiman-Kotler, A.; Harris, K.; Iliadi, K.G.; Persaud, A.; Zhong, Y.; Zhang, Y.; Fang, X.; Boulianne, G.L.; Stewart, B. Regulation of SH3PX1 by dNedd4-long at the Drosophila neuromuscular junction. J. Biol. Chem. 2019, 294, 1739–1752. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-y.; Heidersbach, A.; Kathiriya, I.S.; Garay, B.I.; Ivey, K.N.; Srivastava, D.; Han, Z.; King, I.N. The E3 ubiquitin ligase Nedd4/Nedd4L is directly regulated by microRNA 1. Development 2017, 144, 866–875. [Google Scholar] [CrossRef]
- Duerr, J.; Leitz, D.H.; Szczygiel, M.; Dvornikov, D.; Fraumann, S.G.; Kreutz, C.; Zadora, P.K.; Seyhan Agircan, A.; Konietzke, P.; Engelmann, T.A. Conditional deletion of Nedd4-2 in lung epithelial cells causes progressive pulmonary fibrosis in adult mice. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Scrima, M.; Palaia, L.; Salman, A.N.; Quintiero, A.; Franco, R.; Botti, G.; Pirozzi, P.; Rocco, G.; De Rosa, N.; et al. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am. J. Pathol. 2010, 177, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhong, J.; Yu, P.; Zhao, Q.; Huang, T. YY1-regulated LINC00152 promotes triple negative breast cancer progression by affecting on stability of PTEN protein. Biochem. Biophys. Res. Commun. 2019, 509, 448–454. [Google Scholar] [CrossRef]
- Hasna, J.; Hague, F.; Rodat-Despoix, L.; Geerts, D.; Leroy, C.; Tulasne, D.; Ouadid-Ahidouch, H.; Kischel, P. Orai3 calcium channel and resistance to chemotherapy in breast cancer cells: The p53 connection. Cell Death Differ. 2018, 25, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Fifield, B.-A.; Qemo, I.; Kirou, E.; Cardiff, R.D.; Porter, L.A. The atypical cyclin-like protein Spy1 overrides p53-mediated tumour suppression and promotes susceptibility to breast tumourigenesis. Breast Cancer Res. 2019, 21, 1–18. [Google Scholar] [CrossRef]
- Liu, J.; Wan, L.; Liu, P.; Inuzuka, H.; Liu, J.; Wang, Z.; Wei, W. SCFβ-TRCP-mediated degradation of NEDD4 inhibits tumorigenesis through modulating the PTEN/Akt signaling pathway. Oncotarget 2014, 5, 1026. [Google Scholar] [CrossRef]
- Eide, P.W.; Cekaite, L.; Danielsen, S.A.; Eilertsen, I.A.; Kjenseth, A.; Fykerud, T.A.; Ågesen, T.H.; Bruun, J.; Rivedal, E.; Lothe, R.A. NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cell. Signal. 2013, 25, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Xu, N.; Malyukova, A.; Scarlett, C.; Sun, Y.; Zhang, X.; Ling, D.; Su, S.; Nelson, C.; Chang, D. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013, 20, 503–514. [Google Scholar] [CrossRef]
- Chen, Y.; van de Vijver, M.J.; Hibshoosh, H.; Parsons, R.; Saal, L.H. PTEN and NEDD4 in human breast carcinoma. Pathol. Oncol. Res. 2016, 22, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Li, J.; Wang, L.; Xing, Y.; Li, X.; Ruan, H.; Xi, X.; Xiong, J.; Kuang, R. Inhibition of NEDD4 inhibits cell growth and invasion and induces cell apoptosis in bladder cancer cells. Cell Cycle 2017, 16, 1509–1514. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ke, X.; Li, D.; Wang, Q.; Wang, J.; Liu, X.; Deng, M.; Deng, X.; Xue, Y.; Zhu, Y. NEDD4 promotes cell growth and motility in hepatocellular carcinoma. Cell Cycle 2018, 17, 728–738. [Google Scholar] [CrossRef]
- Ji, J.; Ding, K.; Luo, T.; Xu, R.; Zhang, X.; Huang, B.; Chen, A.; Zhang, D.; Miletic, H.; Bjerkvig, R. PMEPA1 isoform a drives progression of glioblastoma by promoting protein degradation of the Hippo pathway kinase LATS1. Oncogene 2020, 39, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Zhou, J.; Luo, S.; Huang, R.; Zhao, C.; Diao, A. Nedd4 E3 ubiquitin ligase promotes cell proliferation and autophagy. Cell Prolif. 2015, 48, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Yun, Z.; Li, S.; Yan, G.; Kang, Z. NEDD4 triggers FOXA1 ubiquitination and promotes colon cancer progression under microRNA-340-5p suppression and ATF1 upregulation. RNA Biol. 2021, 18, 1981–1995. [Google Scholar] [CrossRef]
- Hang, X.; Zhu, S.; Di, H.; Wu, Z.; Chu, K.; Wang, J.; Xin, H.; Yu, G.; Peng, H.; Miao, X. NEDD4 depletion inhibits hepatocellular carcinoma growth via targeting PTEN. Cell. Physiol. Biochem. 2016, 39, 768–779. [Google Scholar] [CrossRef]
- Huang, Z.J.; Zhu, J.J.; Yang, X.Y.; Biskup, E. NEDD4 promotes cell growth and migration via PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Oncol. Lett. 2017, 14, 2649–2656. [Google Scholar] [CrossRef]
- Weng, M.; Luo, Z.-L.; Wu, X.-L.; Zeng, W.-Z. The E3 ubiquitin ligase NEDD4 is translationally upregulated and facilitates pancreatic cancer. Oncotarget 2017, 8, 20288–20296. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.H.; Seo, E.; Min, S.; Nguyen, Q.A.T.; Choi, J.; Lee, U.J.; Hong, S.S.; Kang, H.; Mansukhani, A.; Jou, I. NEDD 4-induced degradative ubiquitination of phosphatidylinositol 4-phosphate 5-kinase α and its implication in breast cancer cell proliferation. J. Cell. Mol. Med. 2018, 22, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, N.M.; Akkoc, Y.; Kig, C.; Bayraktar, O.; Gozuacik, D.; Kutlu, O. Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 2019, 134, 116–137. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Mishra, S.R.; Behera, B.P.; Mahapatra, K.K.; Panigrahi, D.P.; Bhol, C.S.; Praharaj, P.P.; Sethi, G.; Patra, S.K.; Bhutia, S.K. Autophagy-modulating phytochemicals in cancer therapeutics: Current evidences and future perspectives. Semin. Cancer Biol. 2022, 80, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.T.; Kim, C.; Lee, J.H.; Chinnathambi, A.; Alharbi, S.A.; Shair, O.H.M.; Sethi, G.; Ahn, K.S. Cycloastragenol can negate constitutive STAT3 activation and promote paclitaxel-induced apoptosis in human gastric cancer cells. Phytomedicine 2019, 59, 152907. [Google Scholar] [CrossRef]
- Verma, N.; Manna, S.K. Advanced glycation end products (AGE) potentiates cell death in p53 negative cells via upregulaion of NF-kappa B and impairment of autophagy. J. Cell. Physiol. 2017, 232, 3598–3610. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef]
- Su, J.; Zhou, X.; Yin, X.; Wang, L.; Zhao, Z.; Hou, Y.; Zheng, N.; Xia, J.; Wang, Z. The effects of curcumin on proliferation, apoptosis, invasion, and NEDD4 expression in pancreatic cancer. Biochem. Pharmacol. 2017, 140, 28–40. [Google Scholar] [CrossRef]
- Song, Y.-H.; Zhang, C.-Q.; Chen, F.-F.; Lin, X.-Y. Upregulation of neural precursor cell expressed developmentally downregulated 4-1 is associated with poor prognosis and chemoresistance in lung adenocarcinoma. Chin. Med. J. 2018, 131, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Wang, R.; Sun, A.; Wei, J.; Peng, K.; Dai, Q.; Yang, W.; Lin, Q. The E3 ubiquitin ligase NEDD4 mediates cell migration signaling of EGFR in lung cancer cells. Mol. Cancer 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Huang, C.; Chen, Z.; Huang, H.; Wang, X.; Chen, J. An outlined review for the role of Nedd4-1 and Nedd4-2 in lung disorders. Biomed. Pharmacother. 2020, 125, 109983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qin, Y.; Wu, G.; Wang, J.; Cao, J.; Wang, Y.; Wu, D.; Yang, K.; Zhao, Z.; He, L. PRRG4 promotes breast cancer metastasis through the recruitment of NEDD4 and downregulation of Robo1. Oncogene 2020, 39, 7196–7208. [Google Scholar] [CrossRef]
- Wang, X.; Deng, J.; Yuan, J.; Tang, X.; Wang, Y.; Chen, H.; Liu, Y.; Zhou, L. Curcumin exerts its tumor suppressive function via inhibition of NEDD4 oncoprotein in glioma cancer cells. Int. J. Oncol. 2017, 51, 467–477. [Google Scholar] [CrossRef]
- Sun, A.; Yu, G.; Dou, X.; Yan, X.; Yang, W.; Lin, Q. Nedd4-1 is an exceptional prognostic biomarker for gastric cardia adenocarcinoma and functionally associated with metastasis. Mol. Cancer 2014, 13, 1–10. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Ahn, K.S.; Hsu, A.; Woo, C.C.; Yuan, Y.; Tan, K.H.B.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Koh, A.P.F.; et al. Thymoquinone inhibits bone metastasis of breast cancer cells through abrogation of the CXCR4 signaling axis. Front. Pharm. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Dai, X.; Ahn, K.S.; Wang, L.Z.; Kim, C.; Deivasigamni, A.; Arfuso, F.; Um, J.Y.; Kumar, A.P.; Chang, Y.C.; Kumar, D.; et al. Ascochlorin enhances the sensitivity of doxorubicin leading to the reversal of epithelial-to-mesenchymal transition in hepatocellular carcinoma. Mol. Cancer 2016, 15, 2966–2976. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef]
- Shrestha, P.; Yun, J.-H.; Ko, Y.-J.; Yeon, K.J.; Kim, D.; Lee, H.; Jin, D.-H.; Nam, K.-Y.; Yoo, H.D.; Lee, W. NMR uncovers direct interaction between human NEDD4-1 and p34SEI-1. Biochem. Biophys. Res. Commun. 2017, 490, 984–990. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Buhrmann, C.; Yazdi, M.; Popper, B.; Kunnumakkara, A.B.; Aggarwal, B.B.; Shakibaei, M. Induction of the epithelial-to-mesenchymal transition of human colorectal cancer by human TNF-β (lymphotoxin) and its reversal by resveratrol. Nutrients 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Saghari, S.; Bassiri, F.; Raesi, R.; Zarrabi, A.; Hushmandi, K.; Sethi, G.; Tergaonkar, V. NF-κB as a regulator of cancer metastasis and therapy response: A focus on epithelial-mesenchymal transition. J. Cell Physiol. 2022, 237, 2770–2795. [Google Scholar] [CrossRef]
- Cheng, J.T.; Wang, L.; Wang, H.; Tang, F.R.; Cai, W.Q.; Sethi, G.; Xin, H.W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. [Google Scholar] [CrossRef]
- Lee, J.H.; Chinnathambi, A.; Alharbi, S.A.; Shair, O.H.M.; Sethi, G.; Ahn, K.S. Farnesol abrogates epithelial to mesenchymal transition process through regulating Akt/mTOR pathway. Pharm. Res. 2019, 150, 104504. [Google Scholar] [CrossRef]
- Feng, S.; Yang, G.; Yang, H.; Liang, Z.; Zhang, R.; Fan, Y.; Zhang, G. NEDD4 is involved in acquisition of epithelial-mesenchymal transition in cisplatin-resistant nasopharyngeal carcinoma cells. Cell Cycle 2017, 16, 869–878. [Google Scholar] [CrossRef]
- Chen, L.; Wang, G.; Luo, Y.; Wang, Y.; Xie, C.; Jiang, W.; Xiao, Y.; Qian, G.; Wang, X. Downregulation of LAPTM5 suppresses cell proliferation and viability inducing cell cycle arrest at G0/G1 phase of bladder cancer cells. Int. J. Oncol. 2017, 50, 263–271. [Google Scholar] [CrossRef]
- Qu, M.-H.; Han, C.; Srivastava, A.K.; Cui, T.; Zou, N.; Gao, Z.-Q.; Wang, Q.-E. miR-93 promotes TGF-β-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumor Biol. 2016, 37, 5645–5651. [Google Scholar] [CrossRef]
- Nahand, J.S.; Khanaliha, K.; Mirzaei, H.; Moghoofei, M.; Baghi, H.B.; Esghaei, M.; Khatami, A.R.; Fatemipour, M.; Bokharaei-Salim, F. Possible role of HPV/EBV coinfection in anoikis resistance and development in prostate cancer. BMC Cancer 2021, 21, 1–19. [Google Scholar] [CrossRef]
- Manu, K.A.; Shanmugam, M.K.; Li, F.; Chen, L.; Siveen, K.S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Simvastatin sensitizes human gastric cancer xenograft in nude mice to capecitabine by suppressing nuclear factor-kappa B-regulated gene products. J. Mol. Med. 2014, 92, 267–276. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Hushmandi, K.; Kalantari, M.; Mohammadinejad, R.; Javaheri, T.; Sethi, G. Association of the epithelial-mesenchymal transition (EMT) with cisplatin resistance. Int. J. Mol. Sci. 2020, 21, 4002. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-Y.; Hsu, L.-Y.; Pan, C.-M.; Pikatan, N.W.; Yadav, V.K.; Fong, I.-H.; Chen, C.-H.; Yeh, C.-T.; Chiu, S.-C. The e3 ubiquitin ligase nedd4-1 mediates temozolomide-resistant glioblastoma through pten attenuation and redox imbalance in nrf2–ho-1 axis. Int. J. Mol. Sci. 2021, 22, 10247. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-L.; Chen, H.-J.; Hou, G.-Q.; Zhang, X.-H.; Ge, J.-W. LINC01198 promotes proliferation and temozolomide resistance in a NEDD4-1-dependent manner, repressing PTEN expression in glioma. Aging 2019, 11, 6053. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Luo, M.; Zhang, K.; Zhang, J.; Gao, T.; Connell, D.O.; Yao, F.; Mu, C.; Cai, B.; Shang, Y. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Sun, H.; Ma, H.; Wang, J.; Xia, L.; Zhu, G.; Wang, Z.; Sun, J.; Chen, Z. Phosphatase and tensin homolog deleted on chromosome 10 degradation induced by NEDD4 promotes acquired erlotinib resistance in non–small-cell lung cancer. Tumor Biol. 2017, 39, 1010428317709639. [Google Scholar] [CrossRef]
- Novellasdemunt, L.; Kucharska, A.; Jamieson, C.; Prange-Barczynska, M.; Baulies, A.; Antas, P.; van der Vaart, J.; Gehart, H.; Maurice, M.M.; Li, V.S. NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. EMBO J. 2020, 39, e102771. [Google Scholar] [CrossRef]
- Fukushima, T.; Yoshihara, H.; Furuta, H.; Kamei, H.; Hakuno, F.; Luan, J.; Duan, C.; Saeki, Y.; Tanaka, K.; Iemura, S.-I. Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Bae, S.J.; Kim, M.; Kim, S.-H.; Kwon, Y.E.; Lee, J.-H.; Kim, J.; Chung, C.H.; Lee, W.-J.; Seol, J.H. NEDD4 controls intestinal stem cell homeostasis by regulating the Hippo signalling pathway. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef]
- Nguyen Huu, N.S.; Ryder, W.D.J.; Zeps, N.; Flasza, M.; Chiu, M.; Hanby, A.; Poulsom, R.; Clarke, R.B.; Baron, M. Tumour-promoting activity of altered WWP1 expression in breast cancer and its utility as a prognostic indicator. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2008, 216, 93–102. [Google Scholar] [CrossRef]
- Jung, S.; Li, C.; Jeong, D.; Lee, S.; Ohk, J.; Park, M.; Han, S.; Duan, J.; Kim, C.; Yang, Y. Oncogenic function of p34SEI-1 via NEDD4-1-mediated PTEN ubiquitination/degradation and activation of the PI3K/AKT pathway. Int. J. Oncol. 2013, 43, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Nourashrafeddin, S.; Aarabi, M.; Modarressi, M.H.; Rahmati, M.; Nouri, M. The evaluation of WBP2NL-related genes expression in breast cancer. Pathol. Oncol. Res. 2015, 21, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Yoo, N.J.; Jeong, E.G.; Kim, M.S.; Lee, S.H. Expression of NEDD4-1, a PTEN regulator, in gastric and colorectal carcinomas. APMIS 2008, 116, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, E.; Gul, M.; Melekoglu, R.; Inci Coskun, E.; Sahin, N.; Gul, S.; Bastemur, A.G.; Ciplak, B. Neural precursor cell-expressed developmentally down-regulated 4-like: A new biomarker in the pathophysiology of endometrial cancer. J. Int. Med. Res. 2018, 46, 3709–3716. [Google Scholar] [CrossRef]
- Zhang, Y.; Goodfellow, R.; Li, Y.; Yang, S.; Winters, C.J.; Thiel, K.W.; Leslie, K.K.; Yang, B. NEDD4 ubiquitin ligase is a putative oncogene in endometrial cancer that activates IGF-1R/PI3K/Akt signaling. Gynecol. Oncol. 2015, 139, 127–133. [Google Scholar] [CrossRef]
- Takeuchi, T.; Adachi, Y.; Nagayama, T.; Furihata, M. Nedd4L modulates the transcription of metalloproteinase-1 and-13 genes to increase the invasive activity of gallbladder cancer. Int. J. Exp. Pathol. 2011, 92, 79–86. [Google Scholar] [CrossRef]
- Gao, C.; Pang, L.; Ren, C.; Ma, T. Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Med. Oncol. 2012, 29, 1733–1738. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, S.; Yin, Z.; Sheng, Y.; Yan, Q.; Sun, R.; Lu, M.; Zhang, Z.; Li, Y. The correlation between NEDD4L and HIF-1α levels as a gastric cancer prognostic marker. Int. J. Med. Sci. 2019, 16, 1517–1524. [Google Scholar] [CrossRef]
- He, S.; Deng, J.; Li, G.; Wang, B.; Cao, Y.; Tu, Y. Down-regulation of Nedd4L is associated with the aggressive progression and worse prognosis of malignant glioma. Jpn. J. Clin. Oncol. 2012, 42, 196–201. [Google Scholar] [CrossRef]
- Zhang, H.; Nie, W.; Zhang, X.; Zhang, G.; Li, Z.; Wu, H.; Shi, Q.; Chen, Y.; Ding, Z.; Zhou, X. NEDD4-1 regulates migration and invasion of glioma cells through CNrasGEF ubiquitination in vitro. PLoS ONE 2013, 8, e82789. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, B.; Wang, S.; Wu, Y.; Zhan, W.; Xie, S.; Shi, H.; Yu, R. Regulation of glioma migration and invasion via modification of Rap2a activity by the ubiquitin ligase Nedd4-1. Oncol. Rep. 2017, 37, 2565–2574. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Zhang, J.; Miao, Z.; Ding, Y.; Xu, X.; Zhao, X.; Xu, P.; Wang, Q.; Lin, Y. Suppression of the Smurf1 expression inhibits tumor progression in gliomas. Cell. Mol. Neurobiol. 2018, 38, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Gong, X.; Liu, A.; Lv, X.; Hu, B.; Zhang, H. Downregulation of Nedd4L predicts poor prognosis, promotes tumor growth and inhibits MAPK/ERK signal pathway in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018, 495, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Duan, J.; Fu, W.; Yin, Z.; Sheng, J.; Lei, Z.; Wang, H. Decreased expression of NEDD4L contributes to NSCLC progression and metastasis. Biochem. Biophys. Res. Commun. 2019, 513, 398–404. [Google Scholar] [CrossRef]
- Kito, Y.; Bai, J.; Goto, N.; Okubo, H.; Adachi, Y.; Nagayama, T.; Takeuchi, T. Pathobiological properties of the ubiquitin ligase N edd4 L in melanoma. Int. J. Exp. Pathol. 2014, 95, 24–28. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Cui, M.; Gi, S.; Wang, W.; Han, X. Nedd4L expression is decreased in ovarian epithelial cancer tissues compared to ovarian non-cancer tissue. J. Obstet. Gynaecol. Res. 2015, 41, 1959–1964. [Google Scholar] [CrossRef]
- Hu, X.; Xu, Y.; Fu, Q.; Yu, J.; Huang, J. Nedd4L expression is downregulated in prostate cancer compared to benign prostatic hyperplasia. Eur. J. Surg. Oncol. (EJSO) 2009, 35, 527–531. [Google Scholar] [CrossRef]
- Hellwinkel, O.J.; Asong, L.E.; Rogmann, J.-P.; Sültmann, H.; Wagner, C.; Schlomm, T.; Eichelberg, C. Transcription alterations of members of the ubiquitin–proteasome network in prostate carcinoma. Prostate Cancer Prostatic Dis. 2011, 14, 38–45. [Google Scholar] [CrossRef]
- Hughes, J.R.; Parsons, J.L. The E3 ubiquitin ligase NEDD4L targets OGG1 for ubiquitylation and modulates the cellular DNA damage response. Front. Cell Dev. Biol. 2020, 8, 607060. [Google Scholar] [CrossRef]
- Lønne, G.K.; Masoumi, K.C.; Lennartsson, J.; Larsson, C. Protein kinase Cdelta supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J. Biol. Chem. 2009, 284, 33456–33465. [Google Scholar] [CrossRef]
- Yeung, B.; Ho, K.-C.; Yang, X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS ONE 2013, 8, e61027. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Choi, B.; Mujoo, K.; Fan, X.; Fa, M.; Mukherjee, S.; Owiti, N.; Zhang, N.; An, Z. The E3 ubiquitin ligase NEDD4 negatively regulates HER3/ErbB3 level and signaling. Oncogene 2015, 34, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wan, L.; Liu, J.; Yuan, Z.; Zhang, J.; Guo, J.; Malumbres, M.; Liu, J.; Zou, W.; Wei, W. Cdh1 inhibits WWP2-mediated ubiquitination of PTEN to suppress tumorigenesis in an APC-independent manner. Cell Discov. 2016, 2, 1–13. [Google Scholar] [CrossRef]
- Sun, M.; Cai, J.; Anderson, R.A.; Sun, Y. Type I γ phosphatidylinositol phosphate 5-kinase i5 controls the ubiquitination and degradation of the tumor suppressor mitogen-inducible gene 6. J. Biol. Chem. 2016, 291, 21461–21473. [Google Scholar] [CrossRef]
- Soung, Y.H.; Ford, S.; Yan, C.; Chung, J. The role of arrestin domain-containing 3 in regulating endocytic recycling and extracellular vesicle sorting of integrin β4 in breast cancer. Cancers 2018, 10, 507. [Google Scholar] [CrossRef]
- Suga, J.; Izumiyama, K.; Tanaka, N.; Saji, S. Estradiol promotes rapid degradation of HER3 in ER-positive breast cancer cell line MCF-7. Biochem. Biophys. Rep. 2018, 16, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-F.; Cheng, Y.-K.; Lu, D.-Y.; Wang, S.-L.; Chang, C.-N.; Chang, P.-C.; Yeh, W.-L. Inhibition of estrogen receptor reduces connexin 43 expression in breast cancers. Toxicol. Appl. Pharmacol. 2018, 338, 182–190. [Google Scholar] [CrossRef]
- Jeon, S.-A.; Kim, D.W.; Lee, D.-B.; Cho, J.-Y. NEDD4 plays roles in the maintenance of breast cancer stem cell characteristics. Front. Oncol. 2020, 10, 1680. [Google Scholar] [CrossRef]
- Ren, Y.; Chen, D.; Zhai, Z.; Chen, J.; Li, A.; Liang, Y.; Zhou, J. JAC1 suppresses proliferation of breast cancer through the JWA/p38/SMURF1/HER2 signaling. Cell Death Discov. 2021, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhou, X.; Li, J.; Yao, X.-C.; Liu, W.-L.; Kang, F.-H.; Zou, Z.-X.; Xu, K.-P.; Xu, P.-S.; Tan, G.-S. Identification of a new natural biflavonoids against breast cancer cells induced ferroptosis via the mitochondrial pathway. Bioorganic Chem. 2021, 109, 104744. [Google Scholar] [CrossRef]
- Ahn, Y.; Hwang, C.Y.; Lee, S.-R.; Kwon, K.-S.; Lee, C. The tumour suppressor PTEN mediates a negative regulation of the E3 ubiquitin-protein ligase Nedd4. Biochem. J. 2008, 412, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Nazio, F.; Carinci, M.; Valacca, C.; Bielli, P.; Strappazzon, F.; Antonioli, M.; Ciccosanti, F.; Rodolfo, C.; Campello, S.; Fimia, G.M. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 2016, 215, 841–856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, C.; Yang, X.; Hong, H.; Lu, J.; Hu, W.; Hao, X.; Li, S.; Aikemu, B.; Yang, G. N-myc downstream-regulated gene 1 inhibits the proliferation of colorectal cancer through emulative antagonizing NEDD4-mediated ubiquitylation of p21. J. Exp. Clin. Cancer Res. 2019, 38, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xu, C.Y.; Cui, R.J.; Tang, J.B.; Sun, H.; Yang, Z.K.; Bu, J.Y.; Lin, P.; Huang, N.; Du, Y.D. DNA methylation inhibitor, decitabine, promotes MGC803 gastric cancer cell migration and invasion via the upregulation of NEDD4-1. Mol. Med. Rep. 2015, 12, 8201–8208. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Pieper, R.O.; Li, D.; Wei, P.; Liu, M.; Woo, S.Y.; Aldape, K.D.; Sawaya, R.; Xie, K.; Huang, S. FoxM1B Regulates NEDD4-1 Expression, Leading to Cellular Transformation and Full Malignant Phenotype in Immortalized Human AstrocytesFoxM1B in Astrocytoma Transformation. Cancer Res. 2010, 70, 2951–2961. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-C.; Chen, P.-H.; Ho, K.-H.; Shih, C.-M.; Chou, C.-M.; Cheng, C.-H.; Lee, C.-C. IGF-1-enhanced miR-513a-5p signaling desensitizes glioma cells to temozolomide by targeting the NEDD4L-inhibited Wnt/β-catenin pathway. PLoS ONE 2019, 14, e0225913. [Google Scholar] [CrossRef] [PubMed]
- Bellet, M.M.; Piobbico, D.; Bartoli, D.; Castelli, M.; Pieroni, S.; Brunacci, C.; Chiacchiaretta, M.; Del Sordo, R.; Fallarino, F.; Sidoni, A. NEDD4 controls the expression of GUCD1, a protein upregulated in proliferating liver cells. Cell Cycle 2014, 13, 1902–1911. [Google Scholar] [CrossRef]
- Bai, X.; Jing, L.; Li, Y.; Li, Y.; Luo, S.; Wang, S.; Zhou, J.; Liu, Z.; Diao, A. TMEPAI inhibits TGF-β signaling by promoting lysosome degradation of TGF-β receptor and contributes to lung cancer development. Cell. Signal. 2014, 26, 2030–2039. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Lin, M.; Jinjun, B.; Su, F.; Dan, C.; Yan, C.; Jie, Y.; Jin, Z.; Zi-Chun, H.; Wu, Y. Involvement of general control nonderepressible kinase 2 in cancer cell apoptosis by posttranslational mechanisms. Mol. Biol. Cell 2015, 26, 1044–1057. [Google Scholar] [CrossRef]
- Lévy, F.; Muehlethaler, K.; Salvi, S.; Peitrequin, A.-L.; Lindholm, C.K.; Cerottini, J.-C.; Rimoldi, D. Ubiquitylation of a melanosomal protein by HECT-E3 ligases serves as sorting signal for lysosomal degradation. Mol. Biol. Cell 2005, 16, 1777–1787. [Google Scholar] [CrossRef]
- Aronchik, I.; Kundu, A.; Quirit, J.G.; Firestone, G.L. The antiproliferative response of indole-3-carbinol in human melanoma cells is triggered by an interaction with NEDD4-1 and disruption of wild-type PTEN degradation. Mol. Cancer Res. 2014, 12, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yang, L.; Lei, S.; Tan, W.; Long, J. NEDD4 negatively regulates GITR via ubiquitination in immune microenvironment of melanoma. OncoTargets Ther. 2019, 12, 10629. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Shu, C.; Xu, J.; Chen, D.; Li, J.; Ding, K.; Chen, M.; Li, A.; He, J.; Shu, Y. JP1 suppresses proliferation and metastasis of melanoma through MEK1/2 mediated NEDD4L-SP1-Integrin αvβ3 signaling. Theranostics 2020, 10, 8036–8050. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Amraei, R.; Rahimi, N. NEDD4 regulates ubiquitination and stability of the cell adhesion molecule IGPR-1 via lysosomal pathway. J. Biomed. Sci. 2021, 28, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhu, N.; Chen, S.; Zhao, P.; Ren, H.; Zhu, S.; Tang, H.; Zhu, Y.; Qi, Z. E3 ubiquitin ligase Nedd4 promotes Japanese encephalitis virus replication by suppressing autophagy in human neuroblastoma cells. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Xu, M.; Yin, M.; Hong, J.; Chen, H.; Gao, Y.; Xie, C.; Shen, N.; Gu, S.; Mo, X. Exosomal hsa-miR199a-3p promotes proliferation and migration in neuroblastoma. Front. Oncol. 2019, 9, 459. [Google Scholar] [CrossRef]
- Zhao, R.; Cui, T.; Han, C.; Zhang, X.; He, J.; Srivastava, A.K.; Yu, J.; Wani, A.A.; Wang, Q.-E. DDB2 modulates TGF-β signal transduction in human ovarian cancer cells by downregulating NEDD4L. Nucleic Acids Res. 2015, 43, 7838–7849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Liu, J.; Kang, R.; Tang, D. NEDD4L-mediated LTF protein degradation limits ferroptosis. Biochem. Biophys. Res. Commun. 2020, 531, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Maddika, S.; Kavela, S.; Rani, N.; Palicharla, V.R.; Pokorny, J.L.; Sarkaria, J.N.; Chen, J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 2011, 13, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Hushmandi, K.; Hashemi, M.; Akbari, M.E.; Kubatka, P.; Raei, M.; Koklesova, L.; Shahinozzaman, M.; Mohammadinejad, R.; Najafi, M.; et al. Role of microRNA/epithelial-to-mesenchymal transition axis in the metastasis of bladder cancer. Biomolecules 2020, 10, 1159. [Google Scholar] [CrossRef]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Padala, S.A.; Barsouk, A. Epidemiology of bladder cancer. Med. Sci. 2020, 8, 15. [Google Scholar] [CrossRef]
- Khwairakpam, A.D.; Bordoloi, D.; Thakur, K.K.; Monisha, J.; Arfuso, F.; Sethi, G.; Mishra, S.; Kumar, A.P.; Kunnumakkara, A.B. Possible use of Punica granatum (Pomegranate) in cancer therapy. Pharmacol. Res. 2018, 133, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Braden, A.M.; Stankowski, R.V.; Engel, J.M.; Onitilo, A.A. Breast cancer biomarkers: Risk assessment, diagnosis, prognosis, prediction of treatment efficacy and toxicity, and recurrence. Curr. Pharm. Des. 2014, 20, 4879–4898. [Google Scholar] [CrossRef]
- Libring, S.; Shinde, A.; Chanda, M.K.; Nuru, M.; George, H.; Saleh, A.M.; Abdullah, A.; Kinzer-Ursem, T.L.; Calve, S.; Wendt, M.K. The dynamic relationship of breast cancer cells and fibroblasts in fibronectin accumulation at primary and metastatic tumor sites. Cancers 2020, 12, 1270. [Google Scholar] [CrossRef]
- Thakur, K.K.; Bordoloi, D.; Kunnumakkara, A.B. Alarming burden of triple-negative breast cancer in India. Clin. Breast Cancer 2018, 18, e393–e399. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.K.; Kumar, A.; Banik, K.; Verma, E.; Khatoon, E.; Harsha, C.; Sethi, G.; Gupta, S.C.; Kunnumakkara, A.B. Long noncoding RNAs in triple-negative breast cancer: A new frontier in the regulation of tumorigenesis. J. Cell. Physiol. 2021, 236, 7938–7965. [Google Scholar] [CrossRef] [PubMed]
- Maruthanila, V.; Elancheran, R.; Kunnumakkara, A.; Kabilan, S.; Kotoky, J. Recent development of targeted approaches for the treatment of breast cancer. Breast Cancer 2017, 24, 191–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ahn, K.S.; Shanmugam, M.K.; Wang, H.; Shen, H.; Arfuso, F.; Chinnathambi, A.; Alharbi, S.A.; Chang, Y.; Sethi, G.; et al. Oleuropein induces apoptosis via abrogating NF-κB activation cascade in estrogen receptor-negative breast cancer cells. J. Cell Biochem. 2019, 120, 4504–4513. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Srinivasa, V.; Rangappa, S.; Mervin, L.; Mohan, S.; Paricharak, S.; Baday, S.; Li, F.; Shanmugam, M.K.; Chinnathambi, A.; et al. Trisubstituted-Imidazoles Induce Apoptosis in Human Breast Cancer Cells by Targeting the Oncogenic PI3K/Akt/mTOR Signaling Pathway. PLoS ONE 2016, 11, e0153155. [Google Scholar] [CrossRef]
- Zhang, F.; Dong, W.; Zeng, W.; Zhang, L.; Zhang, C.; Qiu, Y.; Wang, L.; Yin, X.; Zhang, C.; Liang, W. Naringenin prevents TGF-β1 secretion from breast cancer and suppresses pulmonary metastasis by inhibiting PKC activation. Breast Cancer Res. 2016, 18, 1–16. [Google Scholar] [CrossRef]
- Yim, E.-K.; Peng, G.; Dai, H.; Hu, R.; Li, K.; Lu, Y.; Mills, G.B.; Meric-Bernstam, F.; Hennessy, B.T.; Craven, R.J. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 2009, 15, 304–314. [Google Scholar] [CrossRef]
- Luhtala, S.; Staff, S.; Kallioniemi, A.; Tanner, M.; Isola, J. Clinicopathological and prognostic correlations of HER3 expression and its degradation regulators, NEDD4-1 and NRDP1, in primary breast cancer. BMC Cancer 2018, 18, 1–19. [Google Scholar] [CrossRef]
- Guarnieri, A.; Towers, C.; Drasin, D.; Oliphant, M.; Andrysik, Z.; Hotz, T.; Vartuli, R.; Linklater, E.; Pandey, A.; Khanal, S. The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L. Oncogene 2018, 37, 3879–3893. [Google Scholar] [CrossRef] [PubMed]
- Buskwofie, A.; David-West, G.; Clare, C.A. A review of cervical cancer: Incidence and disparities. J. Natl. Med. Assoc. 2020, 112, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Tsikouras, P.; Zervoudis, S.; Manav, B.; Tomara, E.; Iatrakis, G.; Romanidis, C.; Bothou, A.; Galazios, G. Cervical cancer: Screening, diagnosis and staging. J Buon 2016, 21, 320–325. [Google Scholar] [PubMed]
- Parama, D.; Boruah, M.; Yachna, K.; Rana, V.; Banik, K.; Harsha, C.; Thakur, K.K.; Dutta, U.; Arya, A.; Mao, X. Diosgenin, a steroidal saponin, and its analogs: Effective therapies against different chronic diseases. Life Sci. 2020, 260, 118182. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.K.; Jhingran, A.; Klopp, A.H.; Aggarwal, B.B.; Kunnumakkara, A.B.; Broadus, R.R.; Eifel, P.J.; Buchholz, T.A. Expression of nuclear transcription factor kappa B in locally advanced human cervical cancer treated with definitive chemoradiation. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 1331–1336. [Google Scholar] [CrossRef]
- Roy, N.K.; Parama, D.; Banik, K.; Bordoloi, D.; Devi, A.K.; Thakur, K.K.; Padmavathi, G.; Shakibaei, M.; Fan, L.; Sethi, G. An update on pharmacological potential of boswellic acids against chronic diseases. Int. J. Mol. Sci. 2019, 20, 4101. [Google Scholar] [CrossRef] [PubMed]
- Ningegowda, R.; Shivananju, N.S.; Rajendran, P.; Basappa; Rangappa, K.S.; Chinnathambi, A.; Li, F.; Achar, R.R.; Shanmugam, M.K.; Bist, P.; et al. A novel 4,6-disubstituted-1,2,4-triazolo-1,3,4-thiadiazole derivative inhibits tumor cell invasion and potentiates the apoptotic effect of TNFα by abrogating NF-κB activation cascade. Apoptosis 2017, 22, 145–157. [Google Scholar] [PubMed]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Prim. 2015, 1, 15065. [Google Scholar] [CrossRef]
- Buhrmann, C.; Shayan, P.; Banik, K.; Kunnumakkara, A.B.; Kubatka, P.; Koklesova, L.; Shakibaei, M. Targeting NF-κB signaling by calebin a, a compound of turmeric, in multicellular tumor microenvironment: Potential role of apoptosis induction in CRC cells. Biomedicines 2020, 8, 236. [Google Scholar] [CrossRef]
- Ko, J.H.; Um, J.Y.; Lee, S.G.; Yang, W.M.; Sethi, G.; Ahn, K.S. Conditioned media from adipocytes promote proliferation, migration, and invasion in melanoma and colorectal cancer cells. J. Cell Physiol. 2019, 234, 18249–18261. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Femia, M.; Buscarino, V.; Franchi, D.; Garbi, A.; Zanagnolo, V.; Del Grande, M.; Manganaro, L.; Alessi, S.; Giannitto, C. Endometrial cancer: An overview of novelties in treatment and related imaging keypoints for local staging. Cancer Imaging 2018, 18, 1–12. [Google Scholar] [CrossRef]
- Leslie, K.K.; Thiel, K.W.; Goodheart, M.J.; De Geest, K.; Jia, Y.; Yang, S. Endometrial cancer. Obstet. Gynecol. Clin. 2012, 39, 255–268. [Google Scholar] [CrossRef]
- Charo, L.M.; Plaxe, S.C. Recent advances in endometrial cancer: A review of key clinical trials from 2015 to 2019. F1000Research 2019, 8, F1000 Faculty Rev-849. [Google Scholar] [CrossRef] [PubMed]
- Bishayee, A.; Haskell, Y.; Do, C.; Siveen, K.S.; Mohandas, N.; Sethi, G.; Stoner, G.D. Potential benefits of edible berries in the management of aerodigestive and gastrointestinal tract cancers: Preclinical and clinical evidence. Crit. Rev. Food Sci. Nutr. 2016, 56, 1753–1775. [Google Scholar] [CrossRef] [PubMed]
- Manu, K.A.; Shanmugam, M.K.; Rajendran, P.; Li, F.; Ramachandran, L.; Hay, H.S.; Kannaiyan, R.; Swamy, S.N.; Vali, S.; Kapoor, S.; et al. Plumbagin inhibits invasion and migration of breast and gastric cancer cells by downregulating the expression of chemokine receptor CXCR4. Mol. Cancer 2011, 10, 107. [Google Scholar] [CrossRef]
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, G.J.A.; Maciejewski, R.; Polkowski, W.P. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Henamayee, S.; Banik, K.; Sailo, B.L.; Shabnam, B.; Harsha, C.; Srilakshmi, S.; Vgm, N.; Baek, S.H.; Ahn, K.S.; Kunnumakkara, A.B. Therapeutic emergence of rhein as a potential anticancer drug: A review of its molecular targets and anticancer properties. Molecules 2020, 25, 2278. [Google Scholar] [CrossRef] [PubMed]
- Bhuvanalakshmi, G.; Gamit, N.; Patil, M.; Arfuso, F.; Sethi, G.; Dharmarajan, A.; Prem Kumar, A.; Warrier, S. Stemness, pluripotentiality, and Wnt antagonism: sFRP4, a Wnt antagonist mediates pluripotency and stemness in glioblastoma. Cancers 2018, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Swamy, S.G.; Kameshwar, V.H.; Shubha, P.B.; Looi, C.Y.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Sethi, G.; Shivananju, N.S.; Bishayee, A. Targeting multiple oncogenic pathways for the treatment of hepatocellular carcinoma. Target Oncol. 2017, 12, 1–10. [Google Scholar] [CrossRef]
- Dai, X.; Ahn, K.S.; Kim, C.; Siveen, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Shi, J.; Kumar, A.P.; Wang, L.Z.; et al. Ascochlorin, an isoprenoid antibiotic inhibits growth and invasion of hepatocellular carcinoma by targeting STAT3 signaling cascade through the induction of PIAS3. Mol. Oncol. 2015, 9, 818–833. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Chen, K.-F.; Chen, P.-J. Treatment of liver cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a021535. [Google Scholar] [CrossRef]
- Siveen, K.S.; Nguyen, A.H.; Lee, J.H.; Li, F.; Singh, S.S.; Kumar, A.P.; Low, G.; Jha, S.; Tergaonkar, V.; Ahn, K.S.; et al. Negative regulation of signal transducer and activator of transcription-3 signalling cascade by lupeol inhibits growth and induces apoptosis in hepatocellular carcinoma cells. Br. J. Cancer 2014, 111, 1327–1337. [Google Scholar] [CrossRef]
- Lee, J.H.; Mohan, C.D.; Deivasigamani, A.; Jung, Y.Y.; Rangappa, S.; Basappa, S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Garg, M.; et al. Brusatol suppresses STAT3-driven metastasis by downregulating epithelial-mesenchymal transition in hepatocellular carcinoma. J. Adv. Res. 2020, 26, 83–94. [Google Scholar] [CrossRef]
- Girisa, S.; Shabnam, B.; Monisha, J.; Fan, L.; Halim, C.E.; Arfuso, F.; Ahn, K.S.; Sethi, G.; Kunnumakkara, A.B. Potential of zerumbone as an anti-cancer agent. Molecules 2019, 24, 734. [Google Scholar] [CrossRef]
- Tan, S.M.; Li, F.; Rajendran, P.; Kumar, A.P.; Hui, K.M.; Sethi, G. Identification of beta-escin as a novel inhibitor of signal transducer and activator of transcription 3/Janus-activated kinase 2 signaling pathway that suppresses proliferation and induces apoptosis in human hepatocellular carcinoma cells. J. Pharm. Exp. 2010, 334, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Tunissiolli, N.M.; Castanhole-Nunes, M.M.U.; Biselli-Chicote, P.M.; Pavarino, É.C.; da Silva, R.F.; Goloni-Bertollo, E.M. Hepatocellular carcinoma: A comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 863–872. [Google Scholar]
- Lemjabbar-Alaoui, H.; Hassan, O.U.; Yang, Y.-W.; Buchanan, P. Lung cancer: Biology and treatment options. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2015, 1856, 189–210. [Google Scholar] [CrossRef]
- Quinn, B.J.; Dallos, M.; Kitagawa, H.; Kunnumakkara, A.B.; Memmott, R.M.; Hollander, M.C.; Gills, J.J.; Dennis, P.A. Inhibition of lung tumorigenesis by metformin is associated with decreased plasma IGF-I and diminished receptor tyrosine kinase signaling. Cancer Prev. Res. 2013, 6, 801–810. [Google Scholar] [CrossRef]
- Bordoloi, D.; Banik, K.; Padmavathi, G.; Vikkurthi, R.; Harsha, C.; Roy, N.K.; Singh, A.K.; Monisha, J.; Wang, H.; Kumar, A.P. TIPE2 induced the proliferation, survival, and migration of lung cancer cells through modulation of Akt/mTOR/NF-κB signaling cascade. Biomolecules 2019, 9, 836. [Google Scholar] [CrossRef] [PubMed]
- Bordoloi, D.; Harsha, C.; Padmavathi, G.; Banik, K.; Sailo, B.L.; Roy, N.K.; Girisa, S.; Thakur, K.K.; Khwairakpam, A.D.; Chinnathambi, A. Loss of TIPE3 reduced the proliferation, survival and migration of lung cancer cells through inactivation of Akt/mTOR, NF-κB, and STAT-3 signaling cascades. Life Sci. 2022, 293, 120332. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Syn, N.L.; Subhash, V.V.; Any, Y.; Thuya, W.L.; Cheow, E.S.H.; Kong, L.; Yu, F.; Peethala, P.C.; Wong, A.L.; et al. Pan-HDAC inhibition by panobinostat mediates chemosensitization to carboplatin in non-small cell lung cancer via attenuation of EGFR signaling. Cancer Lett. 2018, 417, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Ong, P.S.; Wang, L.; Chia, D.M.; Seah, J.Y.; Kong, L.R.; Thuya, W.L.; Chinnathambi, A.; Lau, J.Y.; Wong, A.L.; Yong, W.P.; et al. A novel combinatorial strategy using Seliciclib(®) and Belinostat(®) for eradication of non-small cell lung cancer via apoptosis induction and BID activation. Cancer Lett. 2016, 381, 49–57. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, K.; Zhi, Y.; Wu, Y.; Chen, B.; Bai, J.; Wang, X. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin. Transl. Med. 2021, 11, e478. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Shanmugam, M.K.; Narula, A.S.; Kim, C.; Lee, J.H.; Namjoshi, O.A.; Blough, B.E.; Sethi, G.; Ahn, K.S. Oxymatrine attenuates tumor growth and deactivates STAT5 signaling in a lung cancer xenograft model. Cancers 2019, 11, 49. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Lee, S.G.; Sethi, G.; Ahn, K.S. Ophiopogonin D, a steroidal glycoside abrogates STAT3 signaling cascade and exhibits anti-cancer activity by causing GSH/GSSG imbalance in lung carcinoma. Cancers 2018, 10, 427. [Google Scholar] [CrossRef]
- Shishodia, S.; Sethi, G.; Ahn, K.S.; Aggarwal, B.B. Guggulsterone inhibits tumor cell proliferation, induces S-phase arrest, and promotes apoptosis through activation of c-Jun N-terminal kinase, suppression of Akt pathway, and downregulation of antiapoptotic gene products. Biochem. Pharm. 2007, 74, 118–130. [Google Scholar] [CrossRef]
- Quirit, J.G.; Lavrenov, S.N.; Poindexter, K.; Xu, J.; Kyauk, C.; Durkin, K.A.; Aronchik, I.; Tomasiak, T.; Solomatin, Y.A.; Preobrazhenskaya, M.N. Indole-3-carbinol (I3C) analogues are potent small molecule inhibitors of NEDD4-1 ubiquitin ligase activity that disrupt proliferation of human melanoma cells. Biochem. Pharmacol. 2017, 127, 13–27. [Google Scholar] [CrossRef]
- Zheng, M.-Z.; Qin, H.-D.; Yu, X.-J.; Zhang, R.-H.; Chen, L.-Z.; Feng, Q.-S.; Zeng, Y.-X. Haplotype of gene Nedd4 binding protein 2 associated with sporadic nasopharyngeal carcinoma in the Southern Chinese population. J. Transl. Med. 2007, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, P.; Scarlett, C.; Malyukova, A.; Liu, B.; Marshall, G.; MacKenzie, K.; Biankin, A.; Liu, T. Histone deacetylase 5 blocks neuroblastoma cell differentiation by interacting with N-Myc. Oncogene 2014, 33, 2987–2994. [Google Scholar] [CrossRef]
- Spel, L.; Nieuwenhuis, J.; Haarsma, R.; Stickel, E.; Bleijerveld, O.B.; Altelaar, M.; Boelens, J.J.; Brummelkamp, T.R.; Nierkens, S.; Boes, M. Nedd4-binding protein 1 and TNFAIP3-interacting protein 1 control MHC-1 display in neuroblastoma. Cancer Res. 2018, 78, 6621–6631. [Google Scholar] [CrossRef]
- Van Arendonk, K.J.; Chung, D.H. Neuroblastoma: Tumor biology and its implications for staging and treatment. Children 2019, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Qiu, L.; Zhang, S.; Han, J. The emerging roles of E3 ubiquitin ligases in ovarian cancer chemoresistance. Cancer Drug Resist. 2021, 4, 365. [Google Scholar] [CrossRef] [PubMed]
- Ong, M.S.; Cai, W.; Yuan, Y.; Leong, H.C.; Tan, T.Z.; Mohammad, A.; You, M.L.; Arfuso, F.; Goh, B.C.; Warrier, S.; et al. ’Lnc’-ing Wnt in female reproductive cancers: Therapeutic potential of long non-coding RNAs in Wnt signalling. Br. J. Pharm. 2017, 174, 4684–4700. [Google Scholar] [CrossRef] [PubMed]
- Ben, Q.; Sun, Y.; Liu, J.; Wang, W.; Zou, D.; Yuan, Y. Nicotine promotes tumor progression and epithelial-mesenchymal transition by regulating the miR-155-5p/NDFIP1 axis in pancreatic ductal adenocarcinoma. Pancreatology 2020, 20, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, Z.; Chikhani, S.; Lui, G.Y.; Sivagurunathan, S.; Richardson, D.R. The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid. Redox Signal. 2013, 18, 874–887. [Google Scholar] [CrossRef]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Pandya, G.; Kirtonia, A.; Sethi, G.; Pandey, A.K.; Garg, M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188423. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Gholami, M.H.; Ang, H.L.; Hashemi, F.; Zarrabi, A.; Zabolian, A.; Hushmandi, K.; Delfi, M.; Khan, H.; Ashrafizadeh, M.; et al. Pre-clinical and clinical applications of small interfering RNAs (siRNA) and co-delivery systems for pancreatic cancer therapy. Cells 2021, 10, 3348. [Google Scholar] [CrossRef]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Frampton, A.E.; Castellano, L.; Colombo, T.; Giovannetti, E.; Krell, J.; Jacob, J.; Pellegrino, L.; Roca-Alonso, L.; Funel, N.; Gall, T.M. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology 2014, 146, 268–277.e18. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, N.; Aggarwal, B.; Newman, R.; Wolff, R.; Kunnumakkara, A.; Abbruzzese, J.; Hong, D.; Camacho, L.; Ng, C.; Kurzrock, R. Curcumin and pancreatic cancer: Phase II clinical trial experience. J. Clin. Oncol. 2007, 25, 4599. [Google Scholar] [CrossRef]
- Komura, K.; Sweeney, C.J.; Inamoto, T.; Ibuki, N.; Azuma, H.; Kantoff, P.W. Current treatment strategies for advanced prostate cancer. Int. J. Urol. 2018, 25, 220–231. [Google Scholar] [CrossRef]
- Heymach, J.V.; Shackleford, T.J.; Tran, H.T.; Yoo, S.-Y.; Do, K.-A.; Wergin, M.; Saintigny, P.; Vollmer, R.T.; Polascik, T.J.; Snyder, D.C. Effect of low-fat diets on plasma levels of NF-κB–regulated inflammatory cytokines and angiogenic factors in men with prostate cancerdiet-mediated modulation of angiogenic factors. Cancer Prev. Res. 2011, 4, 1590–1598. [Google Scholar] [CrossRef]
- Zhang, J.; Ahn, K.S.; Kim, C.; Shanmugam, M.K.; Siveen, K.S.; Arfuso, F.; Samym, R.P.; Deivasigamanim, A.; Lim, L.H.; Wang, L.; et al. Nimbolide-induced oxidative stress abrogates STAT3 signaling cascade and inhibits tumor growth in transgenic adenocarcinoma of mouse prostate model. Antioxid Redox Signal 2016, 24, 575–589. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Baek, S.H.; Ko, J.H.; Lee, S.G.; Yang, W.M.; Um, J.Y.; Sethi, G.; Ahn, K.S. Capsazepine inhibits JAK/STAT3 signaling, tumor growth, and cell survival in prostate cancer. Oncotarget 2017, 8, 17700–17711. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, J.-J.; Zhou, S.-J.; Chen, J.; Hu, Q.; Pu, J.-X.; Lu, J.-L. Diosgenin inhibits the expression of NEDD4 in prostate cancer cells. Am. J. Transl. Res. 2019, 11, 3461–3471. [Google Scholar]
- Sharad, S.; Ravindranath, L.; Haffner, M.C.; Li, H.; Yan, W.; Sesterhenn, I.A.; Chen, Y.; Ali, A.; Srinivasan, A.; McLeod, D.G. Methylation of the PMEPA1 gene, a negative regulator of the androgen receptor in prostate cancer. Epigenetics 2014, 9, 918–927. [Google Scholar] [CrossRef]
- Qi, H.; Grenier, J.; Fournier, A.; Labrie, C. Androgens differentially regulate the expression of NEDD4L transcripts in LNCaP human prostate cancer cells. Mol. Cell. Endocrinol. 2003, 210, 51–62. [Google Scholar] [CrossRef]
- Li, H.; Xu, L.L.; Masuda, K.; Raymundo, E.; McLeod, D.G.; Dobi, A.; Srivastava, S. A Feedback Loop between the Androgen Receptor and a NEDD4-binding Protein, PMEPA1, in Prostate Cancer Cells. J. Biol. Chem. 2008, 283, 28988–28995. [Google Scholar] [CrossRef] [PubMed]
- Sherk, A.B.; Frigo, D.E.; Schnackenberg, C.G.; Bray, J.D.; Laping, N.J.; Trizna, W.; Hammond, M.; Patterson, J.R.; Thompson, S.K.; Kazmin, D. Development of a small-molecule serum-and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res. 2008, 68, 7475–7483. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, Y.; Xu, Y.; Juan, J.; Zhang, Z.; Xu, Z.; Cao, B.; Wang, Q.; Zeng, Y.; Mao, X. The transmembrane protein TMEPAI induces myeloma cell apoptosis by promoting degradation of the c-Maf transcription factor. J. Biol. Chem. 2018, 293, 5847–5859. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Liu, H.; Liu, C. MiR-155 promotes uveal melanoma cell proliferation and invasion by regulating NDFIP1 expression. Technol. Cancer Res. Treat. 2017, 16, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.R.; Lill, N.L.; Boase, N.; Shi, P.P.; Croucher, D.R.; Shan, H.; Qu, J.; Sweezer, E.M.; Place, T.; Kirby, P.A.; et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci. Signal. 2008, 1, ra5. [Google Scholar] [CrossRef]
- Liu, Y.; Oppenheim, R.W.; Sugiura, Y.; Lin, W. Abnormal development of the neuromuscular junction in Nedd4-deficient mice. Dev Biol 2009, 330, 153–166. [Google Scholar] [CrossRef]
- Fouladkou, F.; Lu, C.; Jiang, C.; Zhou, L.; She, Y.; Walls, J.R.; Kawabe, H.; Brose, N.; Henkelman, R.M.; Huang, A.; et al. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J. Biol. Chem. 2010, 285, 6770–6780. [Google Scholar] [CrossRef]
- Kawabe, H.; Neeb, A.; Dimova, K.; Young, S.M., Jr.; Takeda, M.; Katsurabayashi, S.; Mitkovski, M.; Malakhova, O.A.; Zhang, D.E.; Umikawa, M.; et al. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron 2010, 65, 358–372. [Google Scholar] [CrossRef]
- Lu, C.; Thoeni, C.; Connor, A.; Kawabe, H.; Gallinger, S.; Rotin, D. Intestinal knockout of Nedd4 enhances growth of Apc(min) tumors. Oncogene 2016, 35, 5839–5849. [Google Scholar] [CrossRef]

| Cancer Type | Tissues | Results | Reference |
|---|---|---|---|
| Breast Cancer | BC tissues | ↑WWP1 a | [150] |
| BC tissues | ↑NEDD4, P34SEI-1 | [151] | |
| BC tissues | ↑WWP1 | [152] | |
| BC tissues | ↑NEDD4 | [54] | |
| Colorectal Cancer | CRC tissues | ↑NEDD4 | [153] |
| CRC tissues | ↑NEDD4 ↓NEDD4L | [76] | |
| CRC tissues | ↑NEDD4 | [109] | |
| Endometrial Cancer | EC tissues | ↓NEDD4L | [154] |
| EC tissues | ↑NEDD4, FoxM1 | [155] | |
| Gall bladder Cancer | Gallbladder cancer tissues | ↑NEDD4L, MMP-1, MMP-13 | [156] |
| Gastric Cancer | Gastric cancer tissues | ↑NEDD4 | [153] |
| Gastric cancer tissues | ↓NEDD4L | [157] | |
| Gastric cancer tissues | ↑NEDD4 | [126] | |
| Gastric cancer tissues | ↓NEDD4L ↑HIF-1α | [158] | |
| Glioma/Glioblastoma | Glioma tissues | ↓NEDD4L | [159] |
| Glioma tissues | ↑NEDD4 ↓CNrasGEF | [160] | |
| Glioma tissues | ↑NEDD4 | [161] | |
| Glioma tissues | ↑SMURF1 | [162] | |
| Liver Cancer | HCC tissues | ↑NEDD4, p-Akt ↓PTEN | [111] |
| HCC tissues | ↓NEDD4L | [163] | |
| Lung Adenocarcinoma | NSCLC tissues | ↑NEDD4 ↓PTEN | [97] |
| Lung adenocarcinoma tissues | ↑NEDD4, p-Akt ↓PTEN | [121] | |
| NSCLC tissues | ↓NEDD4L | [164] | |
| Melanoma | Melanoma tissues | ↑NEDD4L | [165] |
| Ovarian Cancer | Ovarian cancer tissues | ↓NEDD4L | [166] |
| Prostate Cancer | PC tissues | ↓NEDD4L | [167] |
| PC tissues | ↑NEDD4L, PSMB5, PSMC4 | [168] |
| Cancer Type | In Vitro/ In Vivo | Model/Cell Lines | NEDD4 Family | Mechanism of Action/Outcomes | Reference |
|---|---|---|---|---|---|
| Bladder Carcinoma | In vitro | RT4 cells | NEDD4 (KD) | ↓Cell proliferation, Viability, Migration, Invasion, Notch-1 ↑PTEN, Apoptosis | [105] |
| NEDD4 (OE) | ↑Cell proliferation, Invasion, Migration, Notch-1 ↓Apoptosis, PTEN | ||||
| Bone Cancer | In vitro | U2OS cells | NEDD4L (KD) | ↑OGG1 | [169] |
| Breast Cancer | In vitro | MCF-7, T47D | WWP1 (KD) | ↓Cell growth, Colony formation | [150] |
| In vitro | MDA-MB-231 | NEDD4 (KD) | ↓NEDD4 ↑MKP3 | [170] | |
| In vitro | MCF-7/T47D | WWP1 (KD) | ↑LATS1 ↓Cell number | [171] | |
| NEDD4, ITCH (KD) | ↑LATS1 | ||||
| SMURF1 (KD) | ↓LATS1 | ||||
| In vitro | MCF-7 | NEDD4 (KD) NEDD4 (KD) + NRG-1 | ↑HER3 ↑pAkt, pERK/2, Cell proliferation, Colony number, pHER | [172] | |
| In vivo | MCF-7 (shNEDD4) BALB/c- nu/nu mice xenograft | NEDD4 (KD) + NRG-1 + HER3 mAb | ↓Tumor volume | [172] | |
| In vitro | MDA-MB-231 | WWP2 (KD) | ↓WWP2, pAkt (S473), Cell proliferation, Colony number ↑Cdh1 | [173] | |
| In vitro | MDA-MB-231 (shPIPKIγi5) | NEDD4 (KD) | ↑Mig6 | [174] | |
| In vitro | MDA-MB-231, MCF-7 | NEDD4 (KD) | ↓ITG β4 ubiquitination | [175] | |
| In vitro | MCF-7 (sh-NEDD4) | NEDD4 (KD) + Estradiol | ↓ER, HER3 ↑Cell proliferation | [176] | |
| In vitro | SKBR3 | NEDD4 (OE) | ↓PIP5Kα | [113] | |
| In vitro | MCF-7, BT474 | NEDD4 (Ac) (fulvestrant) | ↓Cx43 | [177] | |
| In vitro | MDA-MB-231, T47D, BT549, ZR-75-1, MCF-7 | NEDD4 (KD) | ↓Cell growth | [54] | |
| MDA-MB-231, T47D | NEDD4 (KD) | ↓IGF-1R, p-AktSer473 ↑PTEN | |||
| In vitro | MDA-MB-231, MDA-MB-436 | NEDD4 (KD) | ↓Cell proliferation, Migration, Mammosphere formation, ALDH1A, CD44 | [178] | |
| In vitro | MDA-MB-231 (PRRG4-OE) | NEDD4 (KD) | ↑Robo1 | [124] | |
| MDA-MB-231 (PRRG4-OE), HCC1954 | NEDD4 (KD) | ↓Cell migration, Invasion | |||
| In vitro | SKBR3 | SMURF1 (KD) | ↑HER2 | [179] | |
| NEDD4 (KD) | ↑SMURF1 | ||||
| In vitro | MCF-7 | NEDD4 (In) by RF-A | ↑VDAC2, apoptosis, Ferroptosis ↓Cell viability | [180] | |
| Cervical Cancer | In vitro | HeLa cells | NEDD4 (OE) | ↓PTEN, PTEN-induced apoptosis | [181] |
| In vitro | HeLa cells | NEDD4 (KD) | ↑Beclin 1 | [119] | |
| In vitro | HeLa cells | NEDD4L (OE) | ↓ULK1 | [182] | |
| NEDD4L (KD) | ↑ULK1 | ||||
| In vitro | HeLa-Cx43 cells | NEDD4 (KD) | ↑Cx43 | [93] | |
| HeLa-CCL2 cells | NEDD4 (OE) | ↓Cx43 | |||
| C33A cells | NEDD4 (OE) | ↓Gap junction, Cx43 | |||
| Colorectal Cancer | In vitro | HCT-15, LoVo | NEDD4 (KD) | ↑Cell morphological alterations, Reorganization of the actin cytoskeleton ↓Cell growth | [102] |
| In vitro | SW1116 | NEDD4 (KD) | ↑p21, NDRG1 | [183] | |
| In vitro | LoVo | NEDD4 (OE) | ↑Cell growth, Vimentin, N-cadherin, snail, ATF-1 ↓E-cadherin, FOXA1, miR-340 | [109] | |
| NEDD4 (KD) | ↑ FOXA1 | ||||
| Caco-2 | NEDD4 (KD) | ↓Cell growth, Cell proliferation, Colony number, Vimentin, N-cadherin, Snail, ATF-1 ↑Apoptosis, Cyto C, PUMA, Apaf-1, Bax, E-cadherin, FOXA1, miR-340 | |||
| NEDD4 (OE) | ↑Cell proliferation, Colony number ↓Apoptosis, Cyto C, PUMA, Apaf-1, Bax, FOXA1 | ||||
| Endometrial Cancer | In vitro | Ishikawa | NEDD4 (OE) | ↑Cell growth, p-ERK, pAkt, IGF-1R | [155] |
| Gall bladder Cancer | In vitro | TGBC1TKB | NEDD4 (KD) | ↓Invasion, MMP-1, MMP-13 | [156] |
| Gastric Cancer | In vitro | AGS, N87 | NEDD4 (KD) | ↓Cell number, Migration, Invasion | [126] |
| In vitro | MGC803 | NEDD4 (Ac) | ↑NEDD4, Migration, Invasion | [184] | |
| Glioma/Glioblastoma | In vitro | NHA-E6/E7/hTERT | NEDD4 (KD) | ↑PTEN | [185] |
| In vitro | U251 | NEDD4 (KD) | ↓Migration, Invasion | [160] | |
| NEDD4 (OE) | ↑Migration, Invasion | ||||
| In vitro | U251, U87 | NEDD4 (KD) | ↓Migration, Invasion | [161] | |
| NEDD4 (OE) | ↑Migration, Invasion ↓Rap2a | ||||
| In vitro | A1207, SNB19 | NEDD4 (In) | ↓p-Akt, Notch-1, Migration, Invasion | [125] | |
| NEDD4 (OE) | ↓Apoptosis ↑Migration, Invasion | ||||
| NEDD4 (KD) | ↑Apoptosis ↓Migration, Invasion, Notch-1, p-Akt | ||||
| In vitro | U87-MG | SMURF1 (KD) | ↓Migration, Invasion, Vimentin, MDM2 ↑E-cadherin, p53, Cleaved Caspase-3, Cleaved PARP | [162] | |
| In vitro | U87-MG, M059K | NEDD4L (OE) | ↓Cell viability ↑p-β-catenin | [186] | |
| U87-MG | NEDD4L (OE) | ↓β-catenin, Cyclin-D1 | |||
| Liver Cancer | In vitro | HepG2 cells | NEDD4 (OE) | ↓GUCD1 | [187] |
| In vitro | Huh7 cells | NEDD4 (KD) | ↓Cell proliferation, Migration, Invasion, p-ERK1/2, p-Akt, p-STAT3 ↑Cytoskeletal changes, S phase cell cycle arrest, PTEN | [110] | |
| In vitro | Huh7, Hep3B, PLC/PRF/5, SMMC7721, LO2 | NEDD4 (KD) | ↑PTEN, E-cadherin ↓Cell proliferation, Migration, p-Akt, Vimentin | [111] | |
| In vitro | SK-hep1 HCCLM3 | NEDD4 (KD) NEDD4 (OE) | ↑Cell number, p-ERK1/2 ↓ Cleaved caspase 3 ↑BIRC3, CASP2, CASP7 | [163] | |
| In vivo | Nude mice (HCCLM3-NEDD4L) | NEDD4L (OE) | ↓Tumor weight ↑p-ERK1/2 | [163] | |
| In vitro | QGY7703, SMMC7721 | NEDD4 (KD) | ↓NEDD4, Cell proliferation, Cell viability, Migration, Invasion, p-Akt ↑Apoptosis, LATS1 | [106] | |
| NEDD4 (OE) | ↑NEDD4, Cell proliferation, Cell viability, Migration, Invasion, p-Akt ↓Apoptosis, LATS1 | ||||
| Lung Cancer | In vitro | NCI-H460 | NEDD4 (OE) | ↓PTEN ↑p-Akt, Cell growth | [97] |
| NCI-H292 | NEDD4 (KD) | ↑PTEN, p21, Gelsolin ↓p-Akt, c-Myc, Cell growth | |||
| In vivo | Nude mice (NCI-H292-shNEDD4) xenografts | NEDD4 (KD) | ↓Tumor volume, p-Akt ↑PTEN | [97] | |
| Nude mice (NCI-H460-NEDD4-HA) xenografts | NEDD4 (OE) | ↑Tumor volume, p-Akt ↓PTEN | |||
| In vitro | A549 | NEDD4 (KD) | Transport of TMEPAI to the lysosome | [188] | |
| In vitro | A549 | NEDD4 (KD) | ↑GCN2 | [189] | |
| In vitro | A549 | NEDD4L (KD) | ↑pSMAD2 | [139] | |
| In vitro | HCC827/ER cells + erlotinib | NEDD4 (KD) | ↑PTEN ↓p-Akt | [146] | |
| NEDD4 (OE) | ↓PTEN ↑p-Akt | ||||
| H1650/ER cells + erlotinib | NEDD4 (KD) + PTEN(OE) | ↑PTEN | |||
| In vivo | Nude mice (HCC827/ER cells) xenograft | NEDD4 (KD) | ↓Tumor growth, Tumor weight | [146] | |
| In vitro | A549 | NEDD4 (KD) | ↓NEDD4, EGF stimulated migration, EGF stimulated cathepsin expression | [122] | |
| NEDD4 (KD) + NEDD4 | ↑EGF stimulated migration | ||||
| In vitro | A549 | NEDD4 (KD) | ↓NEDD4, Cell proliferation, Migration, Invasion, p-Akt, NF-kB, mTOR ↑BAD, PTEN | [121] | |
| In vitro | H1975, HCC827 | NEDD4L (KD) | ↑Cell proliferation, Migration, Invasion, Cell number | [164] | |
| In vivo | Nude mice (HCC827-LUC-shNEDD4) xenografts | NEDD4L (KD) | ↑Tumor growth, Metastasis | [164] | |
| Melanoma | In vitro | Melan-A transduced SK-Mel-37 cells | NEDD4, ITCH (KD) | ↑Melan-A | [190] |
| NEDD4, ITCH (OE) | ↓Melan-A | ||||
| In vitro | G-361 + I3C | NEDD4 (KD) | ↑PTEN, MDM2 ↓p-MDM2, Bcl-2, Caspase 3 | [191] | |
| In vitro | G-361 | NEDD4L (KD) | ↓NEDD4L, Cell growth | [165] | |
| In vivo | BALB/c nude mice (A2058) xenograft | NEDD4L (OE) | ↑Tumor volume | [165] | |
| In vitro | Jurkat cells | NEDD4 (OE) | ↓GITR ↑Foxp3, IL-2 | [192] | |
| NEDD4 (KD) | ↑GITR ↓Foxp3, IL-2 | ||||
| A375 + Jurkat cells | NEDD4 (OE) | ↑Cell survival rate, IL-2, Foxp3 ↓GITR | |||
| In vitro | A375 | NEDD4L (OE) | ↓Cell proliferation, Migration, SP-1 | [193] | |
| In vitro | A375 + erastin | Wt-NEDD4 (OE) | ↓VDAC2/3 protein level ↑K48-linked ubiquitination of VDAC2/3 | [145] | |
| NEDD4 (KD) | ↑VDAC2/3 | ||||
| A375 and G361 | NEDD4 (KD) | ↑Erastin-induced cell death, ROS production, Iron accumulation, GSH depletion, GSSG generation | |||
| NEDD4 (OE) | ↑Cell viability | ||||
| In vivo | Nude mice (A375) xenografts | NEDD4 (KD) | ↓Tumor size, GSH levels ↑MDA levels | [145] | |
| In vitro | A375 | NEDD4 (KD) | ↓NEDD4 ↑IGPR-1 | [194] | |
| SK-MEL-28 | NEDD4 (OE) | ↓IGPR-1 | |||
| Nasopharyngeal Carcinoma | In vitro | CNE1-DDP, CNE2-DDP | NEDD4 (KD) | ↑MET, E-cadherin, ZO-1 ↓Cell attachment and detachment capacity, Migration, Invasion, N-cadherin, Vimentin, Slug | [137] |
| Neuroblastoma | In vitro | BE (2)-C | NEDD4 (KD) | ↑N-Myc, c-Myc protein | [103] |
| In vitro | SK-N-SH | NEDD4 (KD) | ↓NEDD4 mRNA, JEV infection ↑LC3-II, Beclin 1 | [195] | |
| NEDD4 (OE) | ↑JEV NS3 expression | ||||
| In vitro | SK-N-SH | NEDD4 (OE) | ↓Cell proliferation, Migration | [196] | |
| NEDD4 (OE)+ hsa-miR199a-3p mimic | ↑Cell proliferation, Migration | ||||
| Ovarian Cancer | In vitro | NEDD4L, DDB2 (KD) | ↑p-Smad2 | [197] | |
| CP70 | NEDD4L, DDB2 (OE)+ TGF-β | ↑p-Smad2 | |||
| In vitro | NEDD4 (KD) | ↓NEDD4 | [198] | ||
| NEDD4 (KD) + RSL3 | ↑Cell death, MDA, Oxidative stress, LTF protein expression | ||||
| NEDD4 (KD) + Erastin | ↑Cell death, MDA, Oxidative stress, LTF protein expression | ||||
| Pancreatic Cancer | In vitro | MiaPaca-2 | NEDD4 (KD) | ↓NEDD4 ↑N-Myc, c-Myc protein, SIRT2 | [103] |
| In vitro | Patu8988, Panc-1 | NEDD4 (OE) + Curcumin | ↑Cell proliferation, Migration, Invasion ↓Apoptosis, PTEN, p73 | [120] | |
| NEDD4 (KD) + Curcumin | ↓Cell growth, Migration, Invasion ↑Apoptosis, PTEN, p73 | ||||
| In vitro | Panc-1 | NEDD4 (KD) | ↓NEDD4 | [198] | |
| NEDD4 (KD) + RSL3 | ↑Cell death, MDA, Oxidative stress, LTF protein expression | ||||
| NEDD4 (KD) + Erastin | ↑Cell death, MDA, Oxidative stress, LTF protein expression | ||||
| Prostate Cancer | In vitro | DU145 | WWP2 (KD) | ↑PTEN ↓p-Akt, Cell proliferation | [199] |
| WWP2 (KD) + Doxorubicin | ↑Apoptosis | ||||
| In vivo | Nude mice (DU145-shWWP2) xenograft | WWP2 (KD) | ↓Tumor growth | [199] | |
| In vitro | DU145 | NEDD4 (KD) + NRG-1 | ↑Cell proliferation, Migration, p-HER3, p-Akt1, Spheroid formations, HER3 | [172] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayaprakash, S.; Hegde, M.; BharathwajChetty, B.; Girisa, S.; Alqahtani, M.S.; Abbas, M.; Sethi, G.; Kunnumakkara, A.B. Unraveling the Potential Role of NEDD4-like E3 Ligases in Cancer. Int. J. Mol. Sci. 2022, 23, 12380. https://doi.org/10.3390/ijms232012380
Jayaprakash S, Hegde M, BharathwajChetty B, Girisa S, Alqahtani MS, Abbas M, Sethi G, Kunnumakkara AB. Unraveling the Potential Role of NEDD4-like E3 Ligases in Cancer. International Journal of Molecular Sciences. 2022; 23(20):12380. https://doi.org/10.3390/ijms232012380
Chicago/Turabian StyleJayaprakash, Sujitha, Mangala Hegde, Bandari BharathwajChetty, Sosmitha Girisa, Mohammed S. Alqahtani, Mohamed Abbas, Gautam Sethi, and Ajaikumar B. Kunnumakkara. 2022. "Unraveling the Potential Role of NEDD4-like E3 Ligases in Cancer" International Journal of Molecular Sciences 23, no. 20: 12380. https://doi.org/10.3390/ijms232012380
APA StyleJayaprakash, S., Hegde, M., BharathwajChetty, B., Girisa, S., Alqahtani, M. S., Abbas, M., Sethi, G., & Kunnumakkara, A. B. (2022). Unraveling the Potential Role of NEDD4-like E3 Ligases in Cancer. International Journal of Molecular Sciences, 23(20), 12380. https://doi.org/10.3390/ijms232012380






