Discovery of Simple Diacylhydrazine-Functionalized Cinnamic Acid Derivatives as Potential Microtubule Stabilizers
Abstract
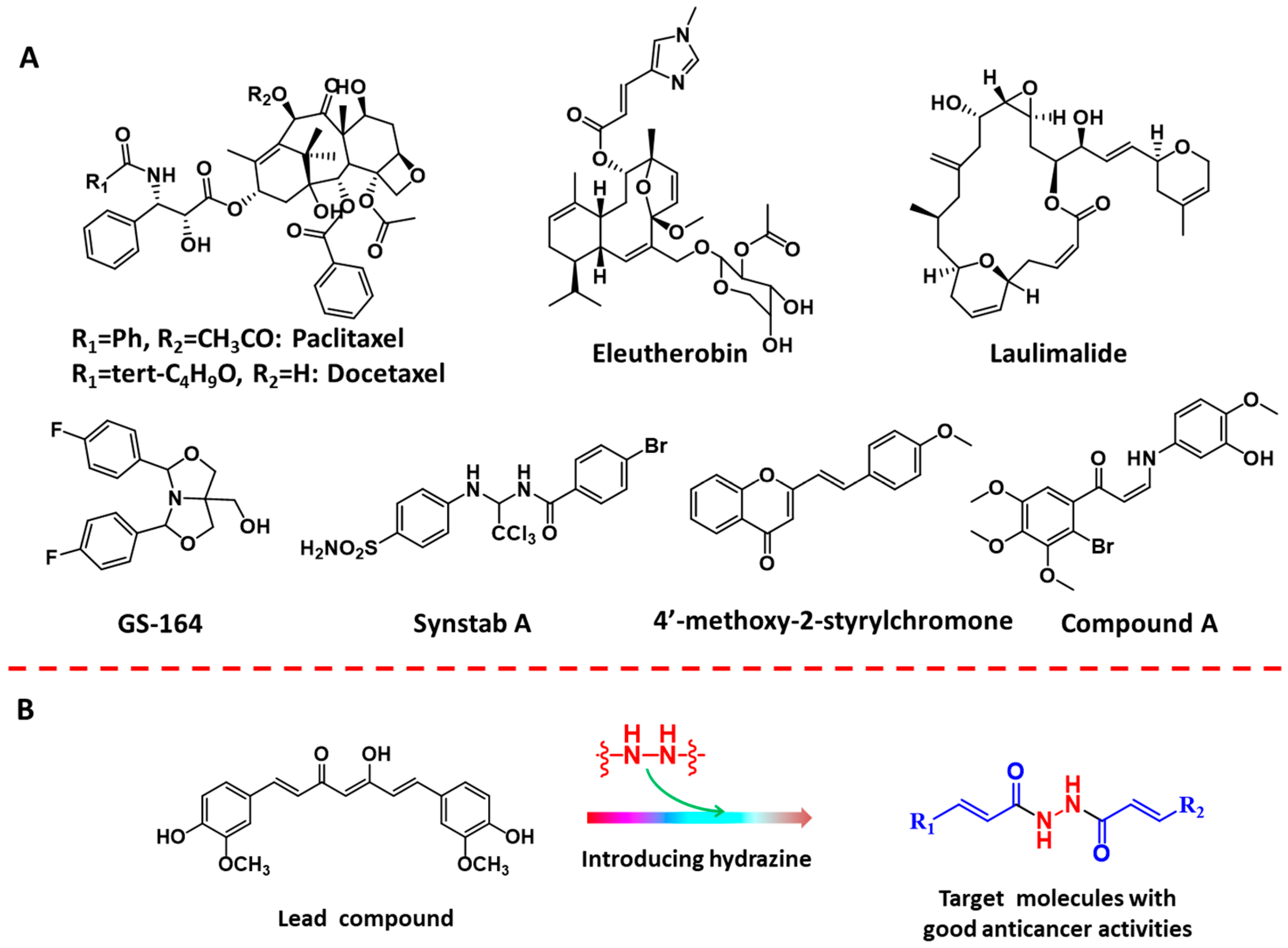
1. Introduction
2. Results
2.1. Chemistry
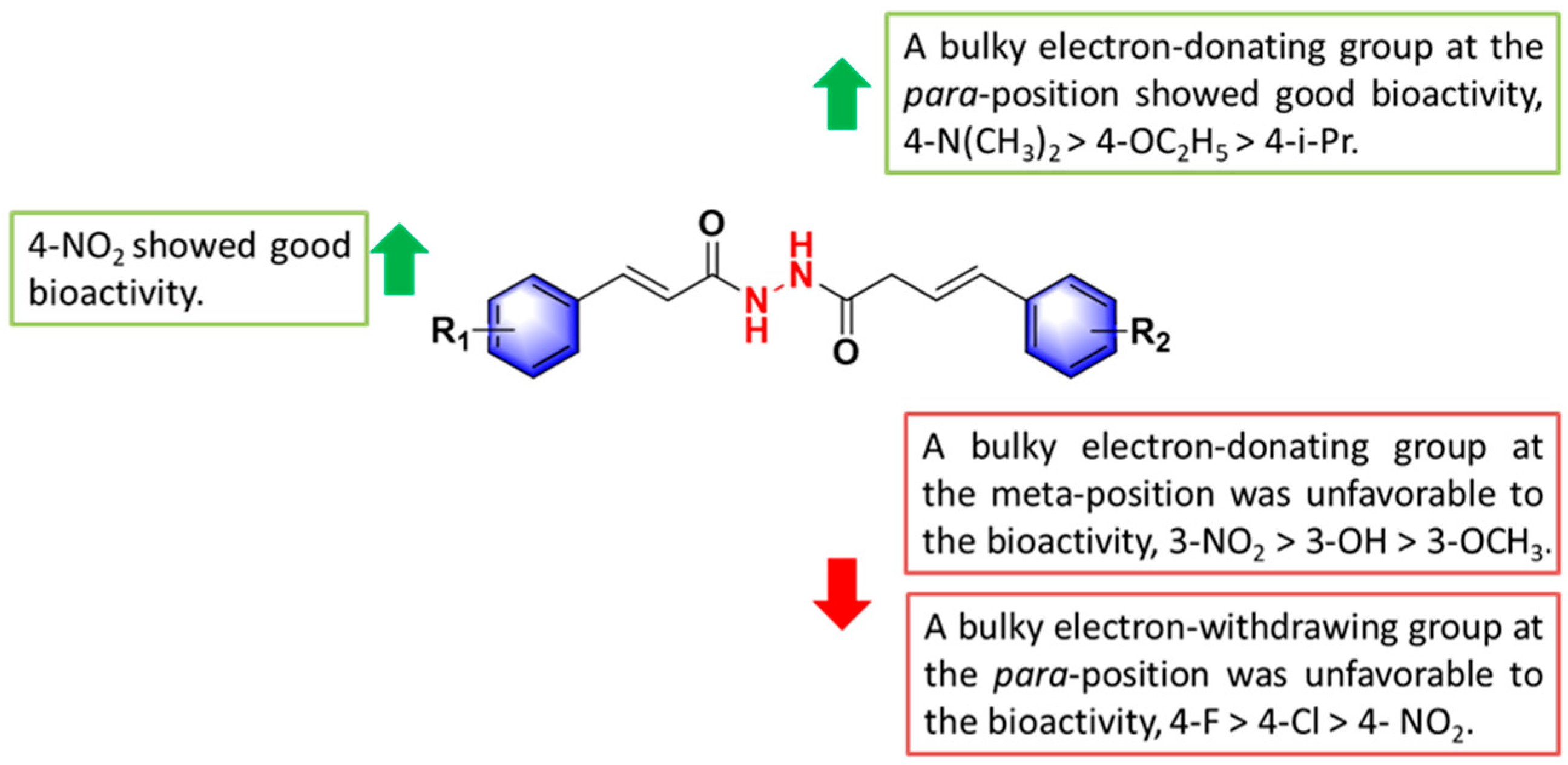
2.2. The Antiproliferative Activity of Title Compounds
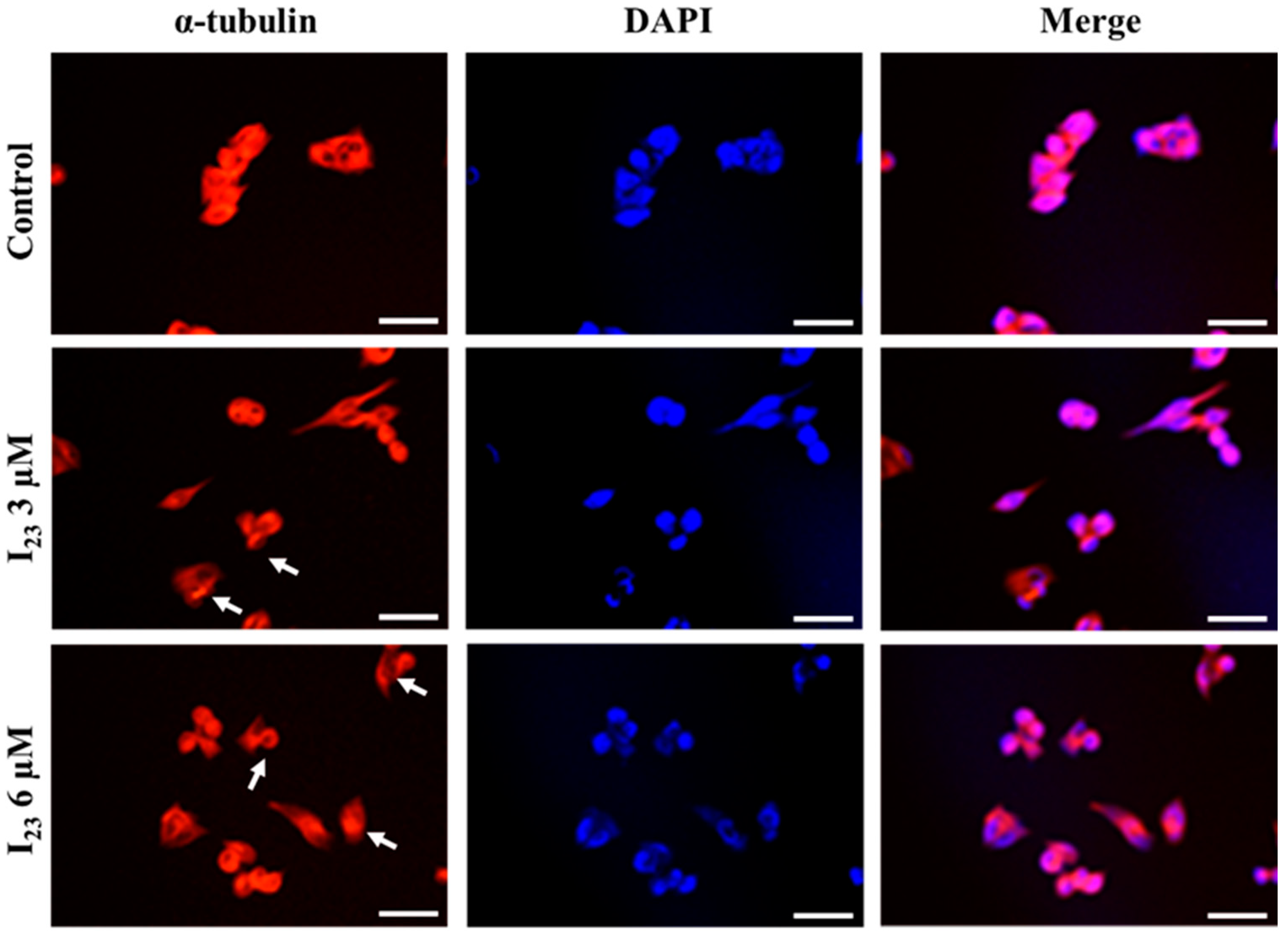
2.3. Immunofluorescence Staining of Tubulin
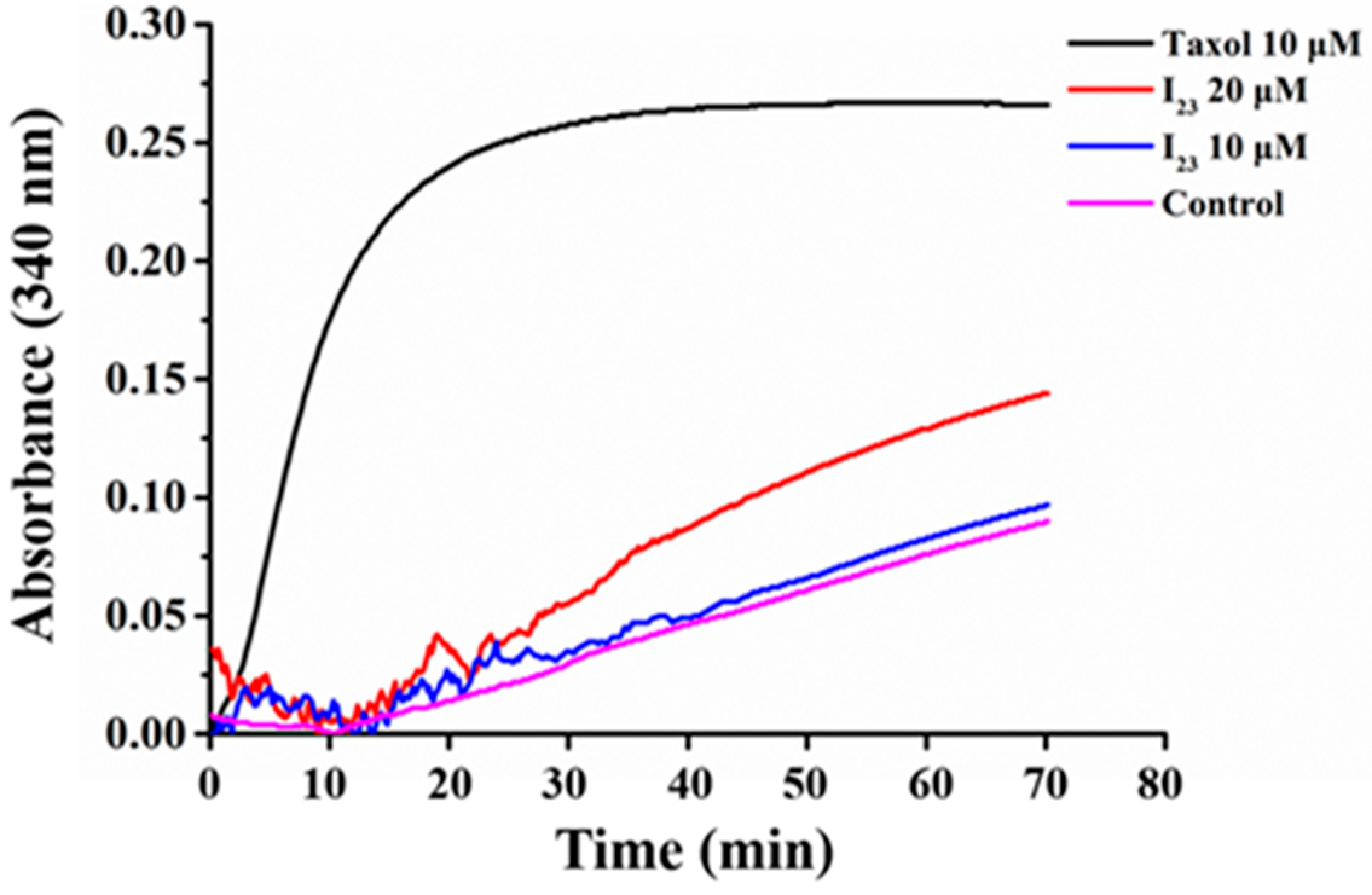
2.4. Effects of Compound I23 on Tubulin Polymerization
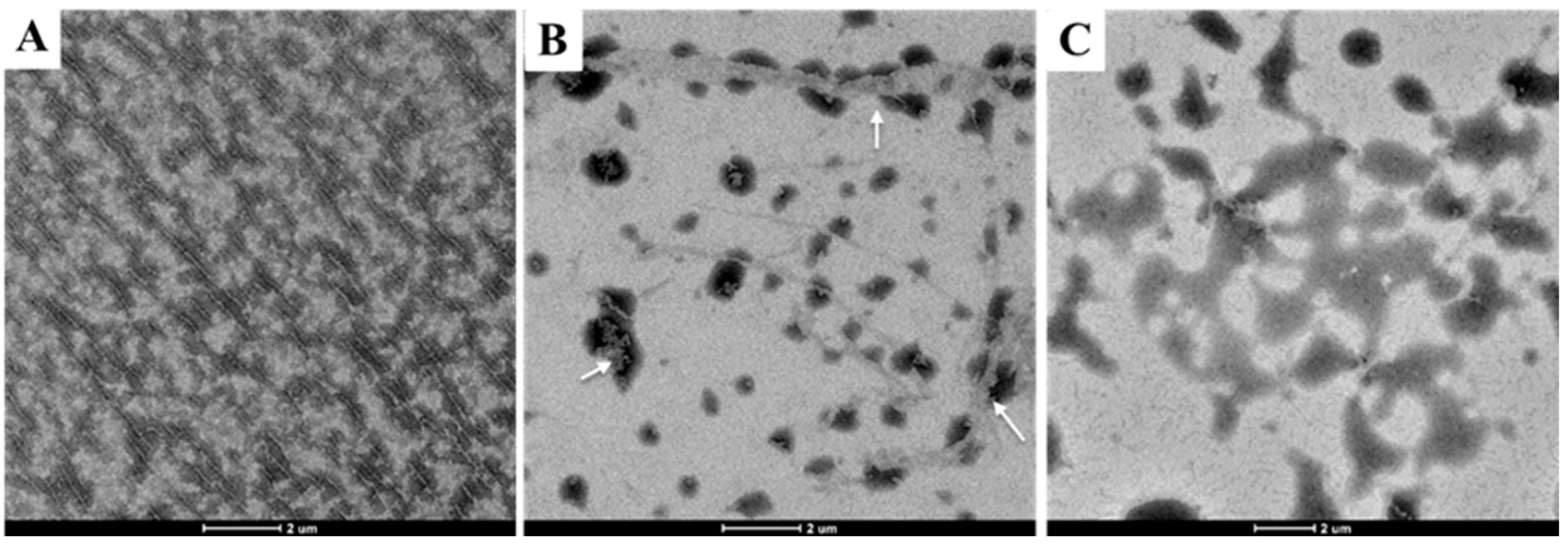
2.5. Tubulin Polymerization Affected by Compound I23 via TEM
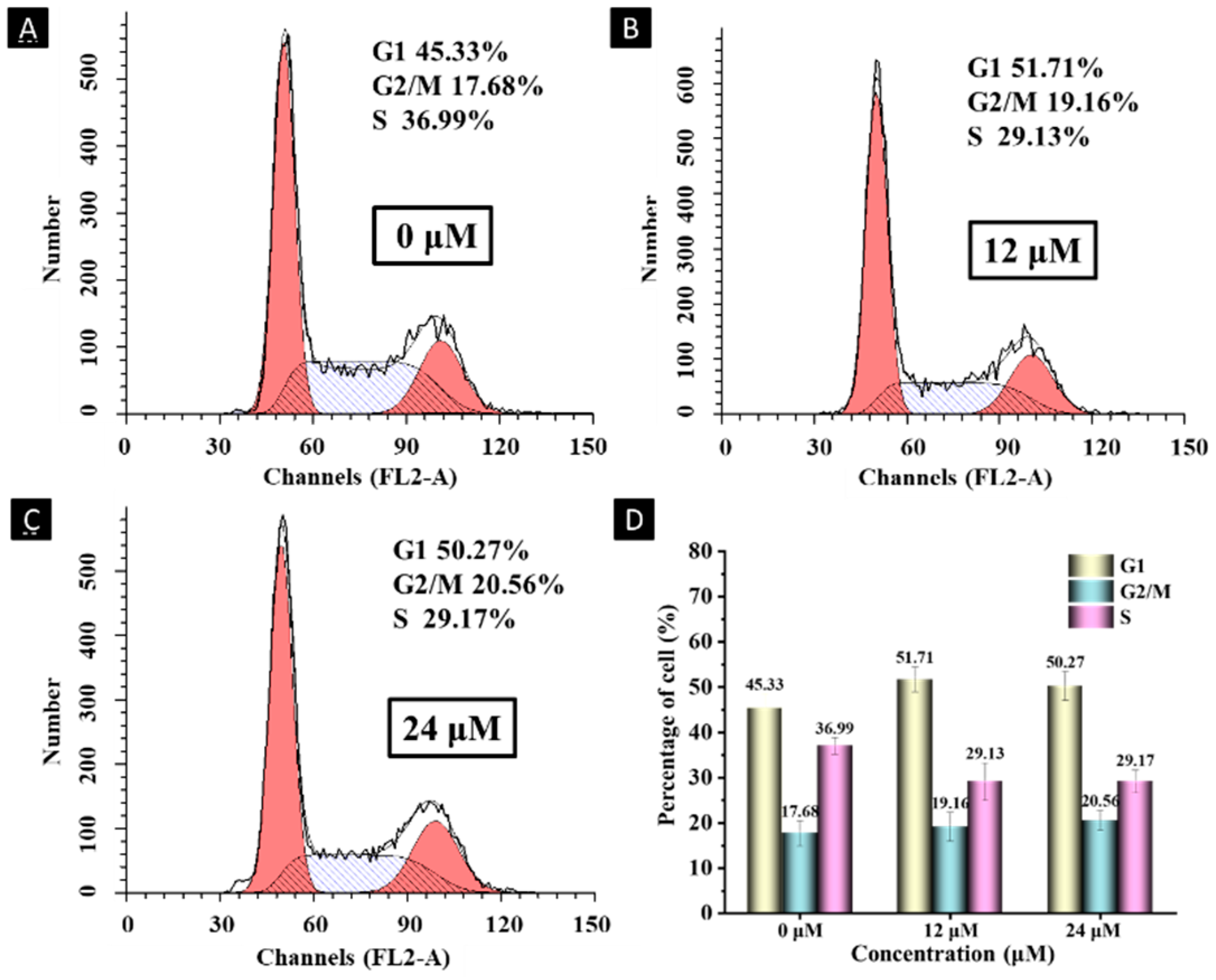
2.6. Cell Cycle Analysis
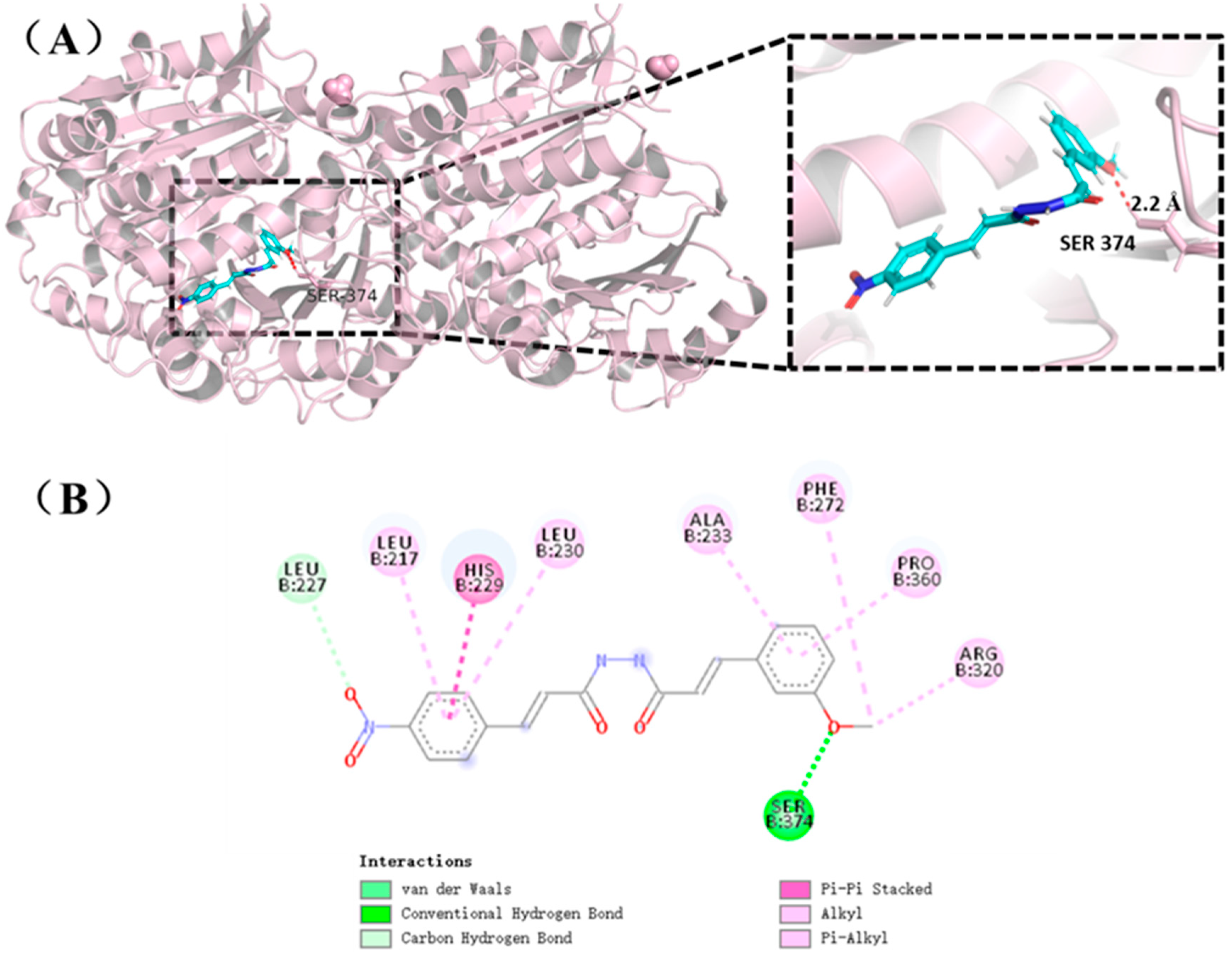
2.7. Molecular Docking of Compound I23 with Tubulin
2.8. Effects on Cell Migration of Compound I23
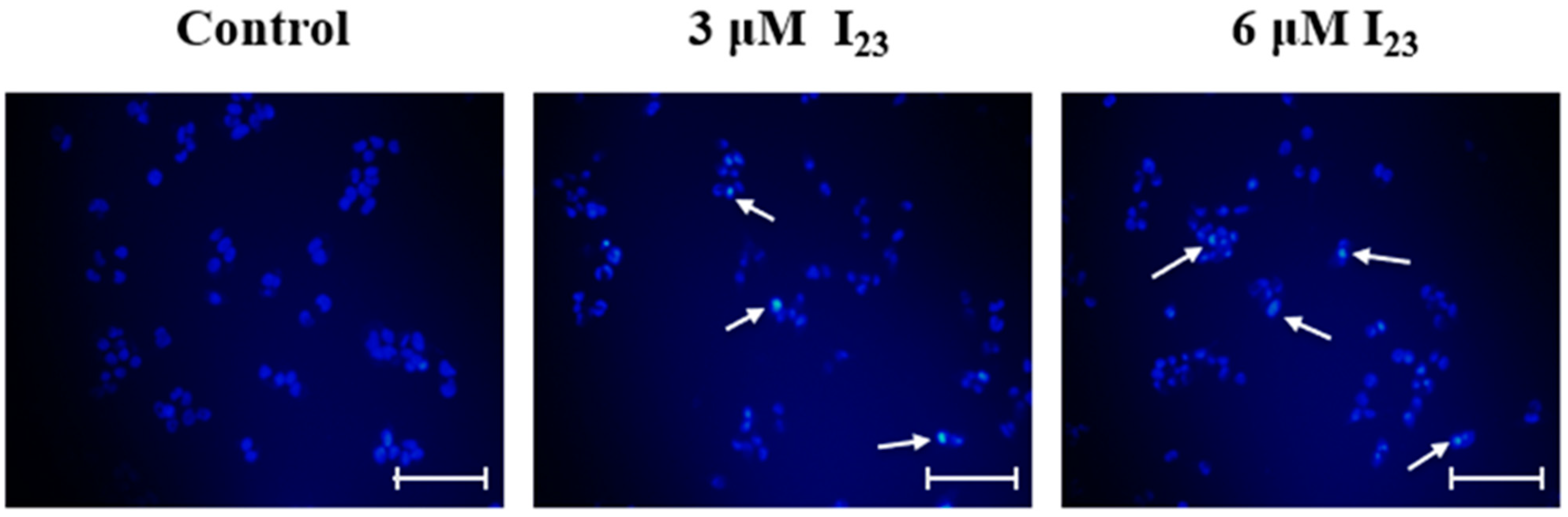
2.9. Apoptosis Effects of HepG2 Cells Caused by Compound I23
2.10. In Silico Drug-likeness Evaluation
3. Experimental Section
3.1. Instruments and Chemicals
3.2. Pharmacology
3.2.1. Cell Culture
3.2.2. MTT Assay
3.2.3. Immunofluorescence Staining Pattern
3.2.4. Tubulin Polymerization Assay In Vitro
3.2.5. Tubulin Polymerization Affected by Target Compounds via TEM
3.2.6. Cell Cycle Analysis
3.2.7. Computational Docking Studies
3.2.8. Scratch Test
3.2.9. Hoechst Apoptosis Experiment
3.2.10. In Silico Pharmacokinetics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Virag, K.; Nyari, T.A. Seasonal variation of cancer mortality in Hungary between 1984 and 2013. Scand J. Public Health 2019, 3, 746–757. [Google Scholar] [CrossRef]
- Nagai, H.; Kim, Y.H. Cancer prevention from the perspective of global cancer burden patterns. J. Thorac. Dis. 2017, 9, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2017, 68, 394–424. [Google Scholar] [CrossRef]
- Fu, B.W.; Wang, N.; Tan, H.Y.; Li, S.; Cheung, F.; Feng, Y.B. Multi-component herbal products in the prevention and treatment of chemotherapy-associated toxicity and side effects: A review on experimental and clinical evidences. Front. Pharmacol. 2018, 9, 1394. [Google Scholar] [CrossRef] [PubMed]
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment. Int. J Oncol. 2019, 54, 407–419. [Google Scholar]
- Seligmann, J.; Twelves, C. Tubulin: An example of targeted chemotherapy. Future Med. Chem. 2013, 5, 339–352. [Google Scholar] [CrossRef]
- Tagliamento, M.; Genova, C.; Rossi, G.; Coco, S.; Rijavec, E.; Dal Bello, M.G.; Boccardo, S.; Grossi, F.; Alama, A. Microtubule-targeting agents in the treatment of non-small cell lung cancer: Insights on new combination strategies and investigational compounds. Expert Opin. Inv. Drugs 2019, 28, 513–523. [Google Scholar] [CrossRef]
- Ferlini, C.; Raspaglio, G.; Cicchillitti, L.; Mozzetti, S.; Prislei, S.; Bartollino, S.; Scambia, G. Looking at drug resistance mechanisms for microtubule interacting drugs: Does TUBB3 work? Curr. Cancer Drug Targets 2007, 7, 704–712. [Google Scholar] [CrossRef]
- Jordan, M.A.; Wilson, L. Microtubules as a target for antiproliferative drugs. Nat. Rev. Cancer 2014, 4, 253–265. [Google Scholar] [CrossRef]
- Kavallaris, M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer 2014, 10, 194–204. [Google Scholar] [CrossRef]
- Čermák, V.; Dostál, V.; Jelínek, M.; Libusová, L.; Kovář, J.; Rösel, D.; Brábek, J. Microtubule-targeting agents and their impact on cancer treatment. Eur. J. Cell Biol. 2020, 99, 151075. [Google Scholar] [CrossRef]
- Chen, S.M.; Meng, L.H.; Ding, J. New microtubule-inhibiting antiproliferative agents. Expert Opin. Inv. Drug 2010, 19, 329–343. [Google Scholar] [CrossRef]
- Zhao, Y.; Fang, W.; Pors, K. Microtubule stabilising agents for cancer chemotherapy. Expert Opin. Ther. Pat. 2009, 19, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Haider, K.; Rahaman, S.; Yar, M.S.; Kamal, A. Tubulin inhibitors as novel antiproliferative agents: An overview on patents (2013–2018). Expert Opin. Ther. Pat. 2019, 29, 623–641. [Google Scholar] [CrossRef]
- Barreca, M.; Stathis, A.; Barraja, P.; Bertoni, F. An overview on anti-tubulin agents for the treatment of lymphoma patients. Pharmacol. Therapeut. 2020, 211, 107552. [Google Scholar] [CrossRef] [PubMed]
- Tangutur, A.D.; Kumar, D.; Krishna, K.V.; Krishna, S. Microtubule targeting agents as cancer chemotherapeutics: An overview of molecular hybrids as stabilizing and destabilizing agents. Curr. Top. Med. Chem. 2017, 17, 2523–2537. [Google Scholar] [CrossRef] [PubMed]
- Zefirova, O.N.; Diikov, A.G.; Zyk, N.V.; Zefirov, N.S. Ligands of the colchicine site of tubulin: A common pharmacophore and new structural classes. Russ. Chem. B 2007, 56, 680–688. [Google Scholar] [CrossRef]
- Borisy, G.G.; Heald, R.; Howard, J.; Janke, C.; Musacchion, A.; Nogals, E. Microtubules: 50 years on from the discovery of tubulin. Nat. Rev. Mol. Cell Biol. 2016, 17, 322–328. [Google Scholar] [CrossRef]
- Li, W. Drugs Targeting Tubulin Polymerization. Pharm. Res.-Dordr. 2012, 29, 2939–2942. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.J.; Xiao, M.; Li, W.; Miller, D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res.-Dordr. 2012, 29, 2943–2971. [Google Scholar] [CrossRef]
- Naaz, F.; Haider, M.R.; Shafi, S.; Yar, M.S. Anti-tubulin agents of natural origin: Targeting taxol, vinca, and colchicine binding domains. Eur. J. Med. Chem. 2019, 171, 310–331. [Google Scholar] [CrossRef]
- Cao, Y.N.; Zheng, L.L.; Wang, D.; Liang, X.X.; Gao, F.; Zhou, X.L. Recent advances in microtubule-stabilizing agents. Eur. J. Med. Chem. 2018, 143, 806–828. [Google Scholar] [CrossRef]
- Zhao, Y.; Mu, X.; Du, G.H. Microtubule-stabilizing agents: New drug discovery and cancer therapy. Pharmacol. Therapeut. 2016, 162, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, D.S. Overview of paclitaxel (Taxol) in advanced lung cancer. Semin. Oncol. 1993, 20, 46–49. [Google Scholar] [PubMed]
- Ojima, I.; Slater, J.C.; Michaud, E.; Kuduk, S.D.; Bounaud, P.Y.; Vrignaud, P.; Vrignaud, P.; Bissery, M.; Veith, J.M.; Pera, P.; et al. Syntheses and structure−activity relationships of the second-generation antiproliferative taxoids: Exceptional activity against drug-resistant cancer cells. J. Med. Chem. 1996, 39, 3889–3896. [Google Scholar] [CrossRef]
- Mooberry, S.L.; Tien, G.; Hernandez, A.H.; Plubrukarn, A.; Davidson, B.S. Laulimalide and isolaulimalide, new paclitaxel-like microtubule-stabilizing agents. Cancer Res. 1999, 59, 653–660. [Google Scholar]
- Shintani, Y.; Tanaka, T.; Nozaki, Y. GS-164, a small synthetic compound, stimulates tubulin polymerization by a similar mechanism to that of Taxol. Cancer Chemother. Pharmacol. 1997, 40, 513–520. [Google Scholar] [CrossRef]
- Haggarty, S.J.; Mayer, T.U.; Miyamoto, D.T.; Fathi, R.; King, R.W.; Mitchison, T.J.; Schreiber, S.L. Dissecting cellular processes using small molecules: Identification of colchicine-like, taxol-like and other small molecules that perturb mitosis. Chem. Biol. 2000, 7, 275–286. [Google Scholar] [CrossRef]
- Marinho, J.; Pedro, M.; Pinto, D.C.G.A.; Silva, A.M.S.; Cavaleiro, J.A.S.; Sunkel, C.E.; Nascimento, M.S.J. 4′-Methoxy-2-styrylchromone a novel microtubule-stabilizing antimitotic agent. Biochem. Pharmacol. 2008, 75, 826–835. [Google Scholar] [CrossRef]
- Reddy, M.V.R.; Akula, B.; Cosenza, S.C.; Lee, C.M.; Mallireddigari, M.R.; Pallela, V.R.; Subbaiah, D.R.C.V.; Udofa, A.; Reddy, E.P. (Z)-1-Aryl-3-arylamino-2-propen-1-ones, highly active stimulators of tubulin polymerization: Synthesis, structure-activity relationship (SAR), tubulin polymerization, and cell growth inhibition studies. J. Med. Chem. 2012, 55, 5174–5187. [Google Scholar] [CrossRef]
- Chakraborti, S.; Das, L.; Kapoor, N.; Das, A.; Dwivedi, V.; Poddar, A.; Chakraborti, G.; Janik, M.E.; Basu, G.; Panda, D.; et al. Curcumin recognizes a unique binding site of tubulin. J. Med. Chem. 2011, 54, 6183–6196. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.K.; Bharne, S.S.; Rathinasamy, K.; Naik, N.R.; Panda, D. Dietary antioxidant curcumin inhibits microtubule assembly through tubulin binding. FEBS J. 2006, 273, 5320–5332. [Google Scholar] [CrossRef]
- Luo, Y.; Qiu, K.M.; Lu, X.; Liu, K.; Fu, J.; Zhu, H.L. Synthesis, biological evaluation, and molecular modeling of cinnamic acyl sulfonamide derivatives as novel antitubulin agents. Bioorg. Med. Chem. 2011, 19, 4730–4738. [Google Scholar] [CrossRef]
- Sinigersky, V.; Wegener, G.; Schopoy, I. Synthesis and properties of a poly(phenylenevinylene) containing 1, 3, 4-oxadiazole rings. Eur. Polym. J. 1993, 29, 617–620. [Google Scholar]
- Zhang, X.N.; Breslav, M.; Grimm, J.; Guan, K.L.; Huang, A.H.; Liu, F.Q.; Maryanoff, C.A.; Palmer, D.; Patel, M.; Qian, Y.; et al. A new procedure for preparation of carboxylic acid hydrazides. J. Org. Chem. 2002, 67, 9471–9474. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Kim, K.H.; Kim, N.D.; Lee, K.Y.; Han, C.K.; Yoon, J.H.; Moon, S.K.; Lee, S.S.; Seong, B.L. Design and biological evaluation of novel tubulin inhibitors as antimitotic agents using a pharmacophore binding model with tubulin. J. Med. Chem. 2006, 49, 5664–5670. [Google Scholar] [CrossRef]
- Greene, T.F.; Wang, S.; Greene, L.M.; Nathwani, S.M.; Pollock, J.K.; Malebari, A.M.; McCabe, T.; Twamly, B.; O’Boyle, N.M.; Zisterer, D.M.; et al. Synthesis and biochemical evaluation of 3-phenoxy-1, 4-diarylazetidin-2-ones as tubulin-targeting antiproliferative agents. J. Med. Chem. 2016, 59, 90–113. [Google Scholar] [CrossRef] [PubMed]
- Chopra, A.; Anderson, A.; Giardina, C. Novel piperazine-based compounds inhibit microtubule dynamics and sensitize colon cancer cells to tumor necrosis factor-induced apoptosis. J. Biol. Chem. 2014, 289, 2978–2991. [Google Scholar] [CrossRef] [PubMed]
- Risinger, A.L.; Li, J.; Bennett, M.J.; Rohena, C.C.; Peng, J.; Schriemer, D.C.; Mooberry, S.L. Taccalonolide Binding to Tubulin Imparts Microtubule Stability and Potent In Vivo Activity. Cancer Res. 2013, 73, 6780–6792. [Google Scholar] [CrossRef]
- Cao, D.; Han, X.L.; Wang, G.C.; Yang, Z.; Peng, F.; Ma, L.; Zhang, R.H.; Ye, H.Y.; Tang, M.H.; Wu, W.S.; et al. Synthesis and biological evaluation of novel pyranochalcone derivatives as a new class of microtubule stabilizing agents. Eur. J. Med. Chem. 2013, 62, 579–589. [Google Scholar] [CrossRef]
- Lu, Y.X.; Wang, Y.; Zhu, W.L. Nonbonding interactions of organic halogens in biological systems: Implications for drug discovery and biomolecular design. Phys. Chem. Chem. Phys. 2010, 12, 4543–4551. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A.J.; Yang, W.T. Revealing Noncovalent Interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef] [PubMed]
- Mahadevi, A.S.; Sastry, G.N. Cooperativity in Noncovalent Interactions. Chem. Rev 2016, 116, 2775–2825. [Google Scholar] [CrossRef] [PubMed]
- Nabeshima, K.; Inoue, T.; Shimao, Y.; Sameshima, T. Matrix metalloproteinases in tumor invasion: Role for cell migration. Pathol. Int. 2002, 52, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, A.; Yang, H.L.; Sharma, R.; Patel, K.D.; Cabral, F. The role of microtubules and their dynamics in cell migration. J. Biol. Chem. 2012, 287, 43359–43369. [Google Scholar] [CrossRef]
- Wang, G.; Liu, W.; Gong, Z.; Huang, Y.; Li, Y.; Peng, Z. Design, synthesis, biological evaluation and molecular docking studies of new chalcone derivatives containing diaryl ether moiety as potential anticancer agents and tubulin polymerization inhibitors. Bioorg. Chem. 2020, 95, 103565. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Yin, Y.; Shuai, W.; Xu, F.J.; Yao, H.; Liu, J.; Cheng, K.G.; Xu, J.Y.; Zhu, Z.Y.; Xu, S.T. Discovery of novel quinazolines as potential anti-tubulin agents occupying three zones of colchicine domain. Bioorg. Chem. 2019, 83, 380–390. [Google Scholar] [CrossRef]
- Le, Y.; Gan, Y.Y.; Fu, Y.H.; Liu, J.M.; Li, W.; Zou, W.; Zhou, Z.X.; Wang, Z.C.; Ouyang, G.P. Design, synthesis and in vitro biological evaluation of quinazolinone derivatives as EGFR inhibitors for antiproliferative treatment. J. Enzym. Inhib. Med. Chem. 2020, 35, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Liang, Y.; Zhou, P.F.; Cheng, J.Y.; Ding, K.L.; Wang, Y. Design, synthesis, antiproliferative activities and biological studies of novel diaryl substituted fused heterocycles as dual ligands targeting tubulin and katanin. Eur. J. Med. Chem. 2019, 178, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Lian, B.P.; Xia, Y.Z.; Shao, Y.Y.; Kong, L.Y. Design, synthesis and biological evaluation of resveratrol-cinnamoyl derivates as tubulin polymerization inhibitors targeting the colchicine binding site. Bioorg. Chem. 2019, 93, 103319. [Google Scholar] [CrossRef]
- Bai, H.H.; Jin, H.; Yang, F.; Zhu, H.Y.; Cai, J.Y. Apigenin induced MCF-7 cell apoptosis-associated reactive oxygen species. Scanning 2014, 36, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Zhao, H.B.; Di, Y.C.; Li, Q.; Shao, D.Y.; Shi, J.L.; Huang, Q.S. Antiproliferative activity of Pinoresinol in vitro: Inducing apoptosis and inhibiting HepG2 invasion. J. Funct. Foods 2018, 45, 206–214. [Google Scholar] [CrossRef]
- Tong, L.Y.; Sun, W.C.; Wu, S.Y.; Han, Y. Characterization of Caerulomycin A as a dual-targeting anticancer agent. Eur. J. Pharmacol. 2022, 922, 174914. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.M.; Ma, X.; Guo, K.R.; Li, J.; Zhang, C.; Wu, L.Q. Dual-functional antitumor conjugates improving the anti-metastasis effect of combretastatin A4 by targeting tubulin polymerization and matrix metalloproteinases. Eur. J. Med. Chem. 2022, 238, 114439. [Google Scholar] [CrossRef] [PubMed]
- Can, S.N.; Lacey, S.; Gur, M.; Carter, A.P.; Yildiz, A. Directionality of dynein is controlled by the angle and length of its stalk. Nature 2019, 566, 407–410. [Google Scholar] [CrossRef]
- Song, Y.L.; Liu, S.S.; Yang, J.; Xie, J.; Zhou, X.; Wu, Z.B.; Liu, L.W.; Wang, P.Y.; Yang, S. Discovery of Epipodophyllotoxin-Derived B2 as Promising XooFtsZ Inhibitor for Controlling Bacterial Cell Division: Structure-Based Virtual Screening, Synthesis, and SAR Study. Int. J. Mol. Sci. 2022, 23, 9119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, H.J.; Gao, X.H.; Feng, Y.M.; Shao, W.B.; Qi, P.Y.; Wu, Z.B.; Liu, L.W.; Wang, P.Y.; Yang, S. The discovery of natural 4′-demethylepipodophyllotoxin from renewable Dysosma versipellis species as a novel bacterial cell division inhibitor for controlling intractable diseases in rice. Ind. Crop Prod. 2021, 174, 114182. [Google Scholar] [CrossRef]
- Hu, X.; Li, L.; Zhang, Q.S.; Wang, Q.Q.; Feng, Z.Z.; Xu, Y.; Xia, Y.; Yu, L.T. Design, synthesis and biological evaluation of a novel tubulin inhibitor SKLB0565 targeting the colchicine binding site. Bioorg. Chem. 2020, 97, 103695. [Google Scholar] [CrossRef] [PubMed]
- Sayeed, I.B.; Vishnuvardhan, M.V.P.S.; Nagarajan, A.; Kantevari, S.; Kamal, A. Imidazopyridine linked triazoles as tubulin inhibitors, effectively triggering apoptosis in lung cancer cell line. Bioorg. Chem. 2018, 80, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.F.; Nayak, V.L.; Budaganaboyina, P.; Mullagiri, K.; Sunkari, S.; Gour, J.; Kamal, A. Synthesis and biological evaluation of imidazo [2,1-b]thiazole-benzimidazole conjugates as microtubule-targeting agents. Bioorg. Chem. 2018, 77, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, J.M.; Meng, J.; Fu, Y.H.; Wu, Z.B.; Ouyang, G.P.; Wang., Z.C. Discovery of facile amides-functionalized rhodanine-3-acetic acid derivatives as potential anticancer agents by disrupting microtubule dynamics. J. Enzym. Inhib. Med. Chem. 2021, 36, 1996–2009. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.L.; Wu, Z.X.; Yi, J.C.; Fu, L.; Yang, Z.J.; Hsieh, C.; Yin, M.Z.; Zeng, X.X.; Wu, C.K.; Lu, A.P.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Zhou, X.; Feng, Y.M.; Qi, P.Y.; Shao, W.B.; Wu, Z.B.; Liu, L.W.; Wang, Y.; Ma, H.D.; Wang, P.Y.; Yang, S. Synthesis and docking study of N-(Cinnamoyl)-N′-(substituted)acryloyl hydrazide derivatives containing pyridinium moieties as a novel class of filamentous temperature-sensitive protein Z inhibitors against the intractable Xanthomonas oryzae pv. oryzae infections in rice. J. Agric. Food Chem. 2020, 68, 8132–8142. [Google Scholar] [CrossRef] [PubMed]
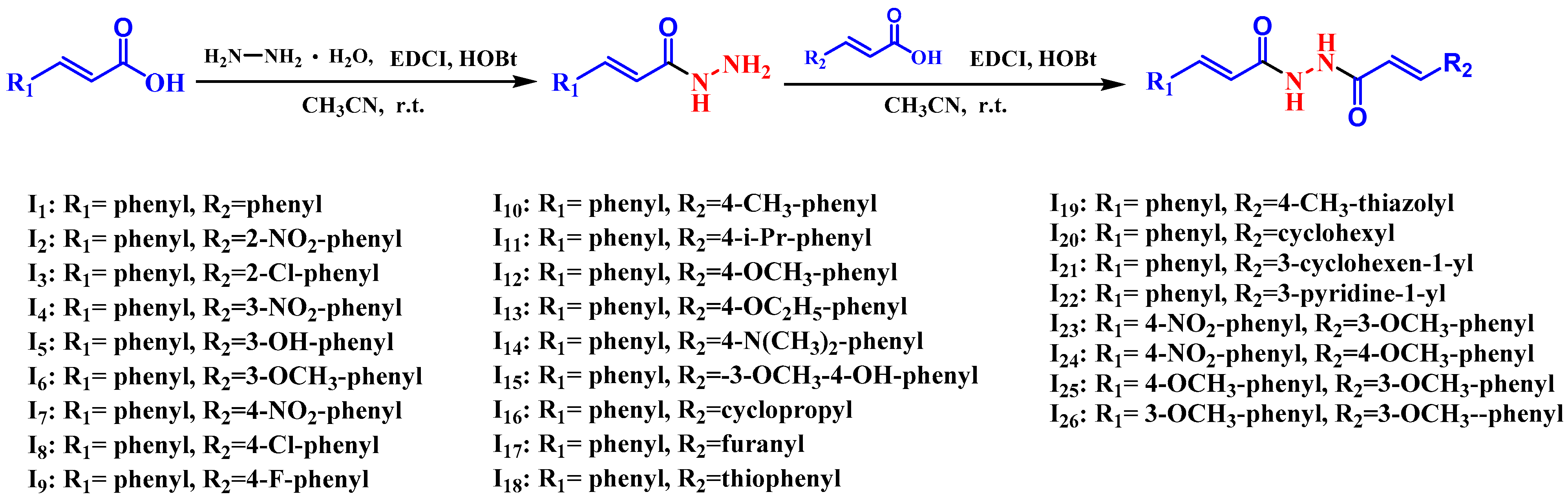



| No. |  | IC50 (μM) b | |||
|---|---|---|---|---|---|
| R1 | R2 | A549 | PC-3 | HepG2 | |
| I1 | -phenyl | -phenyl | 26.7 ± 0.43 | 5.95 ± 1.52 | 8.17 ± 0.46 |
| I2 | -phenyl | 2-NO2-phenyl | 7.55 ± 1.22 | 8.08 ± 2.24 | 9.33 ± 0.95 |
| I3 | -phenyl | 2-Cl-phenyl | 6.48 ± 0.41 | 7.38 ± 0.24 | 9.53 ± 1.74 |
| I4 | -phenyl | 3-NO2-phenyl | 6.05 ± 0.08 | 15.8 ± 1.67 | 6.32 ± 1.00 |
| I5 | -phenyl | 3-OH-phenyl | 5.09 ± 0.08 | 6.68 ± 0.96 | 8.69 ± 1.04 |
| I6 | -phenyl | 3-OCH3-phenyl | 41.0 ± 4.47 | 160 ± 10.9 | 46.8 ± 0.14 |
| I7 | -phenyl | 4-NO2-phenyl | 4.17 ± 0.93 | 8.79 ± 3.19 | 7.63 ± 1.75 |
| I8 | -phenyl | 4-Cl-phenyl | 9.57 ± 1.92 | 63.2 ± 3.86 | 6.98 ± 1.30 |
| I9 | -phenyl | 4-F-phenyl | 24.9 ± 1.54 | 4.68 ± 0.69 | 4.75 ± 0.36 |
| I10 | -phenyl | 4-CH3-phenyl | 19.5 ± 4.95 | 70.3 ± 9.88 | 23.6 ± 2.67 |
| I11 | -phenyl | 4-i-Pr-phenyl | 43.0 ± 7.30 | 19.2 ± 6.57 | 18.6 ± 3.90 |
| I12 | -phenyl | 4-OCH3-phenyl | 10.9 ± 0.25 | 39.4 ± 3.54 | 21.1 ± 4.07 |
| I13 | -phenyl | 4-OC2H5-phenyl | 15.0 ± 4.90 | 22.5 ± 2.64 | 15.4 ± 0.93 |
| I14 | -phenyl | 4-(CH3)2N-phenyl | 12.8 ± 0.60 | 7.81 ± 1.69 | 3.42 ± 0.49 |
| I15 | -phenyl | 3-OCH3-4-OH-phenyl | 35.8 ± 0.59 | >300 | 31.1 ± 7.66 |
| I16 | -phenyl | cyclopropyl | 16.4 ± 0.98 | 80.5 ± 5.43 | 78.5 ± 1.39 |
| I17 | -phenyl | furanyl | 13.0 ± 2.40 | 36.0 ± 0.70 | 34.5 ± 0.28 |
| I18 | -phenyl | thiophenyl | 13.8 ± 0.29 | 15.9 ± 1.14 | 21.1 ± 3.76 |
| I19 | -phenyl | 4-CH3-5-thiazolyl | 26.6 ± 5.08 | 96.4 ± 8.36 | 89.3 ± 8.89 |
| I20 | -phenyl | cyclohexyl | 16.3 ± 3.25 | 21.8 ± 3.15 | >300 |
| I21 | -phenyl | 3-cyclohexen-1-yl | 8.27 ± 0.80 | 21.6 ± 0.89 | 20.4 ± 0.86 |
| I22 | -phenyl | 3-pyridine-1-yl | 60.2 ± 2.37 | 159 ± 5.88 | 43.3 ± 1.10 |
| I23 | 4-NO2-phenyl | 3-OCH3-phenyl | 5.99 ± 1.07 | 4.17 ± 0.57 | 3.36 ± 0.80 |
| I24 | 4-NO2-phenyl | 4-OCH3-phenyl | 5.17 ± 0.41 | 15.7 ± 2.98 | 8.08 ± 2.25 |
| I25 | 4-OCH3-phenyl | 3-OCH3-phenyl | 144 ± 8.95 | 142 ± 6.79 | 80.0 ± 4.04 |
| I26 | 3-OCH3-phenyl | 3-OCH3-phenyl | 11.0 ± 1.72 | 21.0 ± 2.32 | 14.5 ± 2.56 |
| Curcumin | 69.6 ± 1.18 | 73.4 ± 4.99 | 40.2 ± 1.74 | ||
| Colchicine | 1.35 ± 0.08 | 2.12 ± 0.29 | 5.46 ± 1.76 | ||
| Gefitinib | 5.47 ± 1.06 | 5.99 ± 1.47 | 31.5 ± 10.2 | ||
| No. |  | IC50 (μM) b | ||||
|---|---|---|---|---|---|---|
| R1 | R2 | A549 | PC-3 | HepG2 | NRK-52E | |
| I5 | phenyl | 3-OH-phenyl | 5.09 ± 0.08 | 6.68 ± 0.96 | 8.69 ± 1.04 | 50.0 ± 4.40 |
| I7 | phenyl | 4-NO2-phenyl | 4.17 ± 0.93 | 8.79 ± 3.19 | 7.63 ± 1.75 | 21.0 ± 1.75 |
| I14 | phenyl | 4-(CH3)2N-phenyl | 12.8 ± 0.60 | 7.81 ± 1.69 | 3.42 ± 0.93 | 7.00 ± 0.30 |
| I23 | 4-NO2-phenyl | 3-OCH3-phenyl | 5.99 ± 1.07 | 4.17 ± 0.57 | 3.36 ± 0.80 | 6.99 ± 1.60 |
| Curcumin | 69.6 ± 1.18 | 73.4 ± 4.99 | 40.2 ± 1.74 | >300 | ||
| Colchicine | 1.35 ± 0.08 | 2.12 ± 0.29 | 5.46 ± 1.76 | 3.32 ± 0.81 | ||
| Gefitinib | 5.47 ± 1.06 | 5.99 ± 1.47 | 31.5 ± 10.2 | 21.0 ± 2.10 | ||
| Property | Value | Decision |
|---|---|---|
| Absorption | ||
| Caco-2 Permeability | −4.82 | Excellent |
| Madin–Darby canine kidney cells (MDCK) permeability | 0.000185 | Excellent |
| P-glycoprotein (Pgp)-inhibitor | 0.984 | Bad |
| P-glycoprotein (Pgp)-substrate | 0.002 | Excellent |
| Human intestinal absorption (HIA) | 0.021 | Excellent |
| 20% bioavailability (F20%) | 0.002 | Excellent |
| Distribution | ||
| Plasma protein binding (PPB) | 95.57% | Bad |
| Volume distribution (VD) | 0.285 | Excellent |
| Blood–brain barrier (BBB) penetration | 0.038 | Excellent |
| The fraction unbound in plasma (Fu) | 1.635% | Bad |
| Excretion | ||
| Clearance | 7.066 | Excellent |
| The half-life (T1/2) | 0.404 | |
| Toxicity | ||
| hERG blockers | 0.329 | Medium |
| Rat oral acute toxicity | 0.195 | Excellent |
| Maximum recommended daily dose (FDAMDD) | 0.449 | Medium |
| Skin sensitization | 0.916 | Bad |
| Eye corrosion | 0.004 | Excellent |
| Eye irritation | 0.305 | Medium |
| Respiratory toxicity | 0.145 | Excellent |
| Drug-likeness | ||
| Lipinski rule | Accepted | Excellent |
| Pfzer rule | Accepted | Excellent |
| Golden triangle | Accepted | Excellent |
| GSK rule | Accepted | Excellent |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Fu, Y.-H.; Zou, Y.-Y.; Meng, J.; Ou-Yang, G.-P.; Ge, Q.-S.; Wang, Z.-C. Discovery of Simple Diacylhydrazine-Functionalized Cinnamic Acid Derivatives as Potential Microtubule Stabilizers. Int. J. Mol. Sci. 2022, 23, 12365. https://doi.org/10.3390/ijms232012365
Zhou X, Fu Y-H, Zou Y-Y, Meng J, Ou-Yang G-P, Ge Q-S, Wang Z-C. Discovery of Simple Diacylhydrazine-Functionalized Cinnamic Acid Derivatives as Potential Microtubule Stabilizers. International Journal of Molecular Sciences. 2022; 23(20):12365. https://doi.org/10.3390/ijms232012365
Chicago/Turabian StyleZhou, Xiang, Yi-Hong Fu, Ya-Yu Zou, Jiao Meng, Gui-Ping Ou-Yang, Qiang-Sheng Ge, and Zhen-Chao Wang. 2022. "Discovery of Simple Diacylhydrazine-Functionalized Cinnamic Acid Derivatives as Potential Microtubule Stabilizers" International Journal of Molecular Sciences 23, no. 20: 12365. https://doi.org/10.3390/ijms232012365
APA StyleZhou, X., Fu, Y.-H., Zou, Y.-Y., Meng, J., Ou-Yang, G.-P., Ge, Q.-S., & Wang, Z.-C. (2022). Discovery of Simple Diacylhydrazine-Functionalized Cinnamic Acid Derivatives as Potential Microtubule Stabilizers. International Journal of Molecular Sciences, 23(20), 12365. https://doi.org/10.3390/ijms232012365






