Plasma Extracellular Vesicles Play a Role in Immune System Modulation in Minimal Hepatic Encephalopathy
Abstract
1. Introduction
2. Results
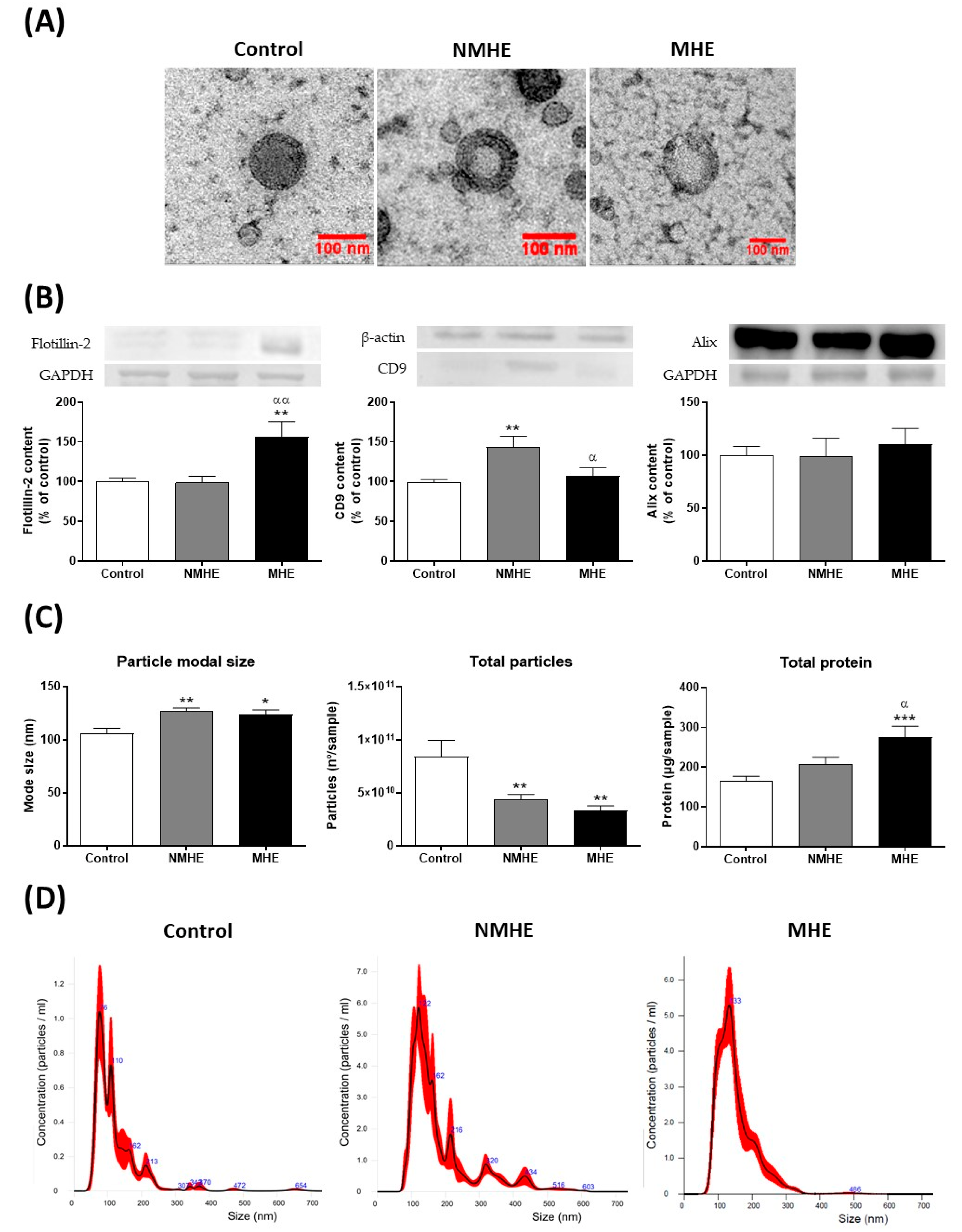
2.1. Cirrhotic Patients Show Lower Number of Plasma EVs which Show Increased Size
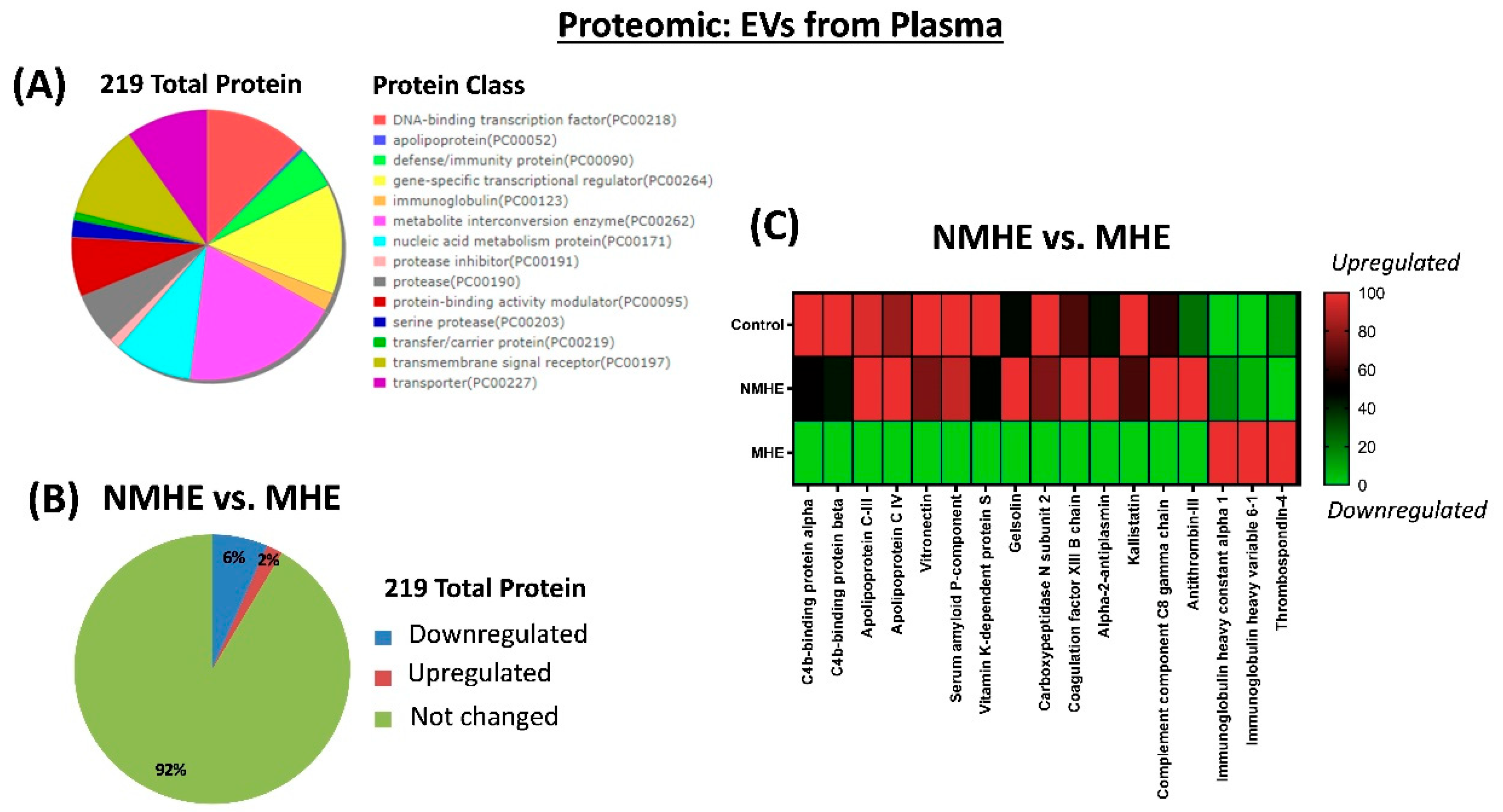
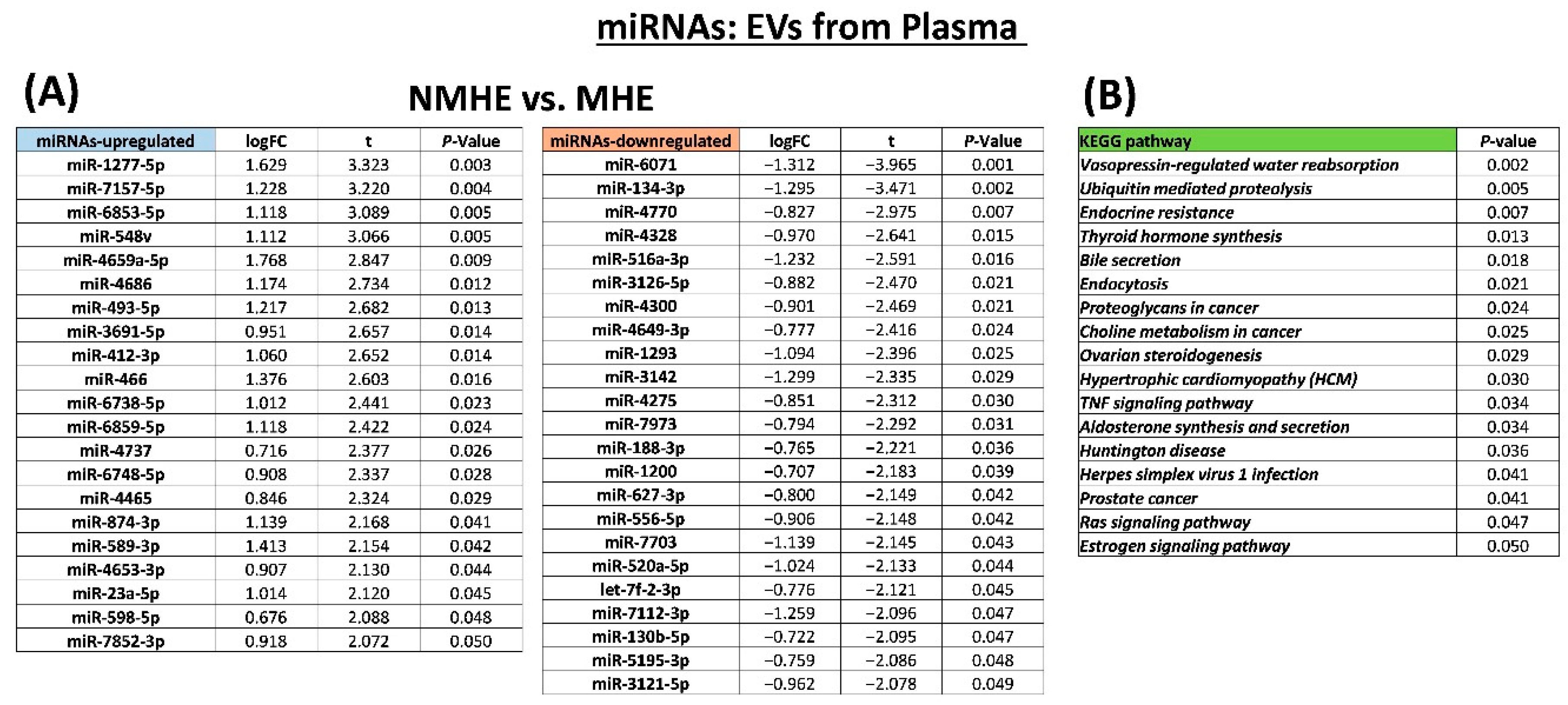
2.2. MHE Alters Protein and miRNAs Cargo of EVs in Plasma
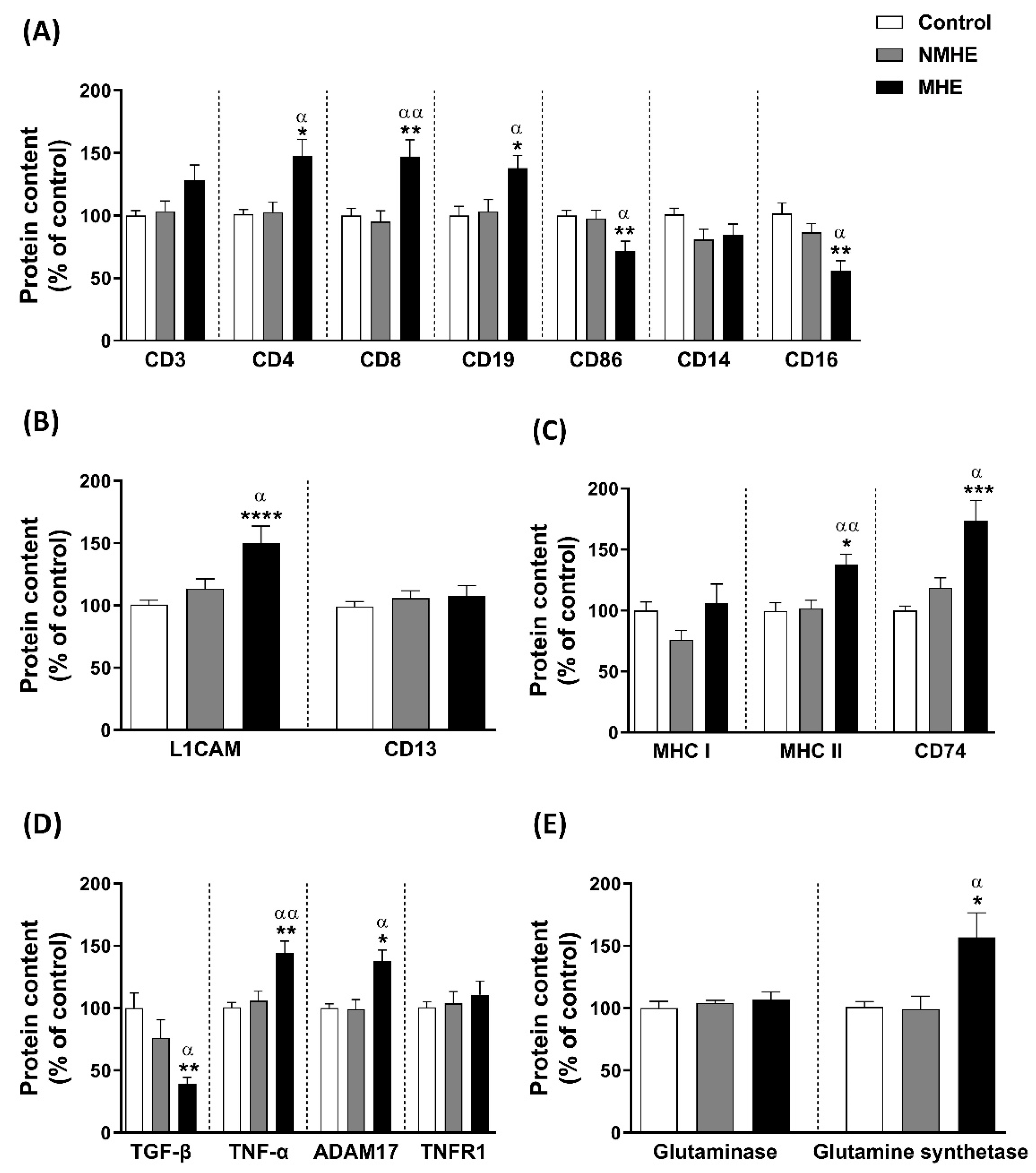
2.3. EVs from Plasma of Patients with MHE Show Altered Content of Immune and Neuronal Cells Markers
2.4. Content of Presenting Antigen Molecules in Plasma EVs
2.5. EVs from Plasma of MHE Patients Show an Increase in Pro-Inflammatory Molecules and a Reduction in Anti-Inflammatory Molecules
2.6. Ammonia Metabolism Enzyme Content in Plasma EVs
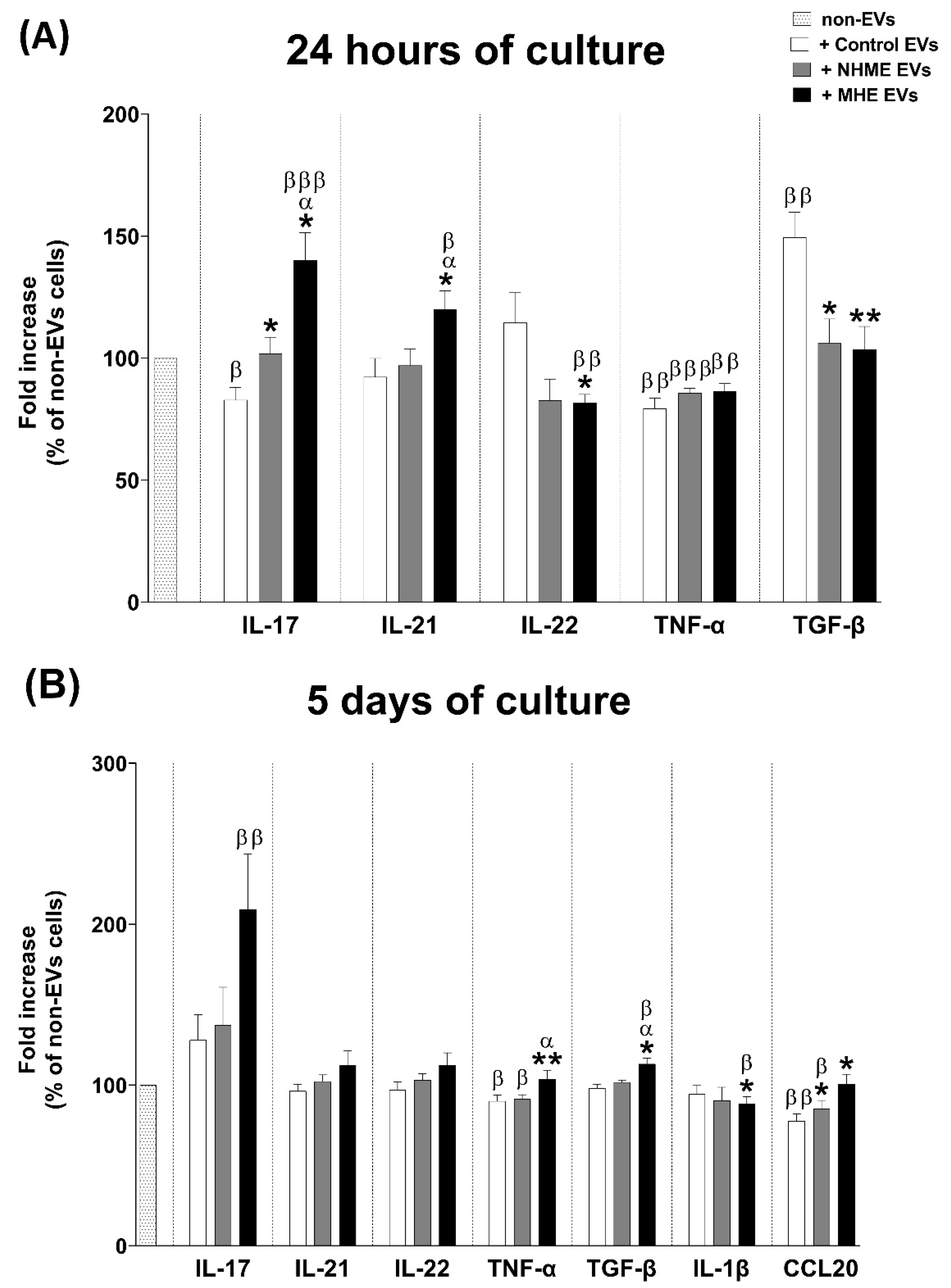
2.7. EVs from Plasma of MHE Patients Modulate the Expression of Pro-Inflammatory Cytokines IL-17, IL-21, and TNF-α, and the Anti-Inflammatory Cytokine TGF-β in Cultured CD4+ T Lymphocytes
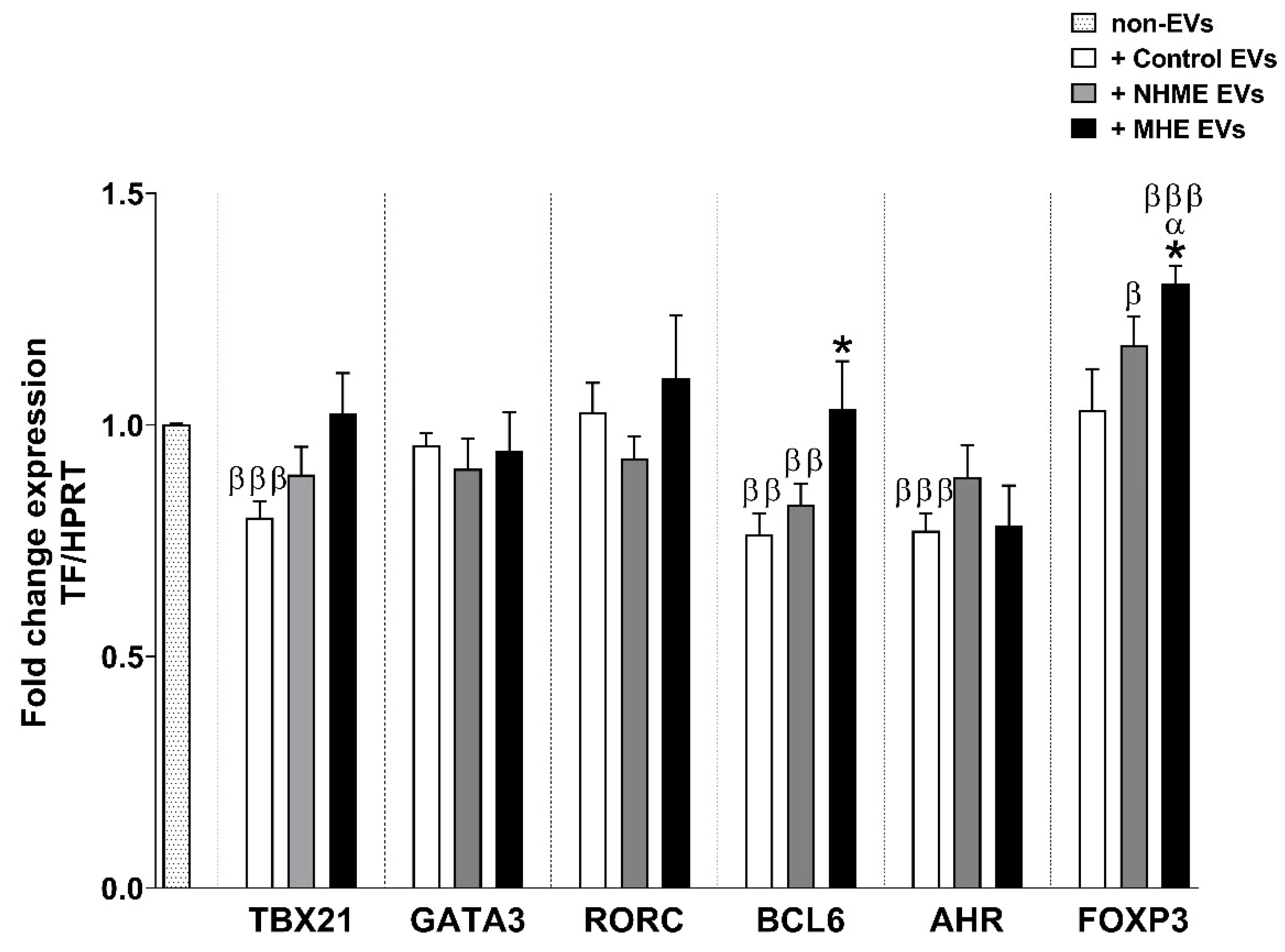
2.8. EVs from Plasma of MHE Patients Increased the Proportion of Th Follicular and Treg Cells and Activation of Th17 Cells
3. Discussion
4. Materials and Methods
4.1. Patients and Controls
4.2. Diagnosis of Minimal Hepatic Encephalopathy (MHE)
4.3. Extracellular Vesicle Isolation
4.4. Nanoparticle Tracking Analysis
4.5. Transmission Electron Microscopy
4.6. Analysis of EVs Protein Cargo by Immunoblotting
4.7. Analysis of EVs TGF-β Cargo by ELISA
4.8. Proteomic Analysis
4.9. miRNA Analysis
4.10. CD4+ T Cell Culture and Treatment with EVs
4.10.1. Determination of Cytokine Production by ELISA
4.10.2. Analysis of Transcription Factors by Quantitative Real-Time PCR (qPCR)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Felipo, V. Hepatic encephalopathy: Effects of liver failure on brain function. Nat. Rev. Neurosci. 2013, 14, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Nardone, R.; Taylor, A.C.; Höller, Y.; Brigo, F.; Lochner, P.; Trinka, E. Minimal hepatic encephalopathy: A review. Neurosci. Res. 2016, 111, 1–12. [Google Scholar] [CrossRef]
- Ridola, L.; Nardelli, S.; Gioia, S.; Riggio, O. Quality of life in patients with minimal hepatic encephalopathy. World J. Gastroenterol. 2018, 24, 5446–5453. [Google Scholar] [CrossRef]
- Felipo, V.; Urios, A.; Montesinos, E.; Molina, I.; Garcia-Torres, M.L.; Civera, M.; del Olmo, J.A.; Ortega, J.; Martinez-Valls, J.; Serra, M.A.; et al. Contribution of hyperammonemia and inflammatory factors to cognitive impairment in minimal hepatic encephalopathy. Metab. Brain Dis. 2012, 27, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Mangas-Losada, A.; García-García, R.; Urios, A.; Escudero-García, D.; Tosca, J.; Giner-Durán, R.; Serra, M.A.; Montoliu, C.; Felipo, V. Minimal hepatic encephalopathy is associated with expansion and activation of CD4+CD28−, Th22 and Tfh and B lymphocytes. Sci. Rep. 2017, 7, 6683. [Google Scholar] [CrossRef]
- Kara, E.E.; Comerford, I.; Fenix, K.A.; Bastow, C.R.; Gregor, C.E.; McKenzie, D.R.; McColl, S.R. Tailored immune responses: Novel effector helper T cell subsets in protective immunity. PLoS Pathog. 2014, 10, e1003905. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Lahesmaa, R. Transcriptional and epigenetic regulation of T-helper lineage specification. Immunol. Rev. 2014, 261, 62–83. [Google Scholar] [CrossRef] [PubMed]
- Ridder, K.; Keller, S.; Dams, M.; Rupp, A.-K.; Schlaudraff, J.; del Turco, D.; Starmann, J.; Macas, J.; Karpova, D.; Devraj, K.; et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014, 12, e1001874. [Google Scholar] [CrossRef] [PubMed]
- Ulivieri, C.; Baldari, C. Regulation of T cell activation and differentiation by extracellular vesicles and their pathogenic role in systemic lupus erythematosus and multiple sclerosis. Molecules 2017, 22, 225. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Kim, J.-H. A Comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int. J. Nanomed. 2021, 16, 1281–1312. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.D.; Wong, W.; Lee, M.M.; Cho, W.C.; Yee, B.K.; Kwan, Y.W.; Tai, W.C. Exosomes in inflammation and inflammatory disease. Proteomics 2019, 19, 1800149. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Altarejos, P.; Cabrera-Pastor, A.; Gonzalez-King, H.; Montoliu, C.; Felipo, V. Extracellular vesicles from hyperammonemic rats induce neuroinflammation and motor incoordination in control rats. Cells 2020, 9, 572. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Dalvi, P.; Abadjian, L.; Tang, N.; Pulliam, L. Blood neuron-derived exosomes as biomarkers of cognitive impairment in HIV. AIDS 2017, 31, F9–F17. [Google Scholar] [CrossRef] [PubMed]
- Hubert, A.; Subra, C.; Jenabian, M.-A.; Tremblay Labrecque, P.-F.; Tremblay, C.; Laffont, B.; Provost, P.; Routy, J.-P.; Gilbert, C. Elevated Abundance, Size, and MicroRNA Content of Plasma Extracellular Vesicles in Viremic HIV-1+ Patients. J Acquir Immune Defic Syndr. 2015, 70, 219–227. [Google Scholar] [CrossRef]
- Welsh, J.A.; Scorletti, E.; Clough, G.F.; Englyst, N.A.; Byrne, C.D. Leukocyte extracellular vesicle concentration is inversely associated with liver fibrosis severity in NAFLD. J. Leukocyte Biol. 2018, 104, 631–639. [Google Scholar] [CrossRef]
- Hemmer, B.; Vergelli, M.; Calabresi, P.; Huang, T.; McFarland, H.F.; Martin, R. Cytokine phenotype of human autoreactive T cell clones specific for the immunodominant myelin basic protein peptide (83–99). J. Neurosci. Res. 1996, 45, 852–862. [Google Scholar] [CrossRef]
- Eguchi, A.; Lazic, M.; Armando, A.M.; Phillips, S.A.; Katebian, R.; Maraka, S.; Quehenberger, O.; Sears, D.D.; Feldstein, A.E. Circulating adipocyte-derived extracellular vesicles are novel markers of metabolic stress. J. Mol. Med. 2016, 94, 1241–1253. [Google Scholar] [CrossRef]
- Boulanger, C.M.; Amabile, N.; Tedgui, A. Circulating microparticles. Hypertension 2006, 48, 180–186. [Google Scholar] [CrossRef]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef]
- Felipo, V.; Hermenegildo, C.; Montoliu, C.; Llansola, M.; Miñana, M.D. Neurotoxicity of ammonia and glutamate: Molecular mechanisms and prevention. Neurotoxicology 1998, 19, 675–681. [Google Scholar] [PubMed]
- ElMlili, N.; Boix, J.; Ahabrach, H.; Rodrigo, R.; Errami, M.; Felipo, V. Chronic hyperammonemia induces tonic activation of NMDA receptors in cerebellum. J. Neurochem. 2010, 112, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Badhwar, A.; Haqqani, A.S. Biomarker potential of brain-secreted extracellular vesicles in blood in Alzheimer’s disease. Alzheimer’s Dement. 2020, 12, e12001. [Google Scholar] [CrossRef] [PubMed]
- Piktel, E.; Levental, I.; Durnaś, B.; Janmey, P.; Bucki, R. Plasma gelsolin: Indicator of inflammation and its potential as a diagnostic tool and therapeutic target. Int. J. Mol. Sci. 2018, 19, 2516. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.Q.; He, Y.H.; Wang, S.B.; Yang, S.; Wang, Y.J.; Nan, C.J.; Bao, Y.F.; Xie, Q.P.; Chen, Y.H. MiR-130b/TNF-α/NF-κB/VEGFA loop inhibits prostate cancer angiogenesis. Clin. Transl. Oncol. 2020, 22, 111–121. [Google Scholar] [CrossRef]
- Gao, W.; Liu, H.; Yuan, J.; Wu, C.; Huang, D.; Ma, Y.; Zhu, J.; Ma, L.; Guo, J.; Shi, H.; et al. Exosomes derived from mature dendritic cells increase endothelial inflammation and atherosclerosis via membrane TNF-α mediated NF-κB pathway. J. Cell. Mol. Med. 2016, 20, 2318–2327. [Google Scholar] [CrossRef]
- Liu, H.; Liang, Z.; Wang, F.; Zheng, X.; Zeng, Z.; He, X.; Gao, X.; Zhi, M.; Wu, X.; Wu, X.; et al. Intestinal CD14+ Macrophages protect CD4+ T cells from activation-induced cell death via exosomal membrane TNF in Crohn’s disease. J. Crohns Colitis 2020, 14, 1619–1631. [Google Scholar] [CrossRef]
- Zhang, H.-G.; Liu, C.; Su, K.; Su, K.; Yu, S.; Zhang, L.; Zhang, S.; Wang, J.; Cao, X.; Grizzle, W.; et al. A Membrane form of TNF-Alpha presented by exosomes delays T cell activation-induced cell death. J. Immunol. 2006, 176, 7385–7393. [Google Scholar] [CrossRef]
- Wahlund, C.J.E.; Güclüler, G.; Hiltbrunner, S.; Veerman, R.E.; Näslund, T.I.; Gabrielsson, S. Exosomes from antigen-pulsed dendritic cells induce stronger antigen-specific immune responses than microvesicles in vivo. Sci. Rep. 2017, 7, 17095. [Google Scholar] [CrossRef]
- Admyre, C.; Bohle, B.; Johansson, S.M.; Focke-Tejkl, M.; Valenta, R.; Scheynius, A.; Gabrielsson, S. B cell–derived exosomes can present allergen peptides and activate allergen-specific t cells to proliferate and produce TH2-like cytokines. J. Allergy Clin. Immun. 2007, 120, 1418–1424. [Google Scholar] [CrossRef]
- Álvarez, V.; Sánchez-Margallo, F.M.; Macías-García, B.; Gómez-Serrano, M.; Jorge, I.; Vázquez, J.; Blázquez, R.; Casado, J.G. The immunomodulatory activity of extracellular vesicles derived from endometrial mesenchymal stem cells on CD4+ T cells is partially mediated by TGFbeta. J. Tissue Eng. Regen. Med. 2018, 12, 2088–2098. [Google Scholar] [CrossRef] [PubMed]
- Crellin, N.K.; Garcia, R.V.; Levings, M.K. Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood 2007, 109, 2014–2022. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Patterson, S.J.; Levings, M.K. The role of the PI3K signaling pathway in CD4+ T cell differentiation and function. Front. Immunol. 2012, 3, 245. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zeng, Z.; He, Z.; Lei, S. Hypoxic pancreatic stellate cell-derived exosomal mirnas promote proliferation and invasion of pancreatic cancer through the PTEN/AKT pathway. Aging 2021, 13, 7120–7132. [Google Scholar] [CrossRef]
- Tao, Z.; Feng, C.; Mao, C.; Ren, J.; Tai, Y.; Guo, H.; Pu, M.; Zhou, Y.; Wang, G.; Wang, M. MiR-4465 directly targets PTEN to inhibit AKT/MTOR pathway–mediated autophagy. Cell Stress Chaperon 2019, 24, 105–113. [Google Scholar] [CrossRef]
- Ragusa, M.; Bosco, P.; Tamburello, L.; Barbagallo, C.; Condorelli, A.G.; Tornitore, M.; Spada, R.S.; Barbagallo, D.; Scalia, M.; Elia, M.; et al. miRNAs Plasma Profiles in Vascular Dementia: Biomolecular Data and Biomedical Implications. Front. Cell Neurosci. 2016, 10, 51. [Google Scholar] [CrossRef]
- Lei, Y.; Yang, M.; Li, H.; Xu, R.; Liu, J. miR-130b regulates PTEN to activate the PI3K/Akt signaling pathway and attenuate oxidative stress-induced injury in diabetic encephalopathy. Int J. Mol. Med. 2021, 48, 141. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, S.; Pan, J.; Wang, Z.; Li, Y.; Xu, X.; Yang, J.; Zhang, X.; Wang, Y.; Liu, M. Ginsenoside Rb1 attenuates microglia activation to improve spinal cord injury via microRNA-130b-5p/TLR4/NF-κB axis. J. Cell Physiol. 2021, 236, 2144–2155. [Google Scholar] [CrossRef]
- Li, Z.H.; Wang, Y.F.; He, D.D.; Zhang, X.M.; Zhou, Y.L.; Yue, H.; Huang, S.; Fu, Z.; Zhang, L.Y.; Mao, Z.Q.; et al. Let-7f-5p suppresses Th17 differentiation via targeting STAT3 in multiple sclerosis. Aging 2019, 11, 4463–4477. [Google Scholar] [CrossRef]
- Weissenborn, K.; Ennen, J.C.; Schomerus, H.; Rückert, N.; Hecker, H. Neuropsychological characterization of hepatic encephalopathy. J. Hepatol. 2001, 34, 768–773. [Google Scholar] [CrossRef]
- Webber, J.; Clayton, A. How pure are your vesicles? J. Extracell. Vesicles 2013, 2, 19861. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1-3.22.29. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Felipo, V.; Minana, M.-D.; Grisolia, S. Long-term ingestion of ammonium increases acetylglutamate and urea levels without affecting the amount of carbamoyl-phosphate synthase. Eur. J. Biochem. 1988, 176, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Shevchenko, A.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.D.; Irizarry, R.A.; WU, Z. Removing Technical Variability in RNA-Seq Data Using Conditional Quantile Normalization. Biostatistics 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Garcia-Garcia, F.; Panadero, J.; Dopazo, J.; Montaner, D. Integrated Gene Set Analysis for MicroRNA Studies. Bioinformatics 2016, 32, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Motulsky, H.M.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef] [PubMed]
| Controls (n = 24) | Patients without MHE (n = 27) | Patients with MHE (n = 23) | |
|---|---|---|---|
| Gender (M/F) | 11/13 | 19/8 | 18/5 |
| Age (years) a | 59 ± 7 | 63 ± 6 | 64 ± 8 |
| Etiology of cirrhosis | |||
| Alcohol | 9 | 11 | |
| HBV/HCV | 0/6 | 2/4 | |
| HBV/HCV + alcohol | 1 | 1 | |
| NASH | 8 | 4 | |
| Others | 3 | 1 | |
| Ascites | 0 | 10 | |
| Child Pugh score (A/B/C) | 25/2/0 | 16/7/0 | |
| MELD score a | 8.2 ± 2 | 9.3 ± 2.6 | |
| Ammonia (uM) a | 13 ± 8 | 15 ± 9 | 40 ± 27 ***/α |
| Platelet count (×109/L) | 241 ± 56 | 115 ± 54 *** | 130 ± 68 *** |
| PHES a | 0.7 ± 1.0 | 0.4 ± 1.0 | −7.1 ± 3.2 ***/ααα |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallego, J.J.; Fiorillo, A.; Casanova-Ferrer, F.; Urios, A.; Ballester, M.-P.; Durbán, L.; Megías, J.; Rubio, T.; Cabrera-Pastor, A.; Escudero-García, D.; et al. Plasma Extracellular Vesicles Play a Role in Immune System Modulation in Minimal Hepatic Encephalopathy. Int. J. Mol. Sci. 2022, 23, 12335. https://doi.org/10.3390/ijms232012335
Gallego JJ, Fiorillo A, Casanova-Ferrer F, Urios A, Ballester M-P, Durbán L, Megías J, Rubio T, Cabrera-Pastor A, Escudero-García D, et al. Plasma Extracellular Vesicles Play a Role in Immune System Modulation in Minimal Hepatic Encephalopathy. International Journal of Molecular Sciences. 2022; 23(20):12335. https://doi.org/10.3390/ijms232012335
Chicago/Turabian StyleGallego, Juan José, Alessandra Fiorillo, Franc Casanova-Ferrer, Amparo Urios, María-Pilar Ballester, Lucia Durbán, Javier Megías, Teresa Rubio, Andrea Cabrera-Pastor, Desamparados Escudero-García, and et al. 2022. "Plasma Extracellular Vesicles Play a Role in Immune System Modulation in Minimal Hepatic Encephalopathy" International Journal of Molecular Sciences 23, no. 20: 12335. https://doi.org/10.3390/ijms232012335
APA StyleGallego, J. J., Fiorillo, A., Casanova-Ferrer, F., Urios, A., Ballester, M.-P., Durbán, L., Megías, J., Rubio, T., Cabrera-Pastor, A., Escudero-García, D., Felipo, V., & Montoliu, C. (2022). Plasma Extracellular Vesicles Play a Role in Immune System Modulation in Minimal Hepatic Encephalopathy. International Journal of Molecular Sciences, 23(20), 12335. https://doi.org/10.3390/ijms232012335









