Impact of Exosomes Released by Different Corneal Cell Types on the Wound Healing Properties of Human Corneal Epithelial Cells
Abstract
1. Introduction
2. Results
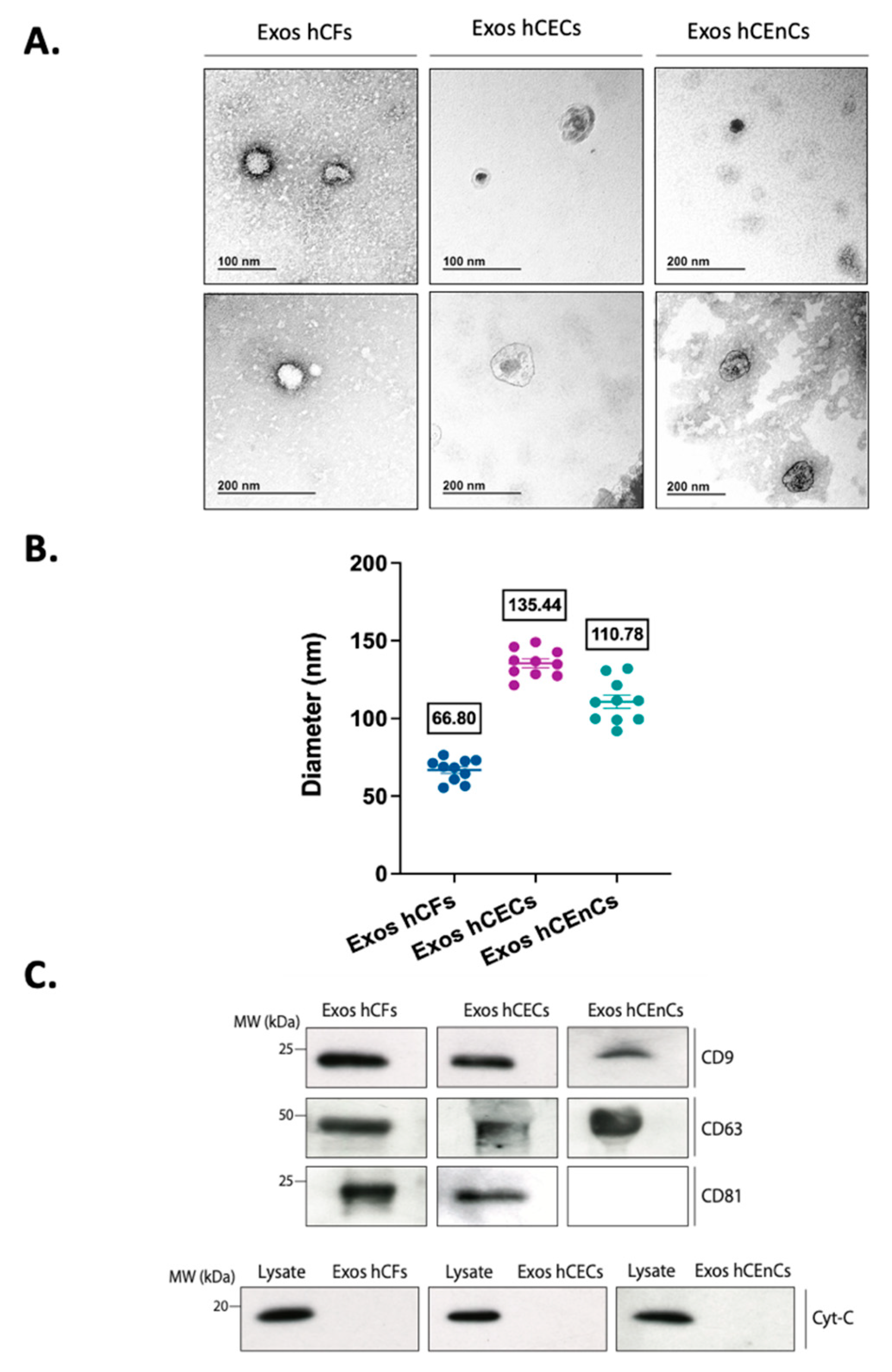
2.1. Characterization of the Exosomes Derived from the Three Different Corneal Cell Types
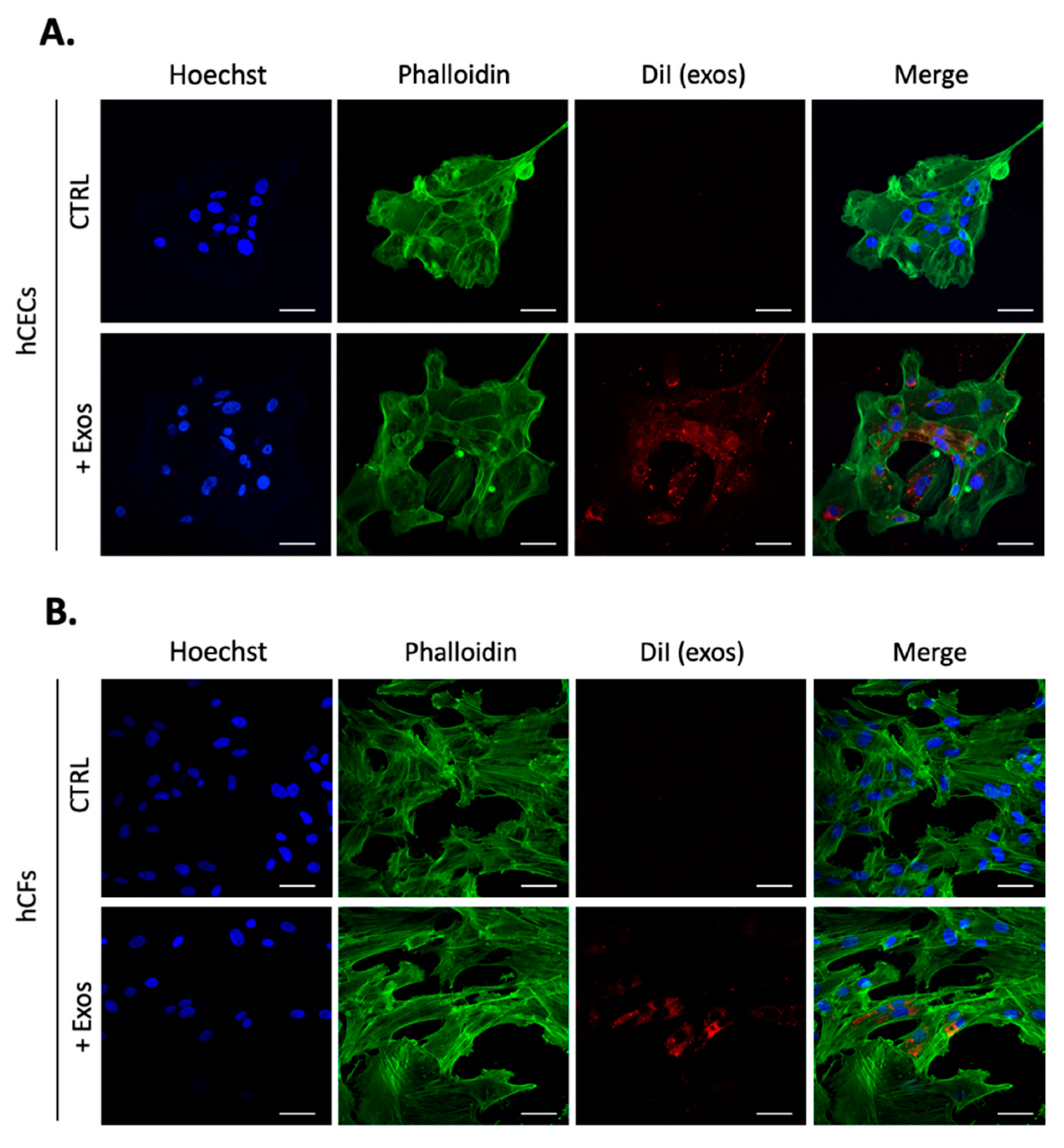
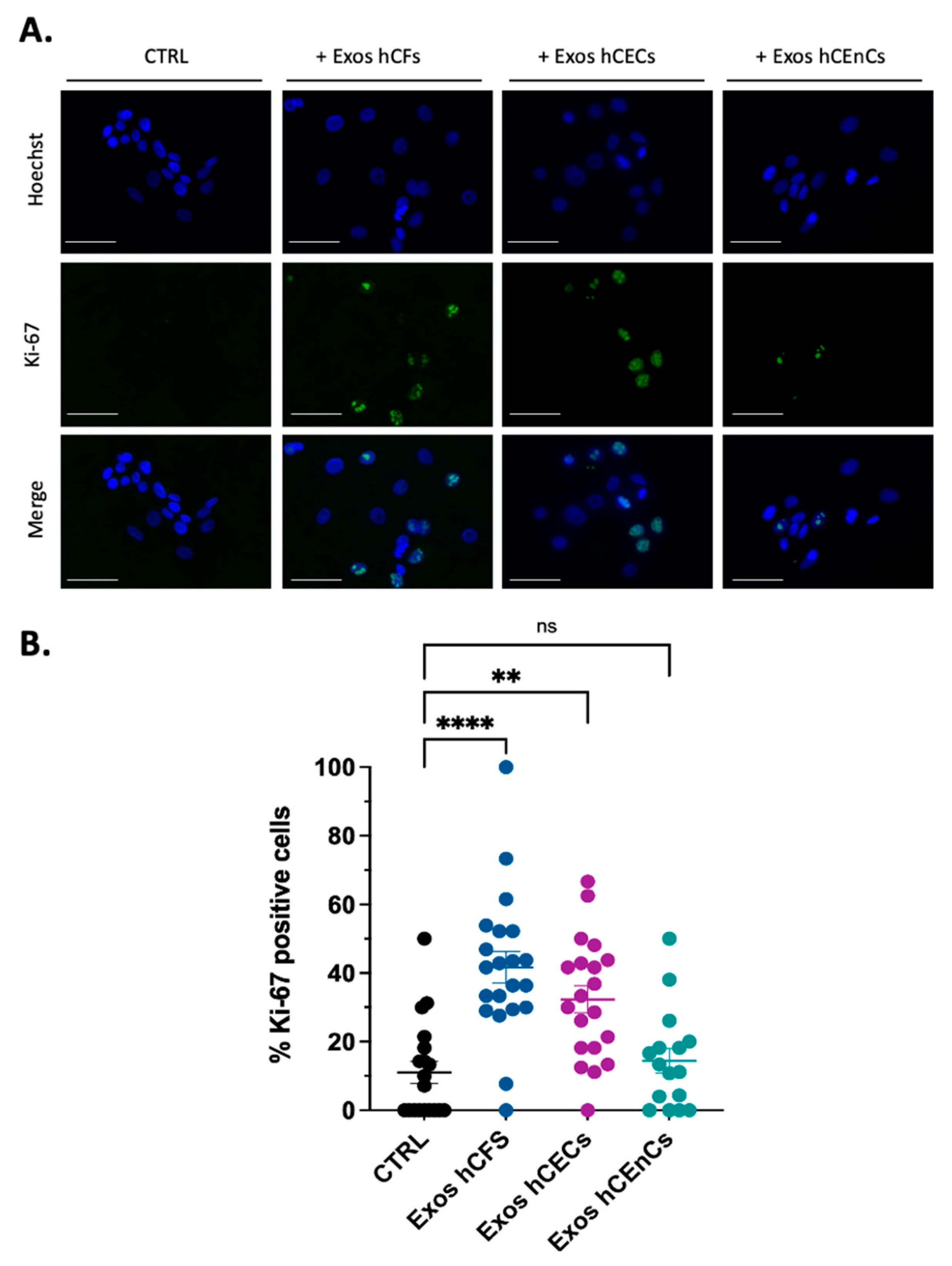
2.2. Human Corneal Exosomes Are Taken Up by hCECs and hCFs and Alter Their Growth Properties
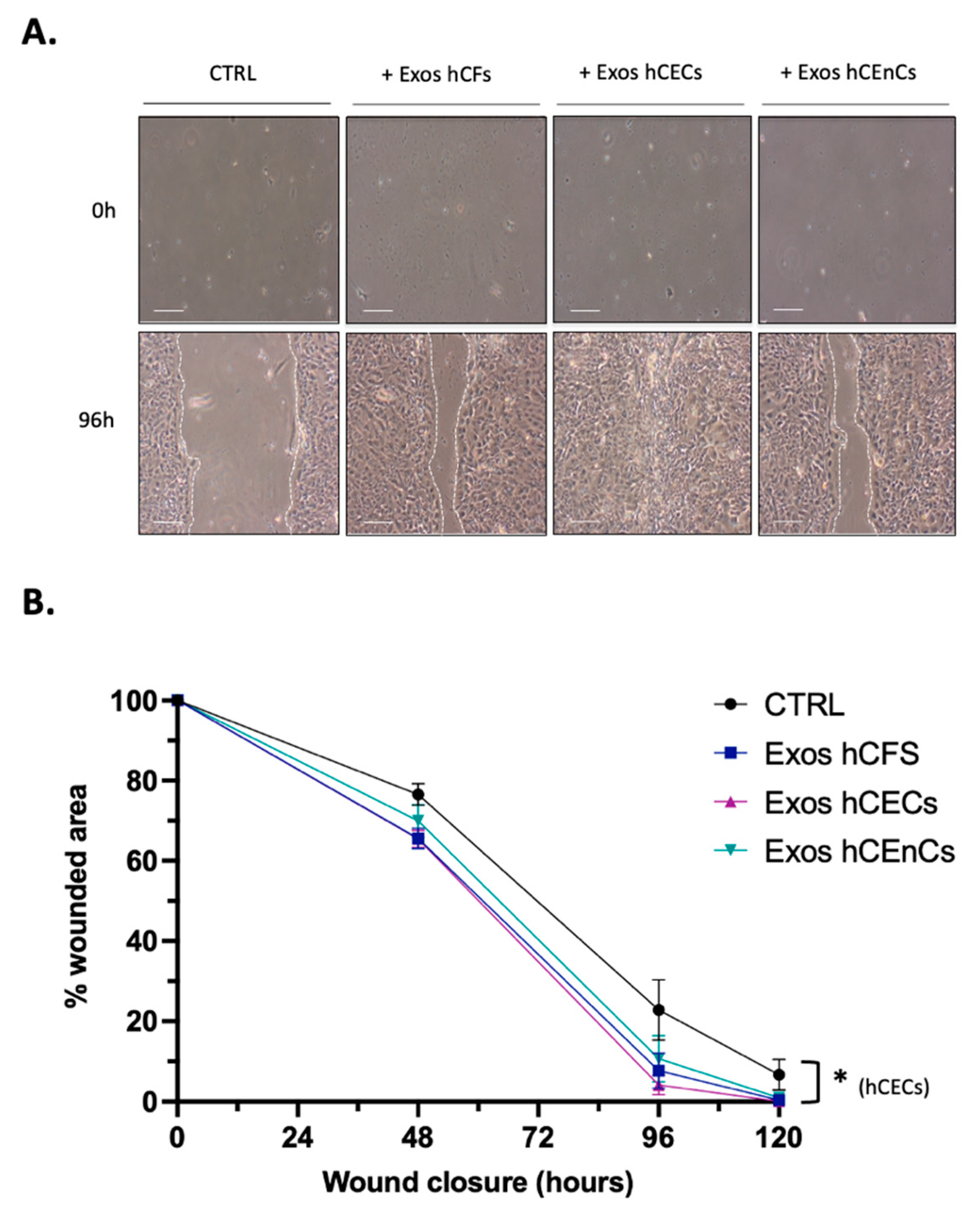
2.3. Human Corneal Exosomes Accelerate Wound Closure of hCECs In Vitro
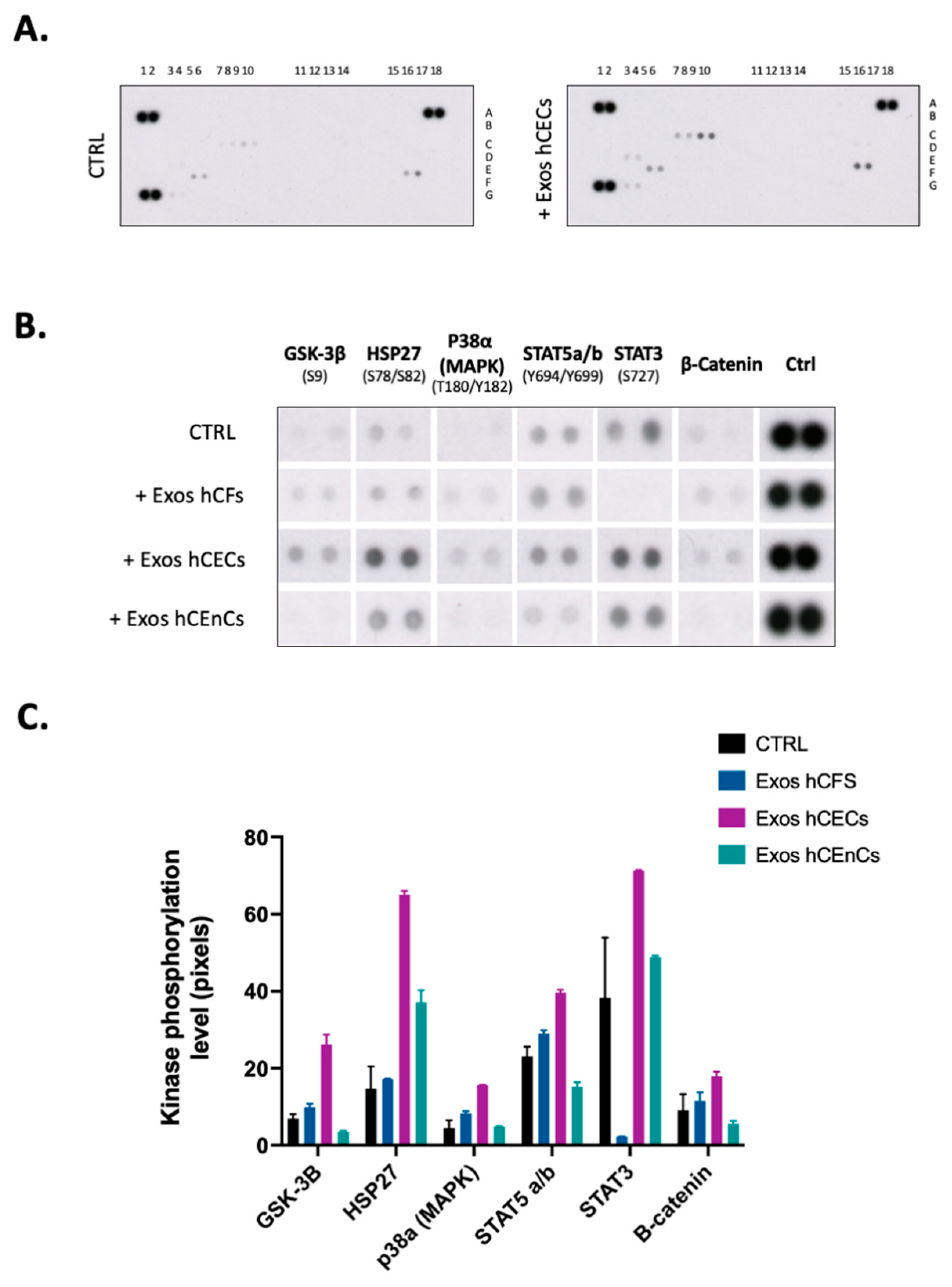
2.4. Signal Transduction Pathways Is Modified by Exosomes in hCECs
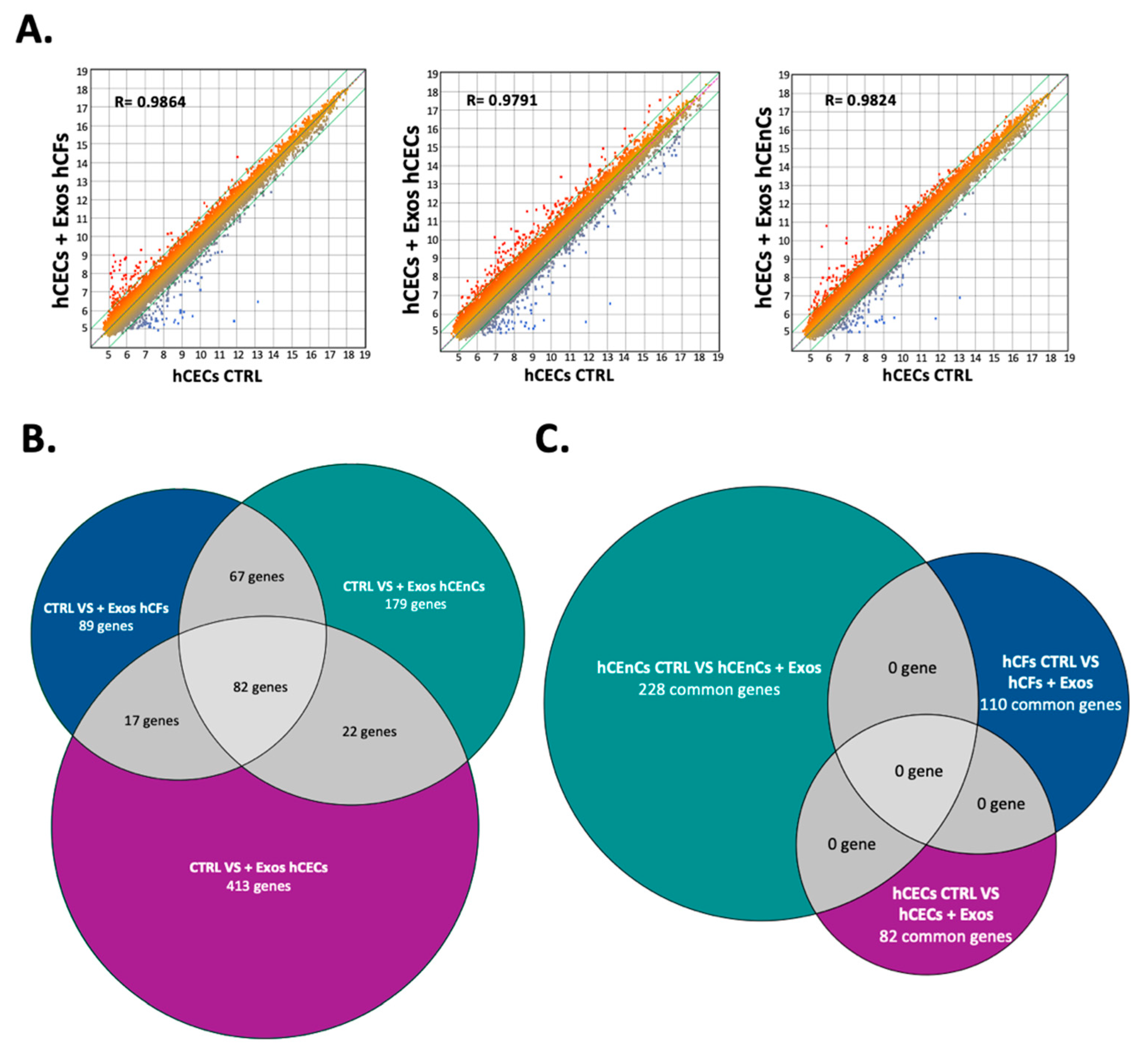
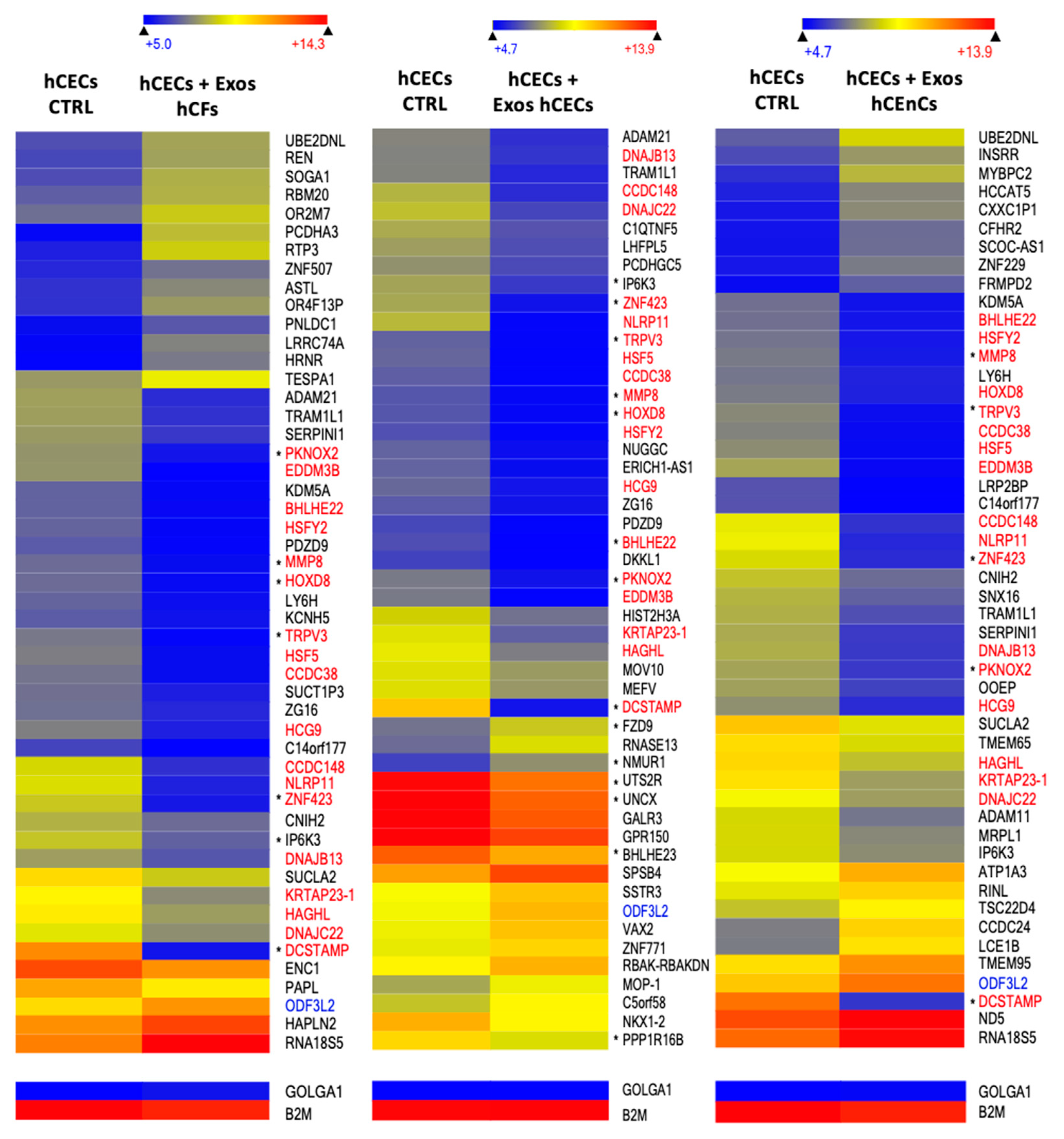
2.5. Exosomes Modify the Gene Expression Pattern in a Cell-Specific Manner
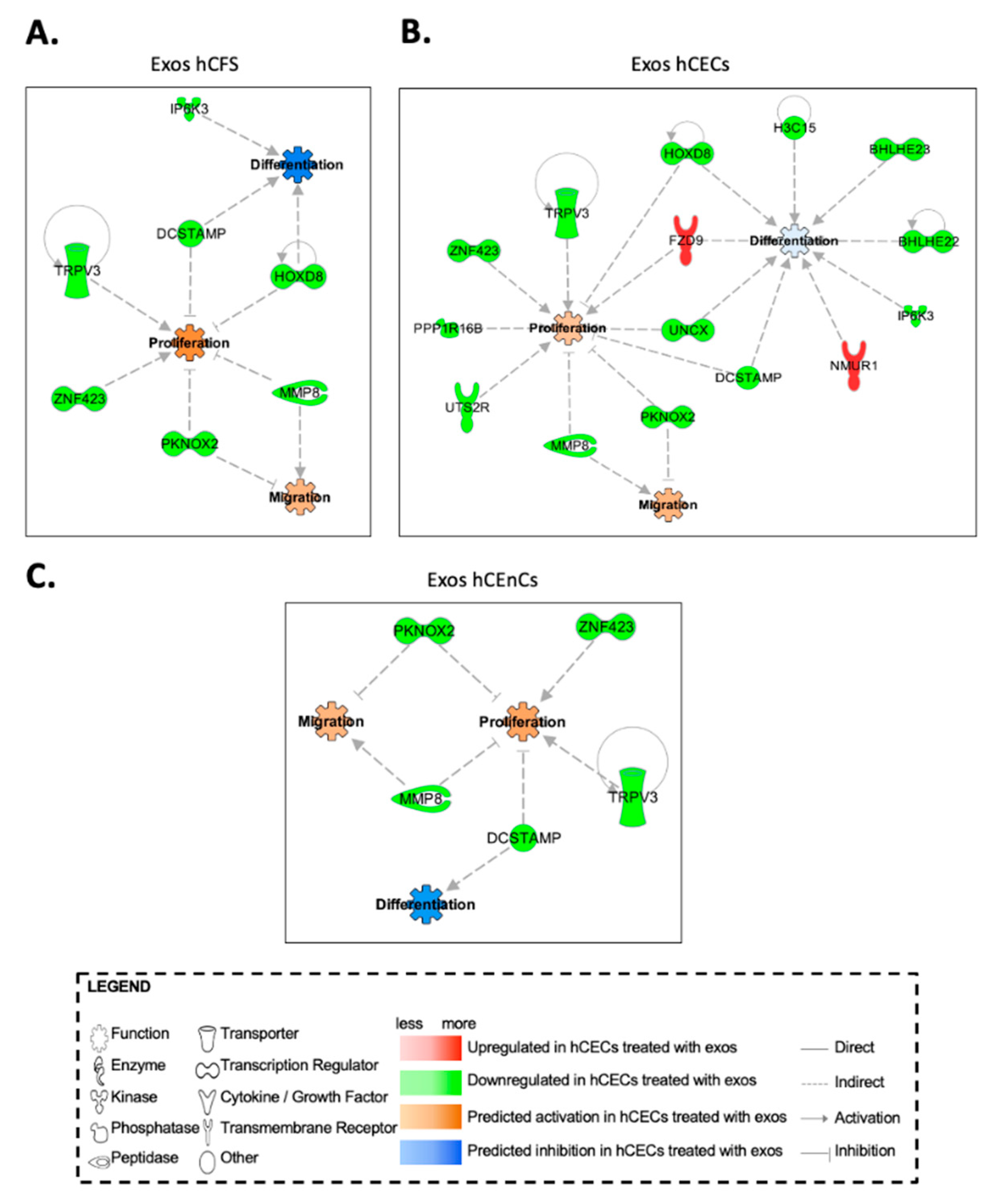
2.6. In Silico Prediction of Biological Functions Affected by Exosomes in hCECs and hCFs through Gene Interaction Network Analyses
3. Discussion
4. Materials and Methods
4.1. Cell Isolation and Culture
4.2. Exosome Enrichment
4.3. Quantification of Exosomes
4.4. Electron Microscopy
4.5. Dynamic Light Scattering (DLS)
4.6. Western Blot Analyses
4.7. Exosomes Uptake
4.8. Ki-67 Immunofluorescences
4.9. Scratch Wound Assay
4.10. Phosphokinase Arrays
4.11. Gene Profiling
4.12. Bioinformatics and Ingenuity Pathway Analyses
4.13. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Y.B.; Gardiner, M.F. Fingernail-induced corneal abrasions: Case series from an ophthalmology emergency department. Cornea 2014, 33, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, Y.H.; Johnson, D.; Mondino, B.J.; Weissman, B.A. Contact lens complications in an urgent-care population: The University of California, Los Angeles, contact lens study. Eye Contact Lens 2012, 38, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.W.; Liu, J.J.; Lee, J.S.; Mohan, R.R.; Mohan, R.R.; Woods, D.J.; He, Y.G.; Wilson, S.E. Proinflammatory chemokine induction in keratocytes and inflammatory cell infiltration into the cornea. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2795–2803. [Google Scholar]
- Ramakrishnan, T.; Constantinou, M.; Jhanji, V.; Vajpayee, R.B. Corneal metallic foreign body injuries due to suboptimal ocular protection. Arch. Environ. Occup. Health 2012, 67, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Vemuganti, G.K.; Murthy, S.I.; Das, S. Update on pathologic diagnosis of corneal infections and inflammations. Middle East Afr. J. Ophthalmol. 2011, 18, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Logothetis, H.D.; Leikin, S.M.; Patrianakos, T. Management of anterior segment trauma. Dis. Mon. 2014, 60, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.T.; Dart, J.K.; Tuft, S.J.; Khaw, P.T. Corneal stem cells in review. Wound Repair Regen. 2001, 9, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, P.K.; Loftus, S.A. Ocular emergencies. Am. Fam. Phys. 2007, 76, 829–836. [Google Scholar]
- Gelston, C.D. Common eye emergencies. Am. Fam. Phys. 2013, 88, 515–519. [Google Scholar]
- Agrawal, V.B.; Tsai, R.J. Corneal epithelial wound healing. Indian J. Ophthalmol. 2003, 51, 5–15. [Google Scholar] [PubMed]
- Meek, K.M.; Knupp, C. Corneal structure and transparency. Prog. Retin. Eye Res. 2015, 49, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sta Iglesia, D.D.; Gala, P.H.; Qiu, T.; Stepp, M.A. Integrin expression during epithelial migration and restratification in the tenascin-C-deficient mouse cornea. J. Histochem. Cytochem. 2000, 48, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.E.; Liu, J.J.; Mohan, R.R. Stromal-epithelial interactions in the cornea. Prog. Retin. Eye Res. 1999, 18, 293–309. [Google Scholar] [CrossRef]
- Lee, J.S.; Liu, J.J.; Hong, J.W.; Wilson, S.E. Differential expression analysis by gene array of cell cycle modulators in human corneal epithelial cells stimulated with epidermal growth factor (EGF), hepatocyte growth factor (HGF), or keratinocyte growth factor (KGF). Curr. Eye Res. 2001, 23, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Ko, M.K.; Kay, E.P. Endothelial mesenchymal transformation mediated by IL-1beta-induced FGF-2 in corneal endothelial cells. Exp. Eye Res. 2012, 95, 35–39. [Google Scholar] [CrossRef]
- Roy, O.; Leclerc, V.B.; Bourget, J.M.; Theriault, M.; Proulx, S. Understanding the process of corneal endothelial morphological change in vitro. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1228–1237. [Google Scholar] [CrossRef]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef]
- Yeates, E.F.; Tesco, G. The Endosome-associated Deubiquitinating Enzyme USP8 Regulates BACE1 Enzyme Ubiquitination and Degradation. J. Biol. Chem. 2016, 291, 15753–15766. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.J.; Ehlers, M.D. Mechanisms and function of dendritic exocytosis. Neuron 2011, 69, 856–875. [Google Scholar] [CrossRef]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Pietrowska, M.; Funk, S.; Gawin, M.; Marczak, L.; Abramowicz, A.; Widlak, P.; Whiteside, T. Isolation of Exosomes for the Purpose of Protein Cargo Analysis with the Use of Mass Spectrometry. Methods Mol. Biol. 2017, 1654, 291–307. [Google Scholar] [PubMed]
- Goetzl, E.J.; Mustapic, M.; Kapogiannis, D.; Eitan, E.; Lobach, I.V.; Goetzl, L.; Schwartz, J.B.; Miller, B.L. Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer’s disease. FASEB J. 2016, 30, 3853–3859. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Han, S.B.; Kim, H.J.; Go, S.I.; Lee, W.S.; Bae, W.K.; Cho, S.H.; Song, E.K.; Lee, O.J.; Kim, H.K.; et al. Exosomal miR-181b-5p Downregulation in Ascites Serves as a Potential Diagnostic Biomarker for Gastric Cancer-associated Malignant Ascites. J. Gastric Cancer 2019, 19, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Panoutsopoulou, K.; Papadimitriou, M.A.; Scorilas, A. Circulating exosomal miRNAs: Clinical significance in human cancers. Expert Rev. Mol. Diagn. 2019, 19, 979–995. [Google Scholar] [CrossRef]
- Fang, Z.; Wang, X.; Wu, J.; Xiao, R.; Liu, J. High serum extracellular vesicle miR-10b expression predicts poor prognosis in patients with acute myeloid leukemia. Cancer Biomark. 2019, 27, 1–9. [Google Scholar] [CrossRef]
- Jain, G.; Stuendl, A.; Rao, P.; Berulava, T.; Pena Centeno, T.; Kaurani, L.; Burkhardt, S.; Delalle, I.; Kornhuber, J.; Hull, M.; et al. A combined miRNA-piRNA signature to detect Alzheimer’s disease. Transl. Psychiatry 2019, 9, 250. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Investig. 2019, 130, 404–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Song, Y.; Han, X.; Qu, P.; Wang, W. Long noncoding RNA PTENP1 affects the recovery of spinal cord injury by regulating the expression of miR-19b and miR-21. J. Cell. Physiol. 2019, 235, 3634–3645. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, J.; Zhou, X.; Xiong, Z.; Zhao, J.; Yu, R.; Huang, F.; Zhang, H.; Chen, L. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci. Rep. 2016, 6, 32993. [Google Scholar] [CrossRef]
- McDonald, M.K.; Tian, Y.; Qureshi, R.A.; Gormley, M.; Ertel, A.; Gao, R.; Aradillas Lopez, E.; Alexander, G.M.; Sacan, A.; Fortina, P.; et al. Functional significance of macrophage-derived exosomes in inflammation and pain. Pain 2014, 155, 1527–1539. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, M.; Gong, A.; Zhang, X.; Wu, X.; Zhu, Y.; Shi, H.; Wu, L.; Zhu, W.; Qian, H.; et al. HucMSC-Exosome Mediated-Wnt4 Signaling Is Required for Cutaneous Wound Healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, L.; Deepthi, S.; Kgk, D.; Malla, R.R. Current Perspectives of Exosomes as Therapeutic Targets and Drug Delivery Vehicles for Pancreatic Cancer. Crit. Rev. Oncog. 2019, 24, 179–190. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, H.; Wang, X.; Rai, A.; Groot, M.; Jin, Y. Exosome-Mediated Small RNA Delivery: A Novel Therapeutic Approach for Inflammatory Lung Responses. Mol. Ther. 2018, 26, 2119–2130. [Google Scholar] [CrossRef]
- Gonzalez, E.; Falcon-Perez, J.M. Cell-derived extracellular vesicles as a platform to identify low-invasive disease biomarkers. Expert Rev. Mol. Diagn. 2015, 15, 907–923. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Han, K.Y.; Tran, J.A.; Chang, J.H.; Azar, D.T.; Zieske, J.D. Potential role of corneal epithelial cell-derived exosomes in corneal wound healing and neovascularization. Sci. Rep. 2017, 7, 40548. [Google Scholar] [CrossRef] [PubMed]
- Samaeekia, R.; Rabiee, B.; Putra, I.; Shen, X.; Park, Y.J.; Hematti, P.; Eslani, M.; Djalilian, A.R. Effect of Human Corneal Mesenchymal Stromal Cell-derived Exosomes on Corneal Epithelial Wound Healing. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5194–5200. [Google Scholar] [CrossRef] [PubMed]
- Ueno, M.; Asada, K.; Toda, M.; Nagata, K.; Sotozono, C.; Kosaka, N.; Ochiya, T.; Kinoshita, S.; Hamuro, J. Concomitant Evaluation of a Panel of Exosome Proteins and MiRs for Qualification of Cultured Human Corneal Endothelial Cells. Investig. Ophthalmol. Vis. Sci. 2016, 57, 4393–4402. [Google Scholar] [CrossRef] [PubMed]
- Zieske, J.D.; Hutcheon, A.E.K.; Guo, X. Extracellular Vesicles and Cell-Cell Communication in the Cornea. Anat. Rec. 2020, 303, 1727–1734. [Google Scholar] [CrossRef]
- Leszczynska, A.; Kulkarni, M.; Ljubimov, A.V.; Saghizadeh, M. Exosomes from normal and diabetic human corneolimbal keratocytes differentially regulate migration, proliferation and marker expression of limbal epithelial cells. Sci. Rep. 2018, 8, 15173. [Google Scholar] [CrossRef]
- Escandon, P.; Liu, A.; Nicholas, S.E.; Khan, A.; Riaz, K.M.; Karamichos, D. Unravelling Novel Roles of Salivary Exosomes in the Regulation of Human Corneal Stromal Cell Migration and Wound Healing. Int. J. Mol. Sci. 2022, 23, 4330. [Google Scholar] [CrossRef]
- Ma, S.; Yin, J.; Hao, L.; Liu, X.; Shi, Q.; Diao, Y.; Yu, G.; Liu, L.; Chen, J.; Zhong, J. Exosomes from Human Umbilical Cord Mesenchymal Stem Cells Treat Corneal Injury via Autophagy Activation. Front. Bioeng. Biotechnol. 2022, 10, 879192. [Google Scholar] [CrossRef]
- Wang, S.; Hou, Y.; Li, X.; Song, Z.; Sun, B.; Li, X.; Zhang, H. Comparison of exosomes derived from induced pluripotent stem cells and mesenchymal stem cells as therapeutic nanoparticles for treatment of corneal epithelial defects. Aging 2020, 12, 19546–19562. [Google Scholar] [CrossRef] [PubMed]
- Edgar, J.R. Q&A: What are exosomes, exactly? BMC Biol. 2016, 14, 46. [Google Scholar]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Atkinson, M.A.; Appella, E.; Corigliano-Murphy, M.A.; Korn, E.D. Enzymatic activity and filament assembly of Acanthamoeba myosin II are regulated by adjacent domains at the end of the tail. FEBS Lett. 1988, 234, 435–438. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Consortium, E.-T.; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, O.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Cunha, G.M.; Na, K.S.; Putra, I.; Lee, H.J.; Hull, S.; Cheng, Y.C.; Blanco, I.J.; Eslani, M.; Djalilian, A.R.; Myung, D. Corneal Wound Healing Effects of Mesenchymal Stem Cell Secretome Delivered Within a Viscoelastic Gel Carrier. Stem Cells Transl. Med. 2019, 8, 478–489. [Google Scholar] [CrossRef]
- Mobaraki, M.; Abbasi, R.; Omidian Vandchali, S.; Ghaffari, M.; Moztarzadeh, F.; Mozafari, M. Corneal Repair and Regeneration: Current Concepts and Future Directions. Front. Bioeng. Biotechnol. 2019, 7, 135. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, X.H.; Luo, C.L.; Zhang, J.M.; He, B.C.; Chen, G. Interleukin-12-anchored exosomes increase cytotoxicity of T lymphocytes by reversing the JAK/STAT pathway impaired by tumor-derived exosomes. Int. J. Mol. Med. 2010, 25, 695–700. [Google Scholar]
- Feng, N.; Jia, Y.; Huang, X. Exosomes from adipose-derived stem cells alleviate neural injury caused by microglia activation via suppressing NF-kB and MAPK pathway. J. Neuroimmunol. 2019, 334, 576996. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, H.; Kong, Y. Exosomes derived from platelet-rich plasma activate YAP and promote the fibrogenic activity of Muller cells via the PI3K/Akt pathway. Exp. Eye Res. 2020, 193, 107973. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Pilgaard, L.; Moos, T.; Duroux, M. A comprehensive overview of exosomes as drug delivery vehicles—Endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta 2014, 1846, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Raiker, R.S.; Jay, S.M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharm. 2015, 12, 3650–3657. [Google Scholar] [CrossRef] [PubMed]
- Meehan, K.; Vella, L.J. The contribution of tumour-derived exosomes to the hallmarks of cancer. Crit. Rev. Clin. Lab. Sci. 2016, 53, 121–131. [Google Scholar] [CrossRef]
- Shen, T.; Zheng, Q.Q.; Shen, J.; Li, Q.S.; Song, X.H.; Luo, H.B.; Hong, C.Y.; Yao, K. Effects of Adipose-derived Mesenchymal Stem Cell Exosomes on Corneal Stromal Fibroblast Viability and Extracellular Matrix Synthesis. Chin. Med. J. 2018, 131, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Wahlgren, J.; Statello, L.; Skogberg, G.; Telemo, E.; Valadi, H. Delivery of Small Interfering RNAs to Cells via Exosomes. Methods Mol. Biol. 2016, 1364, 105–125. [Google Scholar] [PubMed]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Rani, S.; Ryan, A.E.; Griffin, M.D.; Ritter, T. Mesenchymal Stem Cell-derived Extracellular Vesicles: Toward Cell-free Therapeutic Applications. Mol. Ther. 2015, 23, 812–823. [Google Scholar] [CrossRef]
- Guo, S.C.; Tao, S.C.; Yin, W.J.; Qi, X.; Yuan, T.; Zhang, C.Q. Exosomes derived from platelet-rich plasma promote the re-epithelization of chronic cutaneous wounds via activation of YAP in a diabetic rat model. Theranostics 2017, 7, 81–96. [Google Scholar] [CrossRef]
- Hu, Y.; Rao, S.S.; Wang, Z.X.; Cao, J.; Tan, Y.J.; Luo, J.; Li, H.M.; Zhang, W.S.; Chen, C.Y.; Xie, H. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through miR-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics 2018, 8, 169–184. [Google Scholar] [CrossRef]
- Shi, H.; Xu, X.; Zhang, B.; Xu, J.; Pan, Z.; Gong, A.; Zhang, X.; Li, R.; Sun, Y.; Yan, Y.; et al. 3,3′-Diindolylmethane stimulates exosomal Wnt11 autocrine signaling in human umbilical cord mesenchymal stem cells to enhance wound healing. Theranostics 2017, 7, 1674–1688. [Google Scholar] [CrossRef]
- McKay, T.B.; Hutcheon, A.E.K.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Epithelial Cells Promote Myofibroblast Differentiation. Cells 2020, 9, 1080. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Borin, T.F.; Ara, R.; Angara, K.; Cai, J.; Achyut, B.R.; Liu, Y.; Arbab, A.S. Differential in vivo biodistribution of (131)I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomedicine 2019, 21, 102072. [Google Scholar] [CrossRef]
- Oh, M.; Lee, J.; Kim, Y.J.; Rhee, W.J.; Park, J.H. Exosomes Derived from Human Induced Pluripotent Stem Cells Ameliorate the Aging of Skin Fibroblasts. Int. J. Mol. Sci. 2018, 19, 1715. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Liu, Z.; Wu, M.; Sun, M.; Xia, Y.; Wang, Y. Comparison of Proangiogenic Effects of Adipose-Derived Stem Cells and Foreskin Fibroblast Exosomes on Artificial Dermis Prefabricated Flaps. Stem Cells Int. 2020, 2020, 5293850. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Brandao, B.B.; Thomou, T.; Altindis, E.; Kahn, C.R. Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep. 2022, 38, 110277. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Zhou, X.; Li, S.; Jiang, W.; Li, T.; Wang, N.; Xiao, N. Activation of the AKT/GSK-3beta/beta-catenin pathway via photobiomodulation therapy promotes neural stem cell proliferation in neonatal rat models of hypoxic-ischemic brain damage. Ann. Transl. Med. 2022, 10, 55. [Google Scholar] [CrossRef]
- Meng, J.; Liu, H.L.; Ma, D.; Wang, H.Y.; Peng, Y.; Wang, H.L. Upregulation of aurora kinase A promotes vascular smooth muscle cell proliferation and migration by activating the GSK-3beta/beta-catenin pathway in aortic-dissecting aneurysms. Life Sci. 2020, 262, 118491. [Google Scholar] [CrossRef]
- Sun, J.; Yan, P.; Chen, Y.; Chen, Y.; Yang, J.; Xu, G.; Mao, H.; Qiu, Y. MicroRNA-26b inhibits cell proliferation and cytokine secretion in human RASF cells via the Wnt/GSK-3beta/beta-catenin pathway. Diagn. Pathol. 2015, 10, 72. [Google Scholar] [CrossRef]
- Yun, S.H.; Park, J.I. PGC-1alpha Regulates Cell Proliferation and Invasion via AKT/GSK-3beta/beta-catenin Pathway in Human Colorectal Cancer SW620 and SW480 Cells. Anticancer Res. 2020, 40, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yin, J.; Guan, J.; Hu, B.; Niu, X.; Jin, D.; Wang, Y.; Zhang, C. Lithium stimulates human bone marrow derived mesenchymal stem cell proliferation through GSK-3beta-dependent beta-catenin/Wnt pathway activation. FEBS J. 2014, 281, 5371–5389. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Cheng, Y.; Dai, S.; Zou, B.; Guo, X. Suppression of rhomboid domain-containing 1 produces anticancer effects in pancreatic adenocarcinoma through affection of the AKT/GSK-3beta/beta-catenin pathway. Environ. Toxicol. 2022, 37, 1944–1956. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Fan, Z.; Wang, Z.; Dai, Q.; Xiang, Z.; Yuan, F.; Yan, M.; Zhu, Z.; Liu, B.; Li, C. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3beta/beta-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 52. [Google Scholar] [CrossRef]
- Tan, D.; Zhang, Y. Silencing of Nudix type 5 represses proliferation and invasion and enhances chemosensitivity of gastric carcinoma cells by affecting the AKT/GSK-3beta/beta-catenin pathway. Toxicol. Appl. Pharmacol. 2022, 441, 115968. [Google Scholar] [CrossRef]
- Yang, L.; Tan, W.; Wei, Y.; Xie, Z.; Li, W.; Ma, X.; Wang, Q.; Li, H.; Zhang, Z.; Shang, C.; et al. CircLIFR suppresses hepatocellular carcinoma progression by sponging miR-624-5p and inactivating the GSK-3beta/beta-catenin signaling pathway. Cell Death Dis. 2022, 13, 464. [Google Scholar] [CrossRef]
- Zhang, P.; Tian, Q.; Gao, H.; Zhao, A.; Shao, Y.; Yang, J. Inhibition of MAC30 exerts antitumor effects in nasopharyngeal carcinoma via affecting the Akt/GSK-3beta/beta-catenin pathway. J. Biochem. Mol. Toxicol. 2022, 36, e23061. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Y.; Semenov, M.; Han, C.; Baeg, G.H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef]
- Salahshor, S.; Woodgett, J.R. The links between axin and carcinogenesis. J. Clin. Pathol. 2005, 58, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Caldwell, R.B.; Behzadian, M.A. Blockade of VEGF-induced GSK/beta-catenin signaling, uPAR expression and increased permeability by dominant negative p38alpha. Exp. Eye Res. 2012, 100, 101–108. [Google Scholar] [CrossRef]
- Yong, J.; von Bremen, J.; Ruiz-Heiland, G.; Ruf, S. Adiponectin as Well as Compressive Forces Regulate in vitro beta-Catenin Expression on Cementoblasts via Mitogen-Activated Protein Kinase Signaling Activation. Front. Cell Dev. Biol. 2021, 9, 645005. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Feigin, M.E.; Malbon, C.C. p38 mitogen-activated protein kinase regulates canonical Wnt-beta-catenin signaling by inactivation of GSK3beta. J. Cell Sci. 2008, 121 Pt 21, 3598–3607. [Google Scholar] [CrossRef]
- Li, S.; Gallup, M.; Chen, Y.T.; McNamara, N.A. Molecular mechanism of proinflammatory cytokine-mediated squamous metaplasia in human corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2466–2475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, M.; Zhang, W.; Lu, X.Y. Adiponectin stimulates proliferation of adult hippocampal neural stem/progenitor cells through activation of p38 mitogen-activated protein kinase (p38MAPK)/glycogen synthase kinase 3beta (GSK-3beta)/beta-catenin signaling cascade. J. Biol. Chem. 2011, 286, 44913–44920. [Google Scholar] [CrossRef] [PubMed]
- Peake, P.W.; Kriketos, A.D.; Campbell, L.V.; Shen, Y.; Charlesworth, J.A. The metabolism of isoforms of human adiponectin: Studies in human subjects and in experimental animals. Eur. J. Endocrinol. 2005, 153, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, L.; Scherer, P.E. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 1998, 8, 335–338. [Google Scholar] [CrossRef]
- Shikama, Y.; Kurosawa, M.; Furukawa, M.; Ishimaru, N.; Matsushita, K. Involvement of adiponectin in age-related increases in tear production in mice. Aging 2019, 11, 8329–8346. [Google Scholar] [CrossRef]
- Li, Y.; Jin, R.; Li, L.; Hsu, H.H.; You, I.C.; Yoon, H.J.; Yoon, K.C. Therapeutic Effect of Topical Adiponectin-Derived Short Peptides Compared with Globular Adiponectin in Experimental Dry Eye and Alkali Burn. J. Ocul. Pharmacol. Ther. 2020, 36, 88–96. [Google Scholar] [CrossRef]
- Baradaran-Rafii, A.; Ashnagar, A.; Heidari Keshel, S.; Jabbehdari, S.; Baradaran-Rafii, G. Regression of corneal neovascularization: Adiponectin versus bevacizumab eye drops. Eur. J. Ophthalmol. 2021, 31, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cui, L.; Yang, J.M.; Lee, H.S.; Choi, J.S.; Woo, J.M.; Lim, S.K.; Yoon, K.C. The Wound Healing Effects of Adiponectin Eye Drops after Corneal Alkali Burn. Curr. Eye Res. 2016, 41, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Kita, S.; Koyama, Y.; Fukuda, S.; Takeda, H.; Takahashi, M.; Fujishima, Y.; Nagao, H.; Masuda, S.; Tanaka, Y.; et al. Adiponectin/T-cadherin system enhances exosome biogenesis and decreases cellular ceramides by exosomal release. JCI Insight 2018, 3, e99680. [Google Scholar] [CrossRef] [PubMed]
- Acunzo, J.; Andrieu, C.; Baylot, V.; So, A.; Rocchi, P. Hsp27 as a therapeutic target in cancers. Curr. Drug Targets 2014, 15, 423–431. [Google Scholar] [CrossRef]
- Jego, G.; Hazoume, A.; Seigneuric, R.; Garrido, C. Targeting heat shock proteins in cancer. Cancer Lett. 2013, 332, 275–285. [Google Scholar] [CrossRef]
- Han, L.; Jiang, Y.; Han, D.; Tan, W. Hsp27 regulates epithelial mesenchymal transition, metastasis and proliferation in colorectal carcinoma. Oncol. Lett. 2018, 16, 5309–5316. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liu, W.; Zhang, S.; Pu, L.; Deng, B.; Zeng, Q.; Chen, Z.; Wang, X. Downregulation of miR-541 induced by heat stress contributes to malignant transformation of human bronchial epithelial cells via HSP27. Environ. Res. 2020, 184, 108954. [Google Scholar] [CrossRef]
- Sims, J.T.; Ganguly, S.S.; Bennett, H.; Friend, J.W.; Tepe, J.; Plattner, R. Imatinib reverses doxorubicin resistance by affecting activation of STAT3-dependent NF-kappaB and HSP27/p38/AKT pathways and by inhibiting ABCB1. PLoS ONE 2013, 8, e55509. [Google Scholar] [CrossRef] [PubMed]
- Joyce, N.C. Proliferative capacity of the corneal endothelium. Prog. Retin. Eye Res. 2003, 22, 359–389. [Google Scholar] [CrossRef]
- Imanishi, J.; Kamiyama, K.; Iguchi, I.; Kita, M.; Sotozono, C.; Kinoshita, S. Growth factors: Importance in wound healing and maintenance of transparency of the cornea. Prog. Retin. Eye Res. 2000, 19, 113–129. [Google Scholar] [CrossRef]
- Li, D.Q.; Tseng, S.C. Three patterns of cytokine expression potentially involved in epithelial-fibroblast interactions of human ocular surface. J. Cell. Physiol. 1995, 163, 61–79. [Google Scholar] [CrossRef]
- Wilson, S.E.; Chen, L.; Mohan, R.R.; Liang, Q.; Liu, J. Expression of HGF, KGF, EGF and receptor messenger RNAs following corneal epithelial wounding. Exp. Eye Res. 1999, 68, 377–397. [Google Scholar] [CrossRef]
- Stepp, M.A. Corneal integrins and their functions. Exp. Eye Res. 2006, 83, 3–15. [Google Scholar] [CrossRef]
- McKay, T.B.; Yeung, V.; Hutcheon, A.E.K.; Guo, X.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles in the Cornea: Insights from Other Tissues. Anal. Cell. Pathol. 2021, 2021, 9983900. [Google Scholar] [CrossRef]
- Theriault, M.; Gendron, S.P.; Brunette, I.; Rochette, P.J.; Proulx, S. Function-Related Protein Expression in Fuchs Endothelial Corneal Dystrophy Cells and Tissue Models. Am. J. Pathol. 2018, 188, 1703–1712. [Google Scholar] [CrossRef]
- Peh, G.S.; Chng, Z.; Ang, H.P.; Cheng, T.Y.; Adnan, K.; Seah, X.Y.; George, B.L.; Toh, K.P.; Tan, D.T.; Yam, G.H.; et al. Propagation of human corneal endothelial cells: A novel dual media approach. Cell Transplant. 2015, 24, 287–304. [Google Scholar] [CrossRef]
- Bisson, F.; Rochefort, E.; Lavoie, A.; Larouche, D.; Zaniolo, K.; Simard-Bisson, C.; Damour, O.; Auger, F.A.; Guerin, S.L.; Germain, L. Irradiated human dermal fibroblasts are as efficient as mouse fibroblasts as a feeder layer to improve human epidermal cell culture lifespan. Int. J. Mol. Sci. 2013, 14, 4684–4704. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 3, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Le-Bel, G.; Cortez Ghio, S.; Larouche, D.; Germain, L.; Guerin, S.L. Qualitatively Monitoring Binding and Expression of the Transcription Factors Sp1 and NFI as a Useful Tool to Evaluate the Quality of Primary Cultured Epithelial Stem Cells in Tissue Reconstruction. Methods Mol. Biol. 2019, 1879, 43–73. [Google Scholar]
- Couture, C.; Zaniolo, K.; Carrier, P.; Lake, J.; Patenaude, J.; Germain, L.; Guerin, S.L. The tissue-engineered human cornea as a model to study expression of matrix metalloproteinases during corneal wound healing. Biomaterials 2016, 78, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; Couture, C.; Germain, L.; Guerin, S.L. Contribution of the WNK1 kinase to corneal wound healing using the tissue-engineered human cornea as an in vitro model. J. Tissue Eng. Regen. Med. 2019, 13, 1595–1608. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gill, E.E.; Hancock, R.E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desjardins, P.; Berthiaume, R.; Couture, C.; Le-Bel, G.; Roy, V.; Gros-Louis, F.; Moulin, V.J.; Proulx, S.; Chemtob, S.; Germain, L.; et al. Impact of Exosomes Released by Different Corneal Cell Types on the Wound Healing Properties of Human Corneal Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 12201. https://doi.org/10.3390/ijms232012201
Desjardins P, Berthiaume R, Couture C, Le-Bel G, Roy V, Gros-Louis F, Moulin VJ, Proulx S, Chemtob S, Germain L, et al. Impact of Exosomes Released by Different Corneal Cell Types on the Wound Healing Properties of Human Corneal Epithelial Cells. International Journal of Molecular Sciences. 2022; 23(20):12201. https://doi.org/10.3390/ijms232012201
Chicago/Turabian StyleDesjardins, Pascale, Rébecca Berthiaume, Camille Couture, Gaëtan Le-Bel, Vincent Roy, François Gros-Louis, Véronique J. Moulin, Stéphanie Proulx, Sylvain Chemtob, Lucie Germain, and et al. 2022. "Impact of Exosomes Released by Different Corneal Cell Types on the Wound Healing Properties of Human Corneal Epithelial Cells" International Journal of Molecular Sciences 23, no. 20: 12201. https://doi.org/10.3390/ijms232012201
APA StyleDesjardins, P., Berthiaume, R., Couture, C., Le-Bel, G., Roy, V., Gros-Louis, F., Moulin, V. J., Proulx, S., Chemtob, S., Germain, L., & Guérin, S. L. (2022). Impact of Exosomes Released by Different Corneal Cell Types on the Wound Healing Properties of Human Corneal Epithelial Cells. International Journal of Molecular Sciences, 23(20), 12201. https://doi.org/10.3390/ijms232012201








