Characterization and Involvement of Exosomes Originating from Chikungunya Virus-Infected Epithelial Cells in the Transmission of Infectious Viral Elements
Abstract
1. Introduction
2. Results
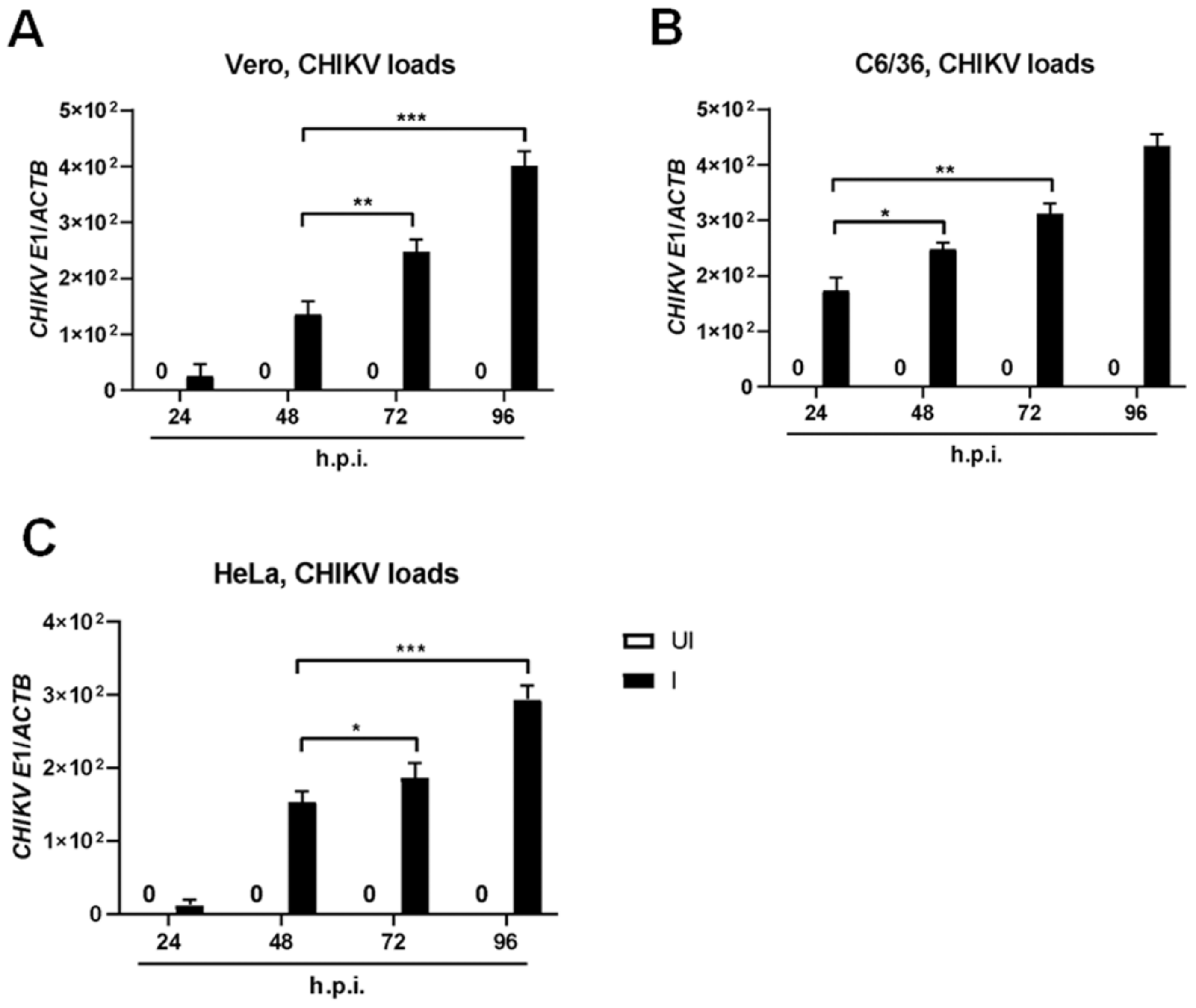
2.1. Susceptibility Profiles of the Established Epithelial Cell Lines to CHIKV Infection
2.2. CHIKV Specifically Facilitates the EV Population
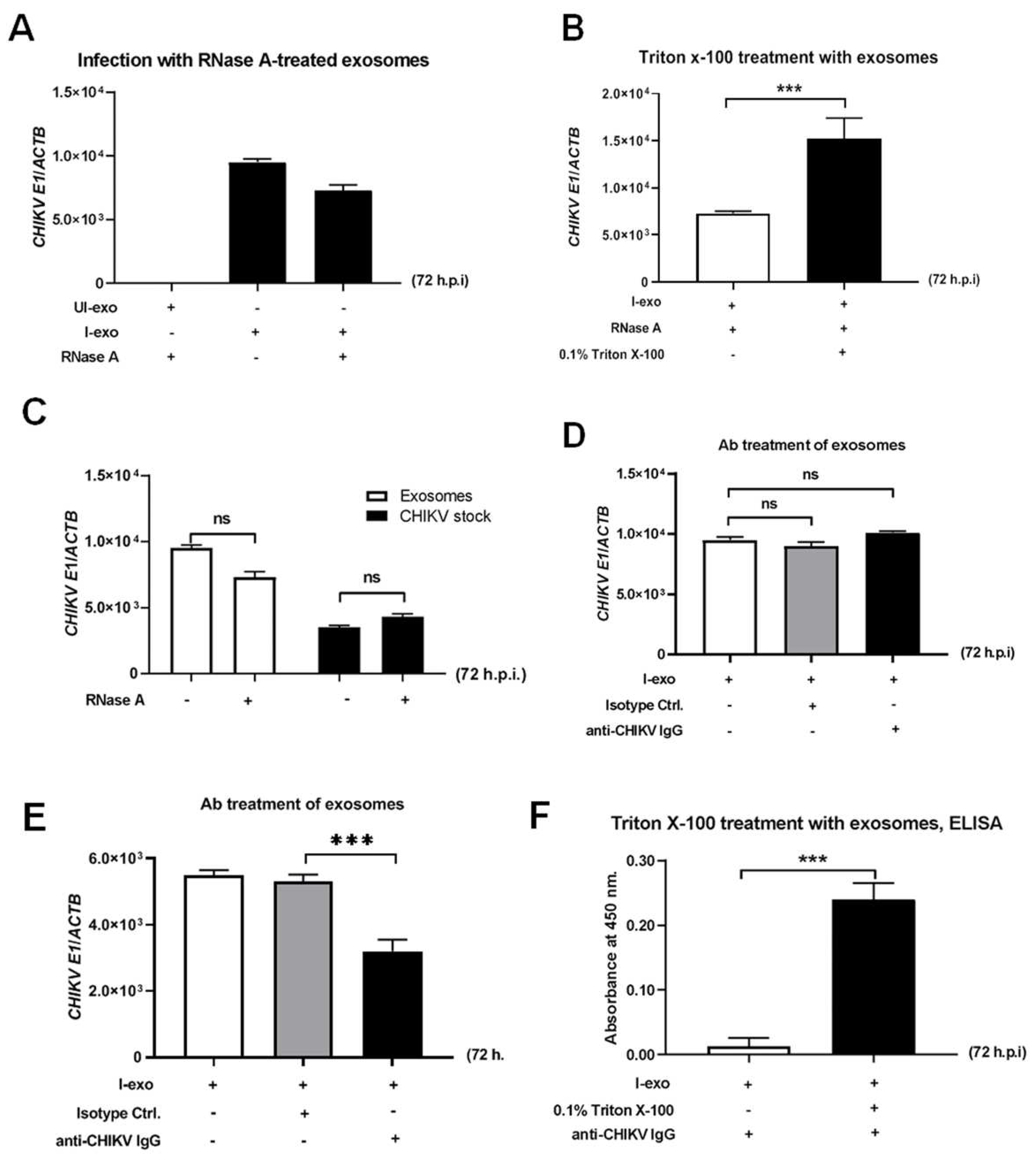
2.3. CHIKV RNA Elements Are Encapsulated by Secreted Exosomes and Are Infectious
2.4. CHIKV RNA Elements and Proteins Are Securely Contained Inside the Exosomes
2.5. CHIKV Infectious Elements Are Transmitted to Neighboring Cells through Exosomes
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cultures
4.2. Preparation of Extracellular Vesicle-Depleted FBS
4.3. Propagation of Laboratory CHIKV Stock
4.4. Infection of CHIKV to Vero Cell Line
4.5. Exosomes Isolation and Enrichment
4.6. Exosome Purification
4.7. Physical Characterization of Isolated Exosomes
4.8. Determination of Exosomes Markers by Immunoblotting
4.9. Detection of CHIKV Genomic Materials in the Cells and Exosomes
4.10. Triton X-100 Treatment of Small EVs
4.11. Exosomes Infectious Assay
4.12. Analysis of EV Transfer CHIKV RNA Elements and Proteins
4.13. Inhibition of Exosome Production by GW4869 Inhibitor
4.14. Transwell Assay
4.15. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwartz, O.; Albert, M.L. Biology and Pathogenesis of Chikungunya Virus. Nat. Rev. Microbiol. 2010, 8, 491–500. [Google Scholar] [CrossRef]
- Mathew, A.J.; Ganapati, A.; Kabeerdoss, J.; Nair, A.; Gupta, N.; Chebbi, P.; Mandal, S.K.; Danda, D. Chikungunya Infection: A Global Public Health Menace. Curr. Allergy Asthma Rep. 2017, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Mateo, L.; Roure, S. Chronic Arthritis in Chikungunya Virus Infection. Reumatol Clin. 2019, 15, 113–116. [Google Scholar] [CrossRef]
- Peters, C.M.M.; Pijnacker, R.; Fanoy, E.B.; Bouwman, L.J.T.; de Langen, L.E.; van den Kerkhof, J.H.T.C.; Reimerink, J.; Pilot, E.; Henry, M.; Oostburg, V.A.; et al. Chikungunya Virus Outbreak in Sint Maarten: Long-Term Arthralgia after a 15-Month Period. J. Vector Borne Dis. 2018, 55, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Kennedy Amaral Pereira, J.; Schoen, R.T. Management of Chikungunya Arthritis. Clin. Rheumatol. 2017, 36, 2179–2186. [Google Scholar] [CrossRef]
- Zaid, A.; Gerardin, P.; Taylor, A.; Mostafavi, H.; Malvy, D.; Mahalingam, S. Chikungunya Arthritis: Implications of Acute and Chronic Inflammation Mechanisms on Disease Management. Arthritis Rheumatol. 2018, 70, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Assunção-Miranda, I.; Cruz-Oliveira, C.; Da Poian, A.T. Molecular Mechanisms Involved in the Pathogenesis of Alphavirus-Induced Arthritis. BioMed Res. Int. 2013, 2013, 973516. [Google Scholar] [CrossRef]
- Lidbury, B.A.; Rulli, N.E.; Suhrbier, A.; Smith, P.N.; McColl, S.R.; Cunningham, A.L.; Tarkowski, A.; van Rooijen, N.; Fraser, R.J.; Mahalingam, S. Macrophage-Derived Proinflammatory Factors Contribute to the Development of Arthritis and Myositis after Infection with an Arthrogenic Alphavirus. J. Infect. Dis. 2008, 197, 1585–1593. [Google Scholar] [CrossRef]
- Chirathaworn, C.; Poovorawan, Y.; Lertmaharit, S.; Wuttirattanakowit, N. Cytokine Levels in Patients with Chikungunya Virus Infection. Asian Pac. J. Trop. Med. 2013, 6, 631–634. [Google Scholar] [CrossRef]
- Chow, A.; Her, Z.; Ong, E.K.S.; Chen, J.-m.; Dimatatac, F.; Kwek, D.J.C.; Barkham, T.; Yang, H.; Rénia, L.; Leo, Y.-S.; et al. Persistent Arthralgia Induced by Chikungunya Virus Infection Is Associated with Interleukin-6 and Granulocyte Macrophage Colony-Stimulating Factor. J. Infect. Dis. 2011, 203, 149–157. [Google Scholar] [CrossRef]
- Reddy, V.; Desai, A.; Krishna, S.S.; Vasanthapuram, R. Molecular Mimicry between Chikungunya Virus and Host Components: A Possible Mechanism for the Arthritic Manifestations. PLoS Negl. Trop. Dis. 2017, 11, e0005238. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.A.C.; Prow, N.A.; Schroder, W.A.; Ellis, J.J.; Cumming, H.E.; Gearing, L.J.; Poo, Y.S.; Taylor, A.; Hertzog, P.J.; Di Giallonardo, F.; et al. Rna-Seq Analysis of Chikungunya Virus Infection and Identification of Granzyme a as a Major Promoter of Arthritic Inflammation. PLoS Pathog. 2017, 13, e1006155. [Google Scholar] [CrossRef]
- McCarthy, M.K.; Morrison, T.E. Chronic Chikungunya Virus Musculoskeletal Disease: What Are the Underlying Mechanisms? Future Microbiol. 2016, 11, 331–334. [Google Scholar] [CrossRef]
- Tanay, A. Chikungunya Virus and Autoimmunity. Curr. Opin. Rheumatol. 2017, 29, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of Extracellular Vesicles (Ev): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current Methods for the Isolation of Extracellular Vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Altan-Bonnet, N.; Perales, C.; Domingo, E. Extracellular Vesicles: Vehicles of En Bloc Viral Transmission. Virus Res. 2019, 265, 143–149. [Google Scholar] [CrossRef]
- Nour, A.M.; Modis, Y. Endosomal Vesicles as Vehicles for Viral Genomes. Trends Cell Biol. 2014, 24, 449–454. [Google Scholar] [CrossRef]
- Chahar, H.S.; Bao, X.; Casola, A. Exosomes and Their Role in the Life Cycle and Pathogenesis of Rna Viruses. Viruses 2015, 7, 3204–3225. [Google Scholar] [CrossRef]
- Meckes, D.G., Jr.; Raab-Traub, N. Microvesicles and Viral Infection. J. Virol. 2011, 85, 12844–12854. [Google Scholar] [CrossRef]
- Vora, A.; Zhou, W.; Londono-Renteria, B.; Woodson, M.; Sherman, M.B.; Colpitts, T.M.; Neelakanta, G.; Sultana, H. Arthropod Evs Mediate Dengue Virus Transmission through Interaction with a Tetraspanin Domain Containing Glycoprotein Tsp29fb. Proc. Natl. Acad. Sci. USA 2018, 115, E6604–E6613. [Google Scholar] [CrossRef]
- Torii, S.; Orba, Y.; Sasaki, M.; Tabata, K.; Wada, Y.; Carr, M.; Hobson-Peters, J.; Hall, R.A.; Takada, A.; Fukuhara, T.; et al. Host Escrt Factors Are Recruited During Chikungunya Virus Infection and Are Required for the Intracellular Viral Replication Cycle. J. Biol. Chem. 2020, 295, 7941–7957. [Google Scholar] [CrossRef]
- Lum, F.M.; Couderc, T.; Chia, B.S.; Ong, R.Y.; Her, Z.; Chow, A.; Leo, Y.S.; Kam, Y.W.; Rénia, L.; Lecuit, M.; et al. Antibody-Mediated Enhancement Aggravates Chikungunya Virus Infection and Disease Severity. Sci. Rep. 2018, 8, 1860. [Google Scholar] [CrossRef]
- Krejbich-Trotot, P.; Denizot, M.; Hoarau, J.J.; Jaffar-Bandjee, M.C.; Das, T.; Gasque, P. Chikungunya Virus Mobilizes the Apoptotic Machinery to Invade Host Cell Defenses. FASEB J. 2011, 25, 314–325. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete Viral Genome Sequence and Discovery of Novel Viruses by Deep Sequencing of Small Rnas: A Generic Method for Diagnosis, Discovery and Sequencing of Viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef]
- Sourisseau, M.; Schilte, C.; Casartelli, N.; Trouillet, C.; Guivel-Benhassine, F.; Rudnicka, D.; Sol-Foulon, N.; Le Roux, K.; Prevost, M.C.; Fsihi, H.; et al. Characterization of Reemerging Chikungunya Virus. PLoS Pathog. 2007, 3, e89. [Google Scholar] [CrossRef] [PubMed]
- Couderc, T.; Chretien, F.; Schilte, C.; Disson, O.; Brigitte, M.; Guivel-Benhassine, F.; Touret, Y.; Barau, G.; Cayet, N.; Schuffenecker, I.; et al. A Mouse Model for Chikungunya: Young Age and Inefficient Type-I Interferon Signaling Are Risk Factors for Severe Disease. PLoS Pathog. 2008, 4, e29. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. The Cell Biology of Chikungunya Virus Infection. Cell Microbiol. 2012, 14, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Thery, C. Exosomes: Secreted Vesicles and Intercellular Communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human Colon Cancer Cell Line Lim1863-Derived Exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef]
- Schageman, J.; Zeringer, E.; Li, M.; Barta, T.; Lea, K.; Gu, J.; Magdaleno, S.; Setterquist, R.; Vlassov, A.V. The Complete Exosome Workflow Solution: From Isolation to Characterization of Rna Cargo. BioMed Res. Int. 2013, 2013, 253957. [Google Scholar] [CrossRef]
- Stapleford, K.A.; Moratorio, G.; Henningsson, R.; Chen, R.; Matheus, S.; Enfissi, A.; Weissglas-Volkov, D.; Isakov, O.; Blanc, H.; Mounce, B.C.; et al. Whole-Genome Sequencing Analysis from the Chikungunya Virus Caribbean Outbreak Reveals Novel Evolutionary Genomic Elements. PLoS Negl. Trop. Dis. 2016, 10, e0004402. [Google Scholar] [CrossRef]
- Bello-Morales, R.; Praena, B.; de la Nuez, C.; Rejas, M.T.; Guerra, M.; Galán-Ganga, M.; Izquierdo, M.; Calvo, V.; Krummenacher, C.; López-Guerrero, J.A. Role of Microvesicles in the Spread of Herpes Simplex Virus 1 in Oligodendrocytic Cells. J. Virol. 2018, 92, 10. [Google Scholar] [CrossRef]
- Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; De Jesús-González, L.A.; Palacios-Rápalo, S.N.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Hurtado-Monzón, A.M.; Gallardo-Flores, C.E.; Alcaraz-Estrada, S.L.; Salas-Benito, J.S.; et al. The Regulation of Flavivirus Infection by Hijacking Exosome-Mediated Cell-Cell Communication: New Insights on Virus-Host Interactions. Viruses 2020, 12, 765. [Google Scholar] [CrossRef]
- York, S.B.; Sun, L.; Cone, A.S.; Duke, L.C.; Cheerathodi, M.R.; Meckes, D.G., Jr. Zika Virus Hijacks Extracellular Vesicle Tetraspanin Pathways for Cell-to-Cell Transmission. mSphere 2021, 6, e0019221. [Google Scholar] [CrossRef] [PubMed]
- Bello-Morales, R.; Ripa, I.; López-Guerrero, J.A. Extracellular Vesicles in Viral Spread and Antiviral Response. Viruses 2020, 12, 623. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Alladi, P.A.; Narayanappa, G.; Vasanthapuram, R.; Desai, A. The Time Course Analysis of Morphological Changes Induced by Chikungunya Virus Replication in Mammalian and Mosquito Cells. Acta Virol. 2018, 62, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Higashi, N.; Matsumoto, A.; Tabata, K.; Nagatomo, Y. Electron Microscope Study of Development of Chikungunya Virus in Green Monkey Kidney Stable (Vero) Cells. Virology 1967, 33, 55–69. [Google Scholar] [CrossRef]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; de Ruiter, P.E.; Willemsen, R.; Demmers, J.A.; Raj, V.S.; Jenster, G.; et al. Exosome-Mediated Transmission of Hepatitis C Virus between Human Hepatoma Huh7.5 Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef]
- Yang, Y.; Han, Q.; Hou, Z.; Zhang, C.; Tian, Z.; Zhang, J. Exosomes Mediate Hepatitis B Virus (Hbv) Transmission and Nk-Cell Dysfunction. Cell Mol. Immunol. 2017, 14, 465–475. [Google Scholar] [CrossRef]
- Chahar, H.S.; Corsello, T.; Kudlicki, A.S.; Komaravelli, N.; Casola, A. Respiratory Syncytial Virus Infection Changes Cargo Composition of Exosome Released from Airway Epithelial Cells. Sci. Rep. 2018, 8, 387. [Google Scholar] [CrossRef]
- Wang, T.; Fang, L.; Zhao, F.; Wang, D.; Xiao, S. Exosomes Mediate Intercellular Transmission of Porcine Reproductive and Respiratory Syndrome Virus. J. Virol. 2018, 92, e0173417. [Google Scholar] [CrossRef]
- Martínez-Rojas, P.P.; Quiroz-García, E.; Monroy-Martínez, V.; Agredano-Moreno, L.T.; Jiménez-García, L.F.; Ruiz-Ordaz, B.H. Participation of Extracellular Vesicles from Zika-Virus-Infected Mosquito Cells in the Modification of Naïve Cells’ Behavior by Mediating Cell-to-Cell Transmission of Viral Elements. Cells 2020, 9, 123. [Google Scholar] [CrossRef]
- Meshram, C.D.; Lukash, T.; Phillips, A.T.; Akhrymuk, I.; Frolova, E.I.; Frolov, I. Lack of Nsp2-Specific Nuclear Functions Attenuates Chikungunya Virus Replication Both in Vitro and in Vivo. Virology 2019, 534, 14–24. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Deng, C.L.; Li, J.Q.; Li, N.; Zhang, Q.Y.; Ye, H.Q.; Yuan, Z.M.; Zhang, B. Infectious Chikungunya Virus (Chikv) with a Complete Capsid Deletion: A New Approach for a Chikv Vaccine. J. Virol. 2019, 93, e0050419. [Google Scholar] [CrossRef]
- Bagno, F.F.; Godói, L.C.; Figueiredo, M.M.; Sérgio, S.A.R.; Moraes, T.F.S.; Salazar, N.C.; Kim, Y.C.; Reyes-Sandoval, A.; da Fonseca, F.G. Chikungunya E2 Protein Produced in E. Coli and Hek293-T Cells-Comparison of Their Performances in Elisa. Viruses 2020, 12, 939. [Google Scholar] [CrossRef]
- Deeba, F.; Haider, M.S.H.; Ahmed, A.; Tazeen, A.; Faizan, M.I.; Salam, N.; Hussain, T.; Alamery, S.F.; Parveen, S. Global Transmission and Evolutionary Dynamics of the Chikungunya Virus. Epidemiol. Infect. 2020, 148, e63. [Google Scholar] [CrossRef]
- Dreux, M.; Garaigorta, U.; Boyd, B.; Décembre, E.; Chung, J.; Whitten-Bauer, C.; Wieland, S.; Chisari, F.V. Short-Range Exosomal Transfer of Viral Rna from Infected Cells to Plasmacytoid Dendritic Cells Triggers Innate Immunity. Cell Host Microbe 2012, 12, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Masrinoul, P.; Puiprom, O.; Tanaka, A.; Kuwahara, M.; Chaichana, P.; Ikuta, K.; Ramasoota, P.; Okabayashi, T. Monoclonal Antibody Targeting Chikungunya Virus Envelope 1 Protein Inhibits Virus Release. Virology 2014, 464–465, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Schnierle, B.S. Cellular Attachment and Entry Factors for Chikungunya Virus. Viruses 2019, 11, 1078. [Google Scholar] [CrossRef] [PubMed]
- Tumkosit, U.; Siripanyaphinyo, U.; Takeda, N.; Tsuji, M.; Maeda, Y.; Ruchusatsawat, K.; Shioda, T.; Mizushima, H.; Chetanachan, P.; Wongjaroen, P.; et al. Anti-Chikungunya Virus Monoclonal Antibody That Inhibits Viral Fusion and Release. J. Virol. 2020, 94, e0025220. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Qian, J.; Ding, J.; Li, J.; Nan, F.; Wang, W.; Qin, Q.; Fei, Y.; Xue, C.; Wang, J.; et al. Detection of Viral Components in Exosomes Derived from Ndv-Infected Df-1 Cells and Their Promoting Ability in Virus Replication. Microb. Pathog. 2019, 128, 414–422. [Google Scholar] [CrossRef]
- Fox, J.M.; Diamond, M.S. Immune-Mediated Protection and Pathogenesis of Chikungunya Virus. J. Immunol. 2016, 197, 4210–4218. [Google Scholar] [CrossRef]
- Petitdemange, C.; Wauquier, N.; Vieillard, V. Control of Immunopathology During Chikungunya Virus Infection. J. Allergy. Clin. Immunol. 2015, 135, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Simizu, B.; Rhim, J.S.; Wiebenga, N.H. Characterization of the Tacaribe Group of Arboviruses. I. Propagation and Plaque Assay of Tacaribe Virus in a Line of African Green Monkey Kidney Cells (Vero). Proc. Soc. Exp. Biol. Med. 1967, 125, 119–123. [Google Scholar] [CrossRef]
- Scherer, W.F.; Syverton, J.T.; Gey, G.O. Studies on the Propagation in Vitro of Poliomyelitis Viruses. Iv. Viral Multiplication in a Stable Strain of Human Malignant Epithelial Cells (Strain Hela) Derived from an Epidermoid Carcinoma of the Cervix. J. Exp. Med. 1953, 97, 695–710. [Google Scholar] [CrossRef]
- Igarashi, A. Isolation of a Singh’s Aedes Albopictus Cell Clone Sensitive to Dengue and Chikungunya Viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef]
- Wikan, N.; Sakoonwatanyoo, P.; Ubol, S.; Yoksan, S.; Smith, D.R. Chikungunya Virus Infection of Cell Lines: Analysis of the East, Central and South African Lineage. PLoS ONE 2012, 7, e31102. [Google Scholar] [CrossRef]
- Chalaem, P.; Chusri, S.; Fernandez, S.; Chotigeat, W.; Anguita, J.; Pal, U.; Promnares, K. Characterization of a Chikungunya Virus Strain Isolated from Banked Patients’ Sera. Virol. J. 2016, 13, 150. [Google Scholar] [CrossRef][Green Version]
- Hurwitz, S.N.; Nkosi, D.; Conlon, M.M.; York, S.B.; Liu, X.; Tremblay, D.C.; Meckes, D.G., Jr. Cd63 Regulates Epstein-Barr Virus Lmp1 Exosomal Packaging, Enhancement of Vesicle Production, and Noncanonical Nf-Kappab Signaling. J. Virol. 2017, 91, e0225116. [Google Scholar] [CrossRef]
- Burassakarn, A.; Srisathaporn, S.; Pientong, C.; Wongjampa, W.; Vatanasapt, P.; Patarapadungkit, N.; Ekalaksananan, T. Exosomes-Carrying Epstein-Barr Virus-Encoded Small Rna-1 Induces Indoleamine 2, 3-Dioxygenase Expression in Tumor-Infiltrating Macrophages of Oral Squamous-Cell Carcinomas and Suppresses T-Cell Activity by Activating Rig-I/Il-6/Tnf-Alpha Pathway. Oral. Oncol. 2021, 117, 105279. [Google Scholar] [CrossRef]
- Kaur, P.; Lee, R.C.; Chu, J.J. Infectious Viral Quantification of Chikungunya Virus-Virus Plaque Assay. Methods Mol. Biol. 2016, 1426, 93–103. [Google Scholar] [CrossRef]
- Agbulos, D.S.; Barelli, L.; Giordano, B.V.; Hunter, F.F. Zika Virus: Quantification, Propagation, Detection, and Storage. Curr. Protoc. Microbiol. 2016, 43, 15.d.4.1–15d.4.16. [Google Scholar] [CrossRef]
- Le, B.C.T.; Ekalaksananan, T.; Thaewnongiew, K.; Phanthanawiboon, S.; Aromseree, S.; Phanitchat, T.; Chuerduangphui, J.; Suwannatrai, A.T.; Alexander, N.; Overgaard, H.J.; et al. Interepidemic Detection of Chikungunya Virus Infection and Transmission in Northeastern Thailand. Am. J. Trop. Med. Hyg. 2020, 103, 1660–1669. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, B.C.T.; Burassakarn, A.; Tongchai, P.; Ekalaksananan, T.; Aromseree, S.; Phanthanawiboon, S.; Polsan, Y.; Alexander, N.; Overgaard, H.J.; Pientong, C. Characterization and Involvement of Exosomes Originating from Chikungunya Virus-Infected Epithelial Cells in the Transmission of Infectious Viral Elements. Int. J. Mol. Sci. 2022, 23, 12117. https://doi.org/10.3390/ijms232012117
Le BCT, Burassakarn A, Tongchai P, Ekalaksananan T, Aromseree S, Phanthanawiboon S, Polsan Y, Alexander N, Overgaard HJ, Pientong C. Characterization and Involvement of Exosomes Originating from Chikungunya Virus-Infected Epithelial Cells in the Transmission of Infectious Viral Elements. International Journal of Molecular Sciences. 2022; 23(20):12117. https://doi.org/10.3390/ijms232012117
Chicago/Turabian StyleLe, Bao Chi Thi, Ati Burassakarn, Panwad Tongchai, Tipaya Ekalaksananan, Sirinart Aromseree, Supranee Phanthanawiboon, Yada Polsan, Neal Alexander, Hans J. Overgaard, and Chamsai Pientong. 2022. "Characterization and Involvement of Exosomes Originating from Chikungunya Virus-Infected Epithelial Cells in the Transmission of Infectious Viral Elements" International Journal of Molecular Sciences 23, no. 20: 12117. https://doi.org/10.3390/ijms232012117
APA StyleLe, B. C. T., Burassakarn, A., Tongchai, P., Ekalaksananan, T., Aromseree, S., Phanthanawiboon, S., Polsan, Y., Alexander, N., Overgaard, H. J., & Pientong, C. (2022). Characterization and Involvement of Exosomes Originating from Chikungunya Virus-Infected Epithelial Cells in the Transmission of Infectious Viral Elements. International Journal of Molecular Sciences, 23(20), 12117. https://doi.org/10.3390/ijms232012117








