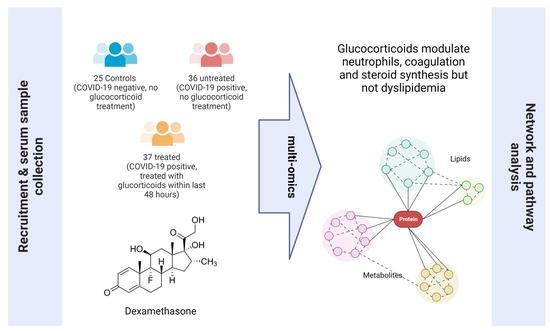
Multi-Omics Reveals Mechanisms of Partial Modulation of COVID-19 Dysregulation by Glucocorticoid Treatment
Abstract
1. Introduction
2. Results
2.1. Population Metadata Overview
2.2. Feature Identification
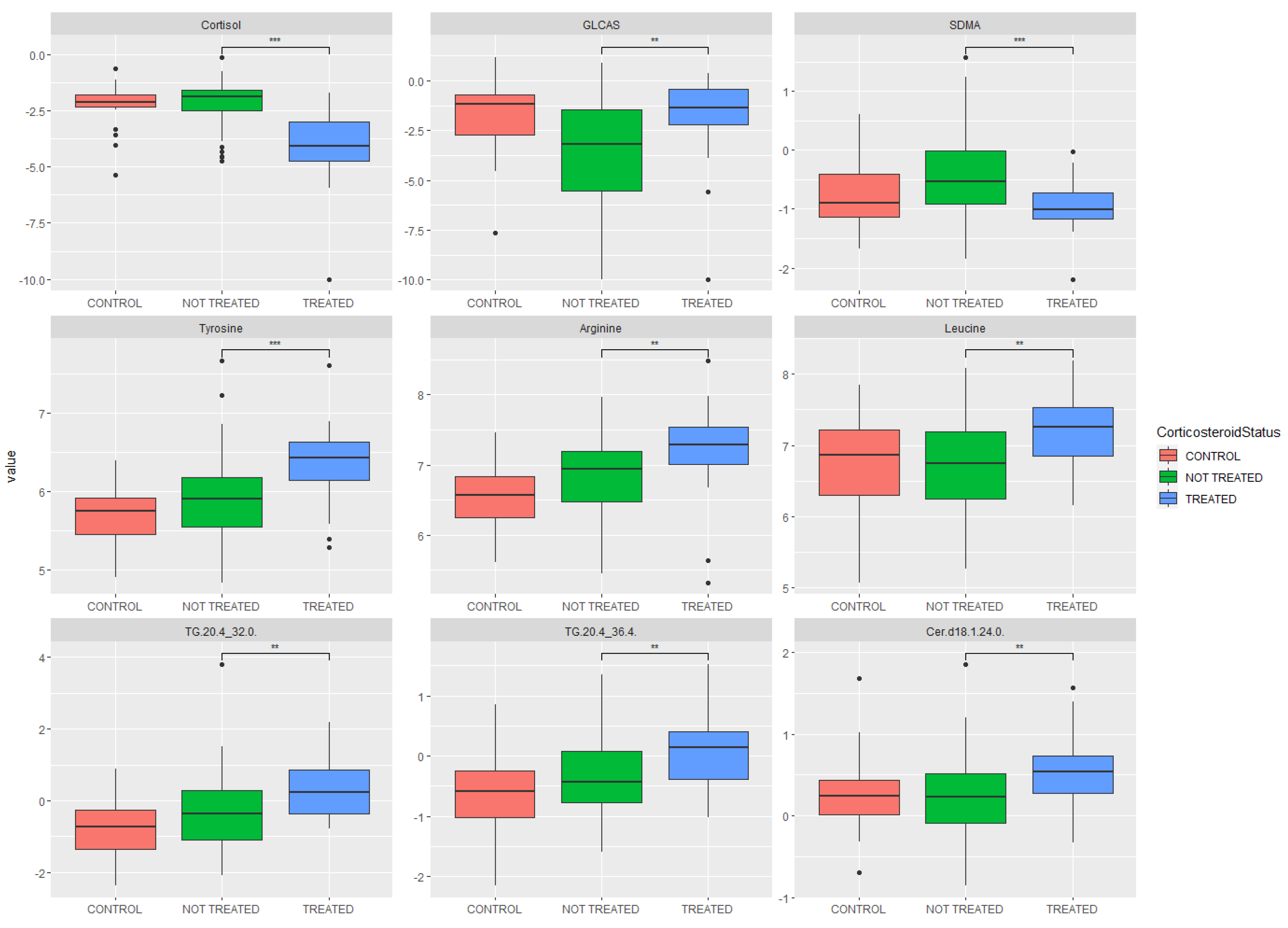
2.3. Metabolomic Analysis: Key Metabolites Altered by Glucocorticoid Treatment
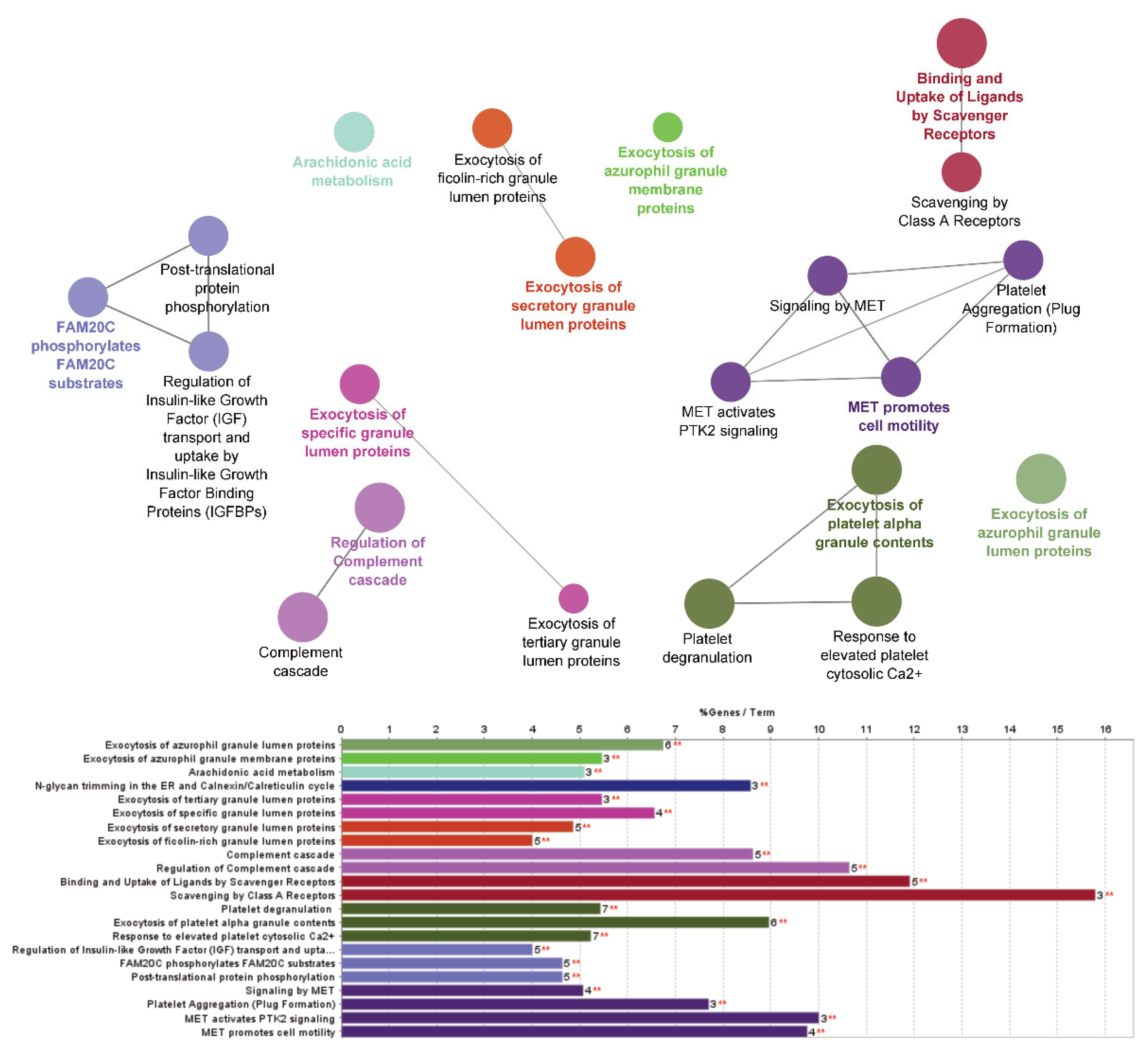
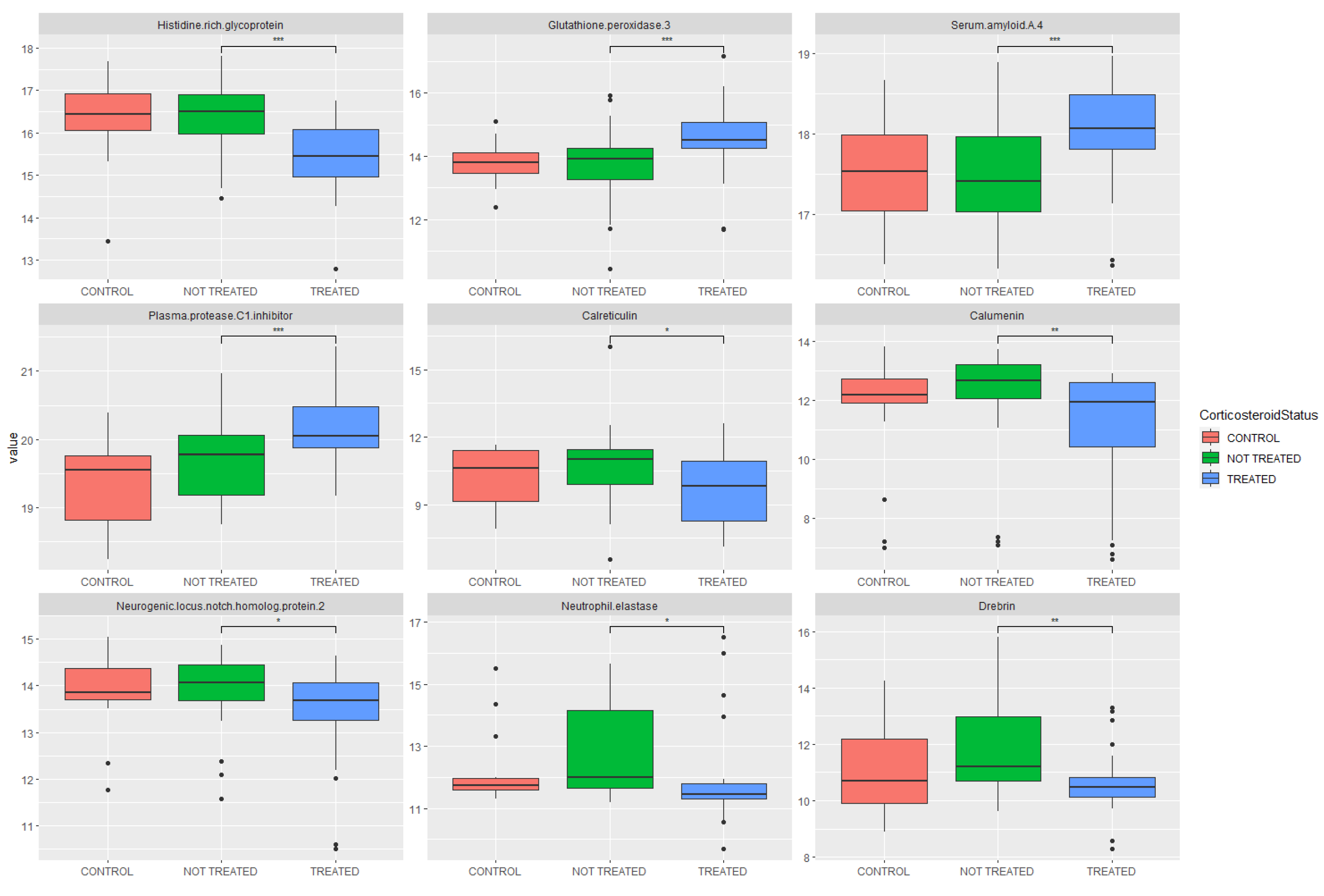
2.4. Proteomic Analysis: Investigation of Pathway Changes Due to Glucocorticoid Treatment
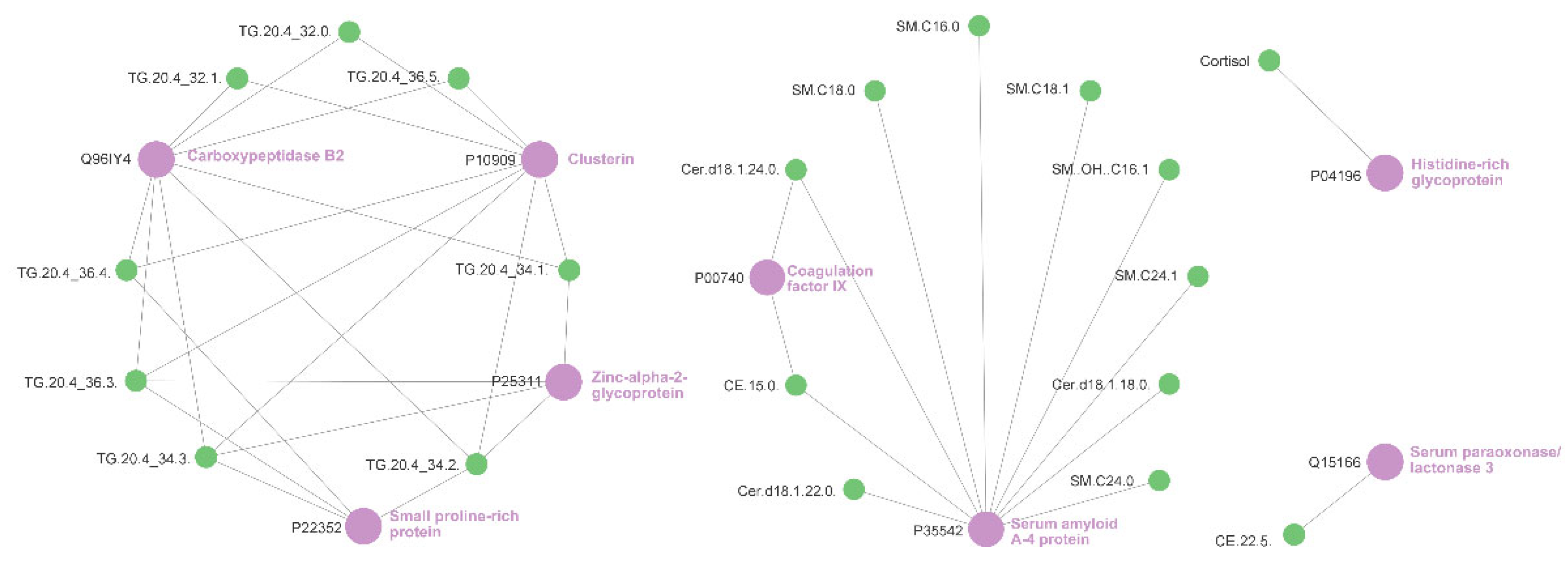
2.5. Relevance Network Analysis of Glucocorticoid Treatment
3. Discussion
4. Materials and Methods
4.1. Participant Recruitment and Ethics
4.2. Sample Collection and Extraction
4.3. Participant Selection
4.4. Serum Instrumentation and Analysis: Metabolomics
4.5. Plasma Instrumentation and Analysis: Proteomics
4.6. Feature Identification
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bechman, K.; Yates, M.; Mann, K.; Nagra, D.; Smith, L.J.; Rutherford, A.I.; Patel, A.; Periselneris, J.; Walder, D.; Dobson, R.J.B.; et al. Inpatient COVID-19 Mortality Has Reduced over Time: Results from an Observational Cohort. PLoS ONE 2022, 17, e0261142. [Google Scholar] [CrossRef] [PubMed]
- Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [CrossRef] [PubMed]
- National Institute for Health and Care Excellence COVID-19 Rapid Guideline: Managing COVID-19. Available online: https://www.nice.org.uk/guidance/NG191 (accessed on 7 August 2022).
- Hox, V.; Lourijsen, E.; Jordens, A.; Aasbjerg, K.; Agache, I.; Alobid, I.; Bachert, C.; Boussery, K.; Campo, P.; Fokkens, W.; et al. Benefits and Harm of Systemic Steroids for Short- And Long-Term Use in Rhinitis and Rhinosinusitis: An EAACI Position Paper. Clin. Transl. Allergy 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Polderman, J.A.W.; Farhang-Razi, V.; van Dieren, S.; Kranke, P.; DeVries, J.H.; Hollmann, M.W.; Preckel, B.; Hermanides, J. Adverse Side-Effects of Dexamethasone in Surgical Patients—An Abridged Cochrane Systematic Review. Anaesthesia 2019, 74, 929–939. [Google Scholar] [CrossRef]
- Wallen, N.; Kita, H.; Weiler, D.; Gleich, G.J. Glucocorticoids Inhibit Cytokine-Mediated Eosinophil Survival. J. Immunol. 1991, 147, 3490–3495. [Google Scholar]
- Alessi, J.; De Oliveira, G.B.; Schaan, B.D.; Telo, G.H. Dexamethasone in the Era of COVID-19: Friend or Foe? An Essay on the Effects of Dexamethasone and the Potential Risks of Its Inadvertent Use in Patients with Diabetes. Diabetol. Metab. Syndr. 2020, 12, 80. [Google Scholar] [CrossRef]
- Harasim-Symbor, E.; Konstantynowicz-Nowicka, K.; Chabowski, A. Additive Effects of Dexamethasone and Palmitate on Hepatic Lipid Accumulation and Secretion. J. Mol. Endocrinol. 2016, 57, 261–273. [Google Scholar] [CrossRef][Green Version]
- Bordag, N.; Klie, S.; Jürchott, K.; Vierheller, J.; Schiewe, H.; Albrecht, V.; Tonn, J.C.; Schwartz, C.; Schichor, C.; Selbig, J. Glucocorticoid (Dexamethasone)-Induced Metabolome Changes in Healthy Males Suggest Prediction of Response and Side Effects. Sci. Rep. 2015, 5, 15954. [Google Scholar] [CrossRef]
- Caterino, M.; Gelzo, M.; Sol, S.; Fedele, R.; Annunziata, A.; Calabrese, C.; Fiorentino, G.; D’Abbraccio, M.; Dell’Isola, C.; Fusco, F.M.; et al. Dysregulation of Lipid Metabolism and Pathological Inflammation in Patients with COVID-19. Sci. Rep. 2021, 11, 2941. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 Infection Alters Kynurenine and Fatty Acid Metabolism, Correlating with IL-6 Levels and Renal Status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma Metabolomic and Lipidomic Alterations Associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Castañé, H.; Iftimie, S.; Baiges-Gaya, G.; Rodríguez-Tomàs, E.; Jiménez-Franco, A.; López-Azcona, A.F.; Garrido, P.; Castro, A.; Camps, J.; Joven, J. Machine Learning and Semi-Targeted Lipidomics Identify Distinct Serum Lipid Signatures in Hospitalized COVID-19-Positive and COVID-19-Negative Patients. Metabolism 2022, 131, 155197. [Google Scholar] [CrossRef]
- Villar, M.; Urra, J.M.; Rodríguez-del-Río, F.J.; Artigas-Jerónimo, S.; Jiménez-Collados, N.; Ferreras-Colino, E.; Contreras, M.; de Mera, I.G.F.; Estrada-Peña, A.; Gortázar, C.; et al. Characterization by Quantitative Serum Proteomics of Immune-Related Prognostic Biomarkers for COVID-19 Symptomatology. Front. Immunol. 2021, 12, 730710. [Google Scholar] [CrossRef]
- Salamanna, F.; Maglio, M.; Landini, M.P.; Fini, M. Platelet Functions and Activities as Potential Hematologic Parameters Related to Coronavirus Disease 2019 (Covid-19). Platelets 2020, 31, 627–632. [Google Scholar] [CrossRef]
- Battaglini, D.; Lopes-Pacheco, M.; Castro-Faria-Neto, H.C.; Pelosi, P.; Rocco, P.R.M. Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19. Front. Immunol. 2022, 13, 857573. [Google Scholar] [CrossRef]
- Spick, M.; Lewis, H.M.; Wilde, M.J.; Hopley, C.; Huggett, J.; Bailey, M.J. Systematic Review with Meta-Analysis of Diagnostic Test Accuracy for COVID-19 by Mass Spectrometry. Metabolism 2021, 126, 154922. [Google Scholar] [CrossRef]
- Lewis, H.-M.; Liu, Y.; Frampas, C.F.; Longman, K.; Spick, M.; Stewart, A.; Sinclair, E.; Kasar, N.; Greener, D.; Whetton, A.D.; et al. Metabolomics Markers of COVID-19 Are Dependent on Collection Wave. Metabolites 2022, 12, 713. [Google Scholar] [CrossRef]
- Sinha, S.; Rosin, N.L.; Arora, R.; Labit, E.; Jaffer, A.; Cao, L.; Farias, R.; Nguyen, A.P.; de Almeida, L.G.N.; Dufour, A.; et al. Dexamethasone Modulates Immature Neutrophils and Interferon Programming in Severe COVID-19. Nat. Med. 2022, 28, 201–211. [Google Scholar] [CrossRef]
- Lacy, P. Mechanisms of Degranulation in Neutrophils. Allergy Asthma Clin. Immunol. 2006, 2, 301-e1. [Google Scholar] [CrossRef]
- Panda, R.; Castanheira, F.V.S.; Schlechte, J.M.; Surewaard, B.G.J.; Shim, H.B.; Zucoloto, A.Z.; Slavikova, Z.; Yipp, B.G.; Kubes, P.; McDonald, B. A Functionally Distinct Neutrophil Landscape in Severe COVID-19 Reveals Opportunities for Adjunctive Therapies. JCI Insight 2022, 7, 152291. [Google Scholar] [CrossRef]
- Sharma, A. Inferring Molecular Mechanisms of Dexamethasone Therapy in Severe COVID-19 from Existing Transcriptomic Data. Gene 2021, 788, 145665. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative Omics for Health and Disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Spick, M.; Lewis, H.-M.; Frampas, C.; Longman, K.; Bailey, M.J. An Integrated Analysis and Comparison of Serum, Saliva and Sebum for COVID-19 Metabolomics. Res. Sq. 2022, 12, 1187. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P. N501y Mutation of Spike Protein in Sars-Cov-2 Strengthens Its Binding to Receptor Ace2. Elife 2021, 10, e69091. [Google Scholar] [CrossRef]
- Siskos, A.P.; Jain, P.; Römisch-Margl, W.; Bennett, M.; Achaintre, D.; Asad, Y.; Marney, L.; Richardson, L.; Koulman, A.; Griffin, J.L.; et al. Interlaboratory Reproducibility of a Targeted Metabolomics Platform for Analysis of Human Serum and Plasma. Anal. Chem. 2017, 89, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-Independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell. Proteom. 2012, 11, O111.016717. [Google Scholar] [CrossRef]
- Alipour, A.; Van Oostrom, A.J.H.H.M.; Izraeljan, A.; Verseyden, C.; Collins, J.M.; Frayn, K.N.; Plokker, T.W.M.; Elte, J.W.F.; Cabezas, M.C. Leukocyte Activation by Triglyceride-Rich Lipoproteins. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 792–797. [Google Scholar] [CrossRef]
- Tobler, A.; Meier, R.; Seitz, M.; Dewald, B.; Baggiolini, M.; Fey, M.F. Glucocorticoids Downregulate Gene Expression of GM-CSF, NAP-1/IL-8, and IL-6, but Not of M-CSF in Human Fibroblasts. Blood 1992, 79, 45–51. [Google Scholar] [CrossRef]
- Smith, P.J.; Cousins, D.J.; Jee, Y.-K.; Staynov, D.Z.; Lee, T.H.; Lavender, P. Suppression of Granulocyte-Macrophage Colony-Stimulating Factor Expression by Glucocorticoids Involves Inhibition of Enhancer Function by the Glucocorticoid Receptor Binding to Composite NF-AT/Activator Protein-1 Elements. J. Immunol. 2001, 167, 2502–2510. [Google Scholar] [CrossRef]
- Schwarz, B.; Sharma, L.; Roberts, L.; Peng, X.; Bermejo, S.; Leighton, I.; Massana, A.C.; Farhadian, S.; Ko, A.I.; Team, Y.I.; et al. Severe SARS-CoV-2 Infection in Humans Is Defined by a Shift in the Serum Lipidome Resulting in Dysregulation of Eicosanoid Immune Mediators. Res. Sq. 2020, rs.3.rs-42999. [Google Scholar] [CrossRef]
- Arnardottir, H.; Pawelzik, S.C.; Öhlund Wistbacka, U.; Artiach, G.; Hofmann, R.; Reinholdsson, I.; Braunschweig, F.; Tornvall, P.; Religa, D.; Bäck, M. Stimulating the Resolution of Inflammation Through Omega-3 Polyunsaturated Fatty Acids in COVID-19: Rationale for the COVID-Omega-F Trial. Front. Physiol. 2021, 11, 1–7. [Google Scholar] [CrossRef]
- Calde, P.C. Eicosanoids. Essays Biochem. 2020, 64, 423–441. [Google Scholar] [CrossRef]
- Karu, N.; Kindt, A.; Lamont, L.; van Gammeren, A.J.; Ermens, A.A.M.; Harms, A.C.; Portengen, L.; Vermeulen, R.C.H.; Dik, W.A.; Langerak, A.W.; et al. Plasma Oxylipins and Their Precursors Are Strongly Associated with COVID-19 Severity and with Immune Response Markers. Metabolites 2022, 12, 619. [Google Scholar] [CrossRef]
- De Man, F.H.; Nieuwland, R.; Van Der Laarse, A.; Romijn, F.; Smelt, A.H.M.; Gevers Leuven, J.A.; Sturk, A. Activated Platelets in Patients with Severe Hypertriglyceridemia: Effects of Triglyceride-Lowering Therapy. Atherosclerosis 2000, 152, 407–414. [Google Scholar] [CrossRef]
- Crothers, K.; DeFaccio, R.; Tate, J.; Alba, P.R.; Goetz, M.B.; Jones, B.; King, J.T.; Marconi, V.; Ohl, M.E.; Rentsch, C.T.; et al. Dexamethasone in Hospitalised Coronavirus-19 Patients Not on Intensive Respiratory Support. Eur. Respir. J. 2021, 60, 2102532. [Google Scholar] [CrossRef]
- Närhi, F.; Moonesinghe, S.R.; Shenkin, S.D.; Drake, T.M.; Mulholland, R.H.; Donegan, C.; Dunning, J.; Fairfield, C.J.; Girvan, M.; Hardwick, H.E.; et al. Implementation of Corticosteroids in Treatment of COVID-19 in the ISARIC WHO Clinical Characterisation Protocol UK: Prospective, Cohort Study. Lancet Digit. Health 2022, 4, e220–e234. [Google Scholar] [CrossRef]
- Whitaker, M.; Elliott, J.; Bodinier, B.; Barclay, W.; Ward, H.; Cooke, G.; Donnelly, C.A.; Chadeau-Hyam, M.; Elliott, P. Variant-Specific Symptoms of COVID-19 among 1,542,510 People in England. medRxiv 2022. [Google Scholar] [CrossRef]
- Ahmed, M.H.; Hassan, A. Dexamethasone for the Treatment of Coronavirus Disease (COVID-19): A Review. SN Compr. Clin. Med. 2020, 2, 2637–2646. [Google Scholar] [CrossRef]
- COVID-19 Mass Spectrometry Coalition COVID-19 Metabolomics and Lipidomics Protocol. Available online: https://covid19-msc.org/metabolomics-and-lipidomics-protocol/ (accessed on 25 May 2021).
- COVID-19 Mass Spectrometry Coalition COVID-19 Proteomics Protocol. Available online: https://covid19-msc.org/proteomics-protocol/ (accessed on 7 August 2022).
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Salie, M.T.; Yang, J.; Ramírez Medina, C.R.; Zühlke, L.J.; Chishala, C.; Ntsekhe, M.; Gitura, B.; Ogendo, S.; Okello, E.; Lwabi, P.; et al. Data-Independent Acquisition Mass Spectrometry in Severe Rheumatic Heart Disease (RHD) Identifies a Proteomic Signature Showing Ongoing Inflammation and Effectively Classifying RHD Cases. Clin. Proteom. 2022, 19, 7. [Google Scholar] [CrossRef]
- Liu, Y.; Buil, A.; Collins, B.C.; Gillet, L.C.; Blum, L.C.; Cheng, L.; Vitek, O.; Mouritsen, J.; Lachance, G.; Spector, T.D.; et al. Quantitative Variability of 342 Plasma Proteins in a Human Twin Population. Mol. Syst. Biol. 2015, 11, 786. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, 11. [Google Scholar] [CrossRef]
- González, I.; Cao, K.A.L.; Davis, M.J.; Déjean, S. Visualising Associations between Paired “omics” Data Sets. BioData Min. 2012, 5, 19. [Google Scholar] [CrossRef]
- Struwe, W.; Emmott, E.; Bailey, M.; Sharon, M.; Sinz, A.; Corrales, F.J.; Thalassinos, K.; Braybrook, J.; Mills, C.; Barran, P. The COVID-19 MS Coalition—Accelerating Diagnostics, Prognostics, and Treatment. Lancet 2020, 395, 1761–1762. [Google Scholar] [CrossRef]
| Parameters | Control Group: Negative for COVID-19 | Not Treated with Glucocorticoids: Positive for COVID-19 | Treated with Glucocorticoids: Positive for COVID-19 | p-Value Glucocorticoids versus Not Treated |
|---|---|---|---|---|
| n | 25 | 36 | 37 | |
| Age (mean, standard deviation; years) | 65.8 ± 21.4 | 62.8 ± 21.2 | 60.6 ± 15.2 | 0.61 |
| Male / Female (n) | 11/14 | 20/16 | 26/11 | 0.19 |
| Treated with Glucocorticoids (n) | 0 | 0 | 37 | 0.00 |
| Treated with Anticoagulants (n) | 3 | 5 | 6 | 0.78 |
| Treated for Hypertension (n) | 8 | 15 | 15 | 0.92 |
| Treated for High Cholesterol (n) | 6 | 2 | 6 | 0.14 |
| Treated for Type 2 Diabetes Mellitus (n) | 8 | 16 | 8 | 0.04 |
| Treated for Ischaemic Heart Disease (n) | 6 | 5 | 6 | 0.78 |
| Ex-Smoker (n) | 9 | 9 | 16 | 0.10 |
| Current Smoker (n) | 0 | 2 | 0 | 0.15 |
| Medical Acute Dependency admission (n) | 4 | 10 | 24 | 0.001 |
| Intensive Care Unit admission (n) | 1 | 0 | 2 | 0.16 |
| Did Not Survive Admission (n) | 1 | 3 | 1 | 0.29 |
| Duration of pre-admission symptoms (mean, standard deviation; days) | 6.9 ± 10.5 | 4.1 ± 4.1 | 7.0 ± 4.3 | 0.004 |
| Time between positive RT-PCR test and sampling (mean, standard deviation; days) | na | 4.3 ± 5.7 | 3.2 ± 3.8 | 0.35 |
| Lymphocytes (mean, standard deviation; cells/μL) | 0.8 ± 0.4 | 0.8 ± 0.6 | 0.7 ± 0.6 | 0.36 |
| C-Reactive Protein (mean, standard deviation; mg/L) | 129.8 ± 94.2 | 127.5 ± 97.8 | 96.3 ± 72.2 | 0.13 |
| Eosinophils (mean, standard deviation; 100/μL) | 0.3 ± 0.3 | 0.1 ± 0.1 | 0.0 ± 0.0 | <0.001 |
| Bilateral Chest X-Ray changes (n) | 2 | 12 | 27 | 0.001 |
| Continuous Positive Airway Pressure (n) | 3 | 4 | 14 | 0.008 |
| Metabolite | p-Value | Fold Change | Increased/Decreased in Glucocorticoid Group |
|---|---|---|---|
| Cortisol | 9.27 × 10−9 | 0.26 | Decreased |
| α-Aminoadipic acid | 4.79 × 10−4 | 1.68 | Increased |
| Tyrosine | 7.95 × 10−4 | 1.31 | Increased |
| Propionylcarnitine | 7.95 × 10−4 | 1.66 | Increased |
| Cer (d18:1/22:0) | 8.61 × 10−4 | 1.28 | Increased |
| Phenylalanine | 9.69 × 10−4 | 1.32 | Increased |
| CE (15:0) | 1.22 × 10−3 | 1.27 | Increased |
| Aconitic acid | 1.27 × 10−3 | 0.70 | Decreased |
| Cer (d18:1/24:0) | 1.37 × 10−3 | 1.28 | Increased |
| SM C18:0 | 1.69 × 10−3 | 1.30 | Increased |
| Carnitine | 2.00 × 10−3 | 1.34 | Increased |
| Arginine | 6.86 × 10−3 | 1.28 | Increased |
| Symmetric dimethylarginine | 6.98 × 10−3 | 0.72 | Decreased |
| Methionine | 7.46 × 10−3 | 1.23 | Increased |
| Cer (d18:2/24:0) | 7.58 × 10−3 | 1.31 | Increased |
| Protein | Uniprot ID | p-Value | Fold Change | Increased/Decreased in Glucocorticoid Group |
|---|---|---|---|---|
| Histidine-rich glycoprotein | P04196 | 3.01 × 10−5 | 0.55 | Decreased |
| Glutathione peroxidase 3 | P22352 | 3.31 × 10−4 | 1.84 | Increased |
| Serum amyloid A-4 protein | P35542 | 6.76 × 10−4 | 1.44 | Increased |
| Plasma protease C1 inhibitor | P05155 | 8.02 × 10−4 | 1.35 | Increased |
| Calreticulin | P27797 | 1.39 × 10−3 | 0.46 | Decreased |
| Calumenin | O43852 | 2.20 × 10−3 | 0.50 | Decreased |
| Laminin subunit beta-1 | P07942 | 2.47 × 10−3 | 0.85 | Decreased |
| Neurogenic locus notch homolog protein 2 | Q04721 | 2.71 × 10−3 | 0.74 | Decreased |
| Neutrophil elastase | P08246 | 3.19 × 10−3 | 0.54 | Decreased |
| Serpin B3 | P29508 | 3.22 × 10−3 | 0.61 | Decreased |
| Drebrin | Q16643 | 3.76 × 10−3 | 0.46 | Decreased |
| Coagulation factor IX | P00740 | 4.22 × 10−3 | 1.30 | Increased |
| Cytoplasmic aconitate hydratase | P21399 | 5.35 × 10−3 | 0.78 | Decreased |
| Transgelin | Q01995 | 5.50 × 10−3 | 0.75 | Decreased |
| Carboxypeptidase B2 | Q96IY4 | 5.82 × 10−3 | 1.35 | Increased |
| Pathway Name | # Entities Identified | p-Value | p-Value FDR Corrected |
|---|---|---|---|
| Steroid hormone biosynthesis | 2 | 5.45 × 10−9 | 2.29 × 10−7 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 2 | 6.89 × 10−4 | 9.65 × 10−3 |
| Phenylalanine metabolism | 2 | 6.89 × 10−4 | 9.65 × 10−3 |
| Tyrosine metabolism | 1 | 3.80 × 10−3 | 3.19 × 10−2 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 1 | 3.80 × 10−3 | 3.19 × 10−2 |
| Aminoacyl-tRNA biosynthesis | 20 | 6.62 × 10−3 | 4.63 × 10−2 |
| Arginine biosynthesis | 3 | 1.20 × 10−2 | 5.70 × 10−2 |
| Valine, leucine and isoleucine biosynthesis | 4 | 1.20 × 10−2 | 5.70 × 10−2 |
| Sphingolipid metabolism | 1 | 1.22 × 10−2 | 5.70 × 10−2 |
| Valine, leucine and isoleucine degradation | 4 | 1.43 × 10−2 | 6.00 × 10−2 |
| Pathway Name | # Entities Identified | p-Value | p-Value FDR Corrected |
|---|---|---|---|
| Neutrophil degranulation | 17 | 1.34 × 10−8 | 1.75 × 10−7 |
| Innate immune system | 23 | 1.34 × 10−8 | 1.75 × 10−7 |
| Exocytosis of platelet alpha granule contents | 6 | 2.75 × 10−6 | 3.30 × 10−5 |
| Platelet activation, signalling and aggregation | 10 | 2.75 × 10−6 | 3.30 × 10−5 |
| Binding and uptake of ligands by scavenger receptors | 5 | 4.32 × 10−6 | 4.75 × 10−5 |
| Regulation of complement cascade | 5 | 2.16 × 10−5 | 1.95 × 10−4 |
| Platelet degranulation | 7 | 2.75 × 10−6 | 3.30 × 10−5 |
| Exocytosis of azurophil granule lumen proteins | 6 | 1.29 × 10−5 | 1.29 × 10−4 |
| Response to elevated platelet cytosolic Ca2+ | 7 | 2.75 × 10−6 | 3.30 × 10−5 |
| Complement cascade | 5 | 2.16 × 10−5 | 1.95 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spick, M.; Campbell, A.; Baricevic-Jones, I.; von Gerichten, J.; Lewis, H.-M.; Frampas, C.F.; Longman, K.; Stewart, A.; Dunn-Walters, D.; Skene, D.J.; et al. Multi-Omics Reveals Mechanisms of Partial Modulation of COVID-19 Dysregulation by Glucocorticoid Treatment. Int. J. Mol. Sci. 2022, 23, 12079. https://doi.org/10.3390/ijms232012079
Spick M, Campbell A, Baricevic-Jones I, von Gerichten J, Lewis H-M, Frampas CF, Longman K, Stewart A, Dunn-Walters D, Skene DJ, et al. Multi-Omics Reveals Mechanisms of Partial Modulation of COVID-19 Dysregulation by Glucocorticoid Treatment. International Journal of Molecular Sciences. 2022; 23(20):12079. https://doi.org/10.3390/ijms232012079
Chicago/Turabian StyleSpick, Matt, Amy Campbell, Ivona Baricevic-Jones, Johanna von Gerichten, Holly-May Lewis, Cecile F. Frampas, Katie Longman, Alexander Stewart, Deborah Dunn-Walters, Debra J. Skene, and et al. 2022. "Multi-Omics Reveals Mechanisms of Partial Modulation of COVID-19 Dysregulation by Glucocorticoid Treatment" International Journal of Molecular Sciences 23, no. 20: 12079. https://doi.org/10.3390/ijms232012079
APA StyleSpick, M., Campbell, A., Baricevic-Jones, I., von Gerichten, J., Lewis, H.-M., Frampas, C. F., Longman, K., Stewart, A., Dunn-Walters, D., Skene, D. J., Geifman, N., Whetton, A. D., & Bailey, M. J. (2022). Multi-Omics Reveals Mechanisms of Partial Modulation of COVID-19 Dysregulation by Glucocorticoid Treatment. International Journal of Molecular Sciences, 23(20), 12079. https://doi.org/10.3390/ijms232012079