Abstract
Pancreatic cancer is one of the most lethal malignant tumors. However, the molecular mechanisms responsible for its progression are little known. This study aimed to understand the regulatory role of CD44V3 in pancreatic cancer. A Kaplan–Meier analysis was performed to reveal the correlation between CD44/CD44V3 expression and the prognosis of pancreatic cancer patients. CD44V3 and U2AF1 were knocked down using shRNAs. The proliferation, migration, invasion, and stemness of two pancreatic cell lines, BxPC-3 and AsPC-1, were examined. The expression of CD44V3, cancer-associated markers, and the activation of AKT signaling were detected by qRT-PCR and Western blot. Both CD44 and CD44V3 expression levels were associated with a poor prognosis in pancreatic cancer patients. Interestingly, the expression of CD44V3, instead of CD44, was greatly increased in tumor tissues. CD44V3 knockdown inhibited the proliferation, migration, invasion, and stemness of cancer cells. CD44V3 splicing was regulated by U2AF1 and downregulation of U2AF1 enhanced CD44V3 expression, which promoted pancreatic cancer progression. CD44V3 is an important cancer-promoting factor, which may serve as a potential candidate for pancreatic cancer intervention.
1. Introduction
Pancreatic cancer, one of the most lethal malignant tumors, caused more than 432,000 deaths in 2018 worldwide [1]. The main risk factors for pancreatic cancer include a family history of chronic pancreatitis, smoking, diabetes, age, and some genetic disorders [2,3,4,5]. Although massive efforts have been invested in past decades, surgery, radiation, chemotherapy, and other conventional approaches have showed little effect on the treatment of this malignant tumor [6,7,8,9]. To date, surgical resection provides the best chance of cure. However, more than 80% of patients develop advanced unresectable pancreatic cancer, and less than 5% of patients survive five years after diagnosis [10]. Hence, it is very urgent to understand the biological mechanisms underlying the development and progression of pancreatic cancer.
Cancer stem cells are a subset of the cell population that is able to dictate the heterogeneity, proliferation, metastasis, and multi-drug resistance in tumors [11,12]. The hyaluronic acid receptor CD44 is a cancer stem cell marker involved in multiple regulation of almost all cancer types, such as cell survival, proliferation, motility, and tumor microenvironment remodeling [13,14,15]. In human glioblastoma multiforme, CD44 enhanced the cancer stem cell phenotypes and promoted therapeutic resistance, whose expression level was correlated with a poor survival rate [14]. High CD44 levels have been linked to cell proliferation and tumorigenesis in everything from solid tumors to hematologic malignancies, and siRNA knockdown of CD44 could significantly inhibit tumorigenicity [11]. Furthermore, CD44 is one of the most commonly used cancer stem cell surface markers in sorting different subpopulations of tumor tissues [16,17,18].
The CD44 gene contains 20 exons, half of which are constant exons expressed in all isoforms, while the other half are variable exons (V1-10) undergoing extensive alternative splicing by spliceosome [19,20]. Alternative splicing can produce many CD44 isoforms that possess various tissue-specific effects during cancer progression [20]. CD44V3 contains exon V3 that encodes a heparin sulfate site, which allows it to bind heparin sulfate binding growth factors, such as epidermal growth factor (EGF) and fibroblast growth factors (FGFs) [21,22,23]. CD44 V8-10 can stabilize xCT, a cysteine glutamate transporter, and then enhance cystine uptake and intracellular antioxidant synthesis [24]. The inclusion of variant exons of CD44 is regulated by oncogenic signals, which might contribute to the expression of a large variety of CD44 variants in advanced stage tumors [25,26].
In the current study, the regulatory roles of CD44 in pancreatic cancer progression and prognosis were investigated. Although CD44 levels correlated with poor prognosis, in comparison with adjacent normal tissues, its expression level was decreased in tumor tissues. However, the expression of CD44V3 was significantly increased in tumor tissues and correlated with the poor prognosis of patients. Functional studies indicated that the elevation of CD44V3 was regulated by U2AF1, an important component of the spliceosome, which then enhanced the proliferation and invasion ability and promoted epithelial-mesenchymal transition (EMT) and stemness of cancer cells [27,28]. The tumor promoting effect of CD44V3 might be mediated by the AKT signaling pathway. This finding suggests that targeting the CD44V3 isoform might pave the way for a novel treatment strategy for pancreatic cancer.
2. Results
2.1. CD44 Is Correlated with Poor Prognosis of Pancreatic Cancer Patients
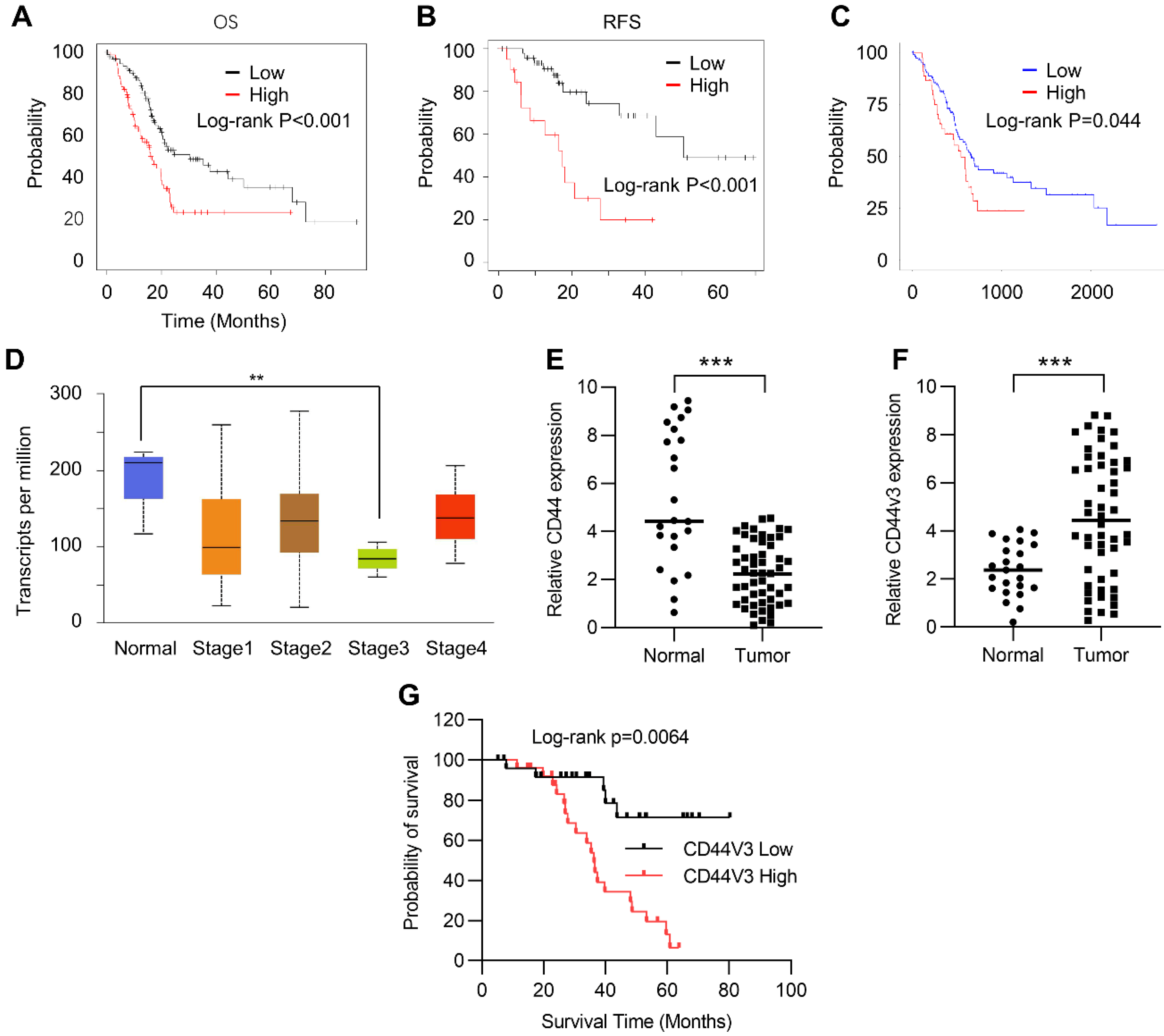
In order to study the role of CD44 in the progression of pancreatic cancer, we first investigated the correlation between the prognosis and the expression level of CD44 in cancer tissues using the Kaplan–Meier Plotter (Table S1). Kaplan–Meier plots of overall survival (OS) and relapse-free survival (RFS) in pancreatic cancer patients were stratified according to their CD44 levels. The results showed that high CD44 expression levels positively correlated with poor OS and RFS in pancreatic cancer patients (Figure 1A,B). We further analyzed this correlation in the TCGA pancreatic cancer database using Ualcan (http://ualcan.path.uab.edu/, accessed on 8 May 2021) and arrived at a similar result: high CD44 expression predicted poor prognosis (Figure 1C). Next, we examined CD44 expression in normal and different stages of pancreatic tumor tissues using the Kaplan–Meier Plotter. Unexpectedly, CD44 levels were higher in adjacent normal tissues than in pancreatic tumor tissues (Figure 1D). A similar result was observed in normal and pancreatic tumor tissues detected by RT-qPCR (Figure 1E). To explore this contradiction, we examined the expression of CD44V3, which has been demonstrated to have a critical role in regulating the development and occurrence of various tumors, in tumor and normal tissues by RT-qPCR. Our data showed that CD44V3 expression levels were markedly increased in tumor tissues compared to adjacent normal tissues (Figure 1F). Moreover, pancreatic cancer patients with high CD44V3 levels showed poor OS, which showed a more potent correlation than that of CD44 (Figure 1A,G). All this data suggests that CD44V3 might be one of the important players in regulating tumor progression and prognosis of pancreatic cancer patients.
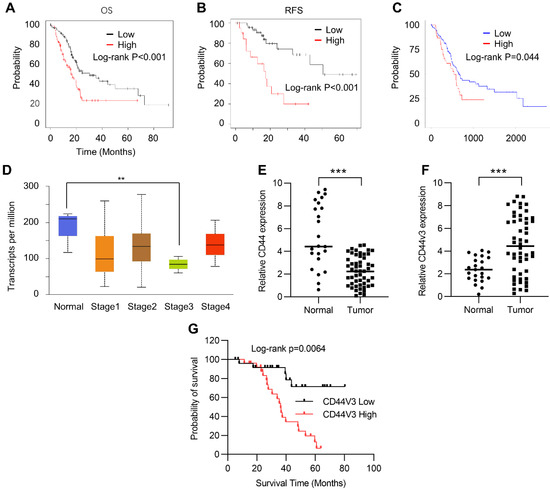
Figure 1.
CD44 was correlated with a poor prognosis of pancreatic cancer. (A) Kaplan–Meier plots of overall survival in pancreatic cancer patients stratified according to their CD44 levels. The Kaplan–Meier plotter was used to analyze the data. (B) Kaplan–Meier plots of release-free survival in pancreatic cancer patients stratified according to their CD44 levels. The Kaplan–Meier plotter was used to analyze the data. (C) Kaplan–Meier plots of overall survival in pancreatic cancer patients stratified according to their CD44 levels. The data was analyzed in TCGA dataset from ualcan (http://ualcan.path.uab.edu, accessed on 8 May 2021). (D) CD44 levels in normal and pancreatic tumor tissues at various stages. The data was analyzed using the TCGA dataset from ualcan (http://ualcan.path.uab.edu, accessed on 8 May 2021). (E) The CD44 levels in normal and pancreatic tumor tissues were analyzed by qPCR. (F) The CD44V3 levels in normal and pancreatic tumor tissues were analyzed by qPCR. (G) Kaplan–Meier plots of overall survival in pancreatic cancer patients stratified according to their CD44V3 levels. The data are presented as mean ± S.D. ** p < 0.01; *** p < 0.001.
2.2. U2AF1 Regulates CD44V3 Splicing
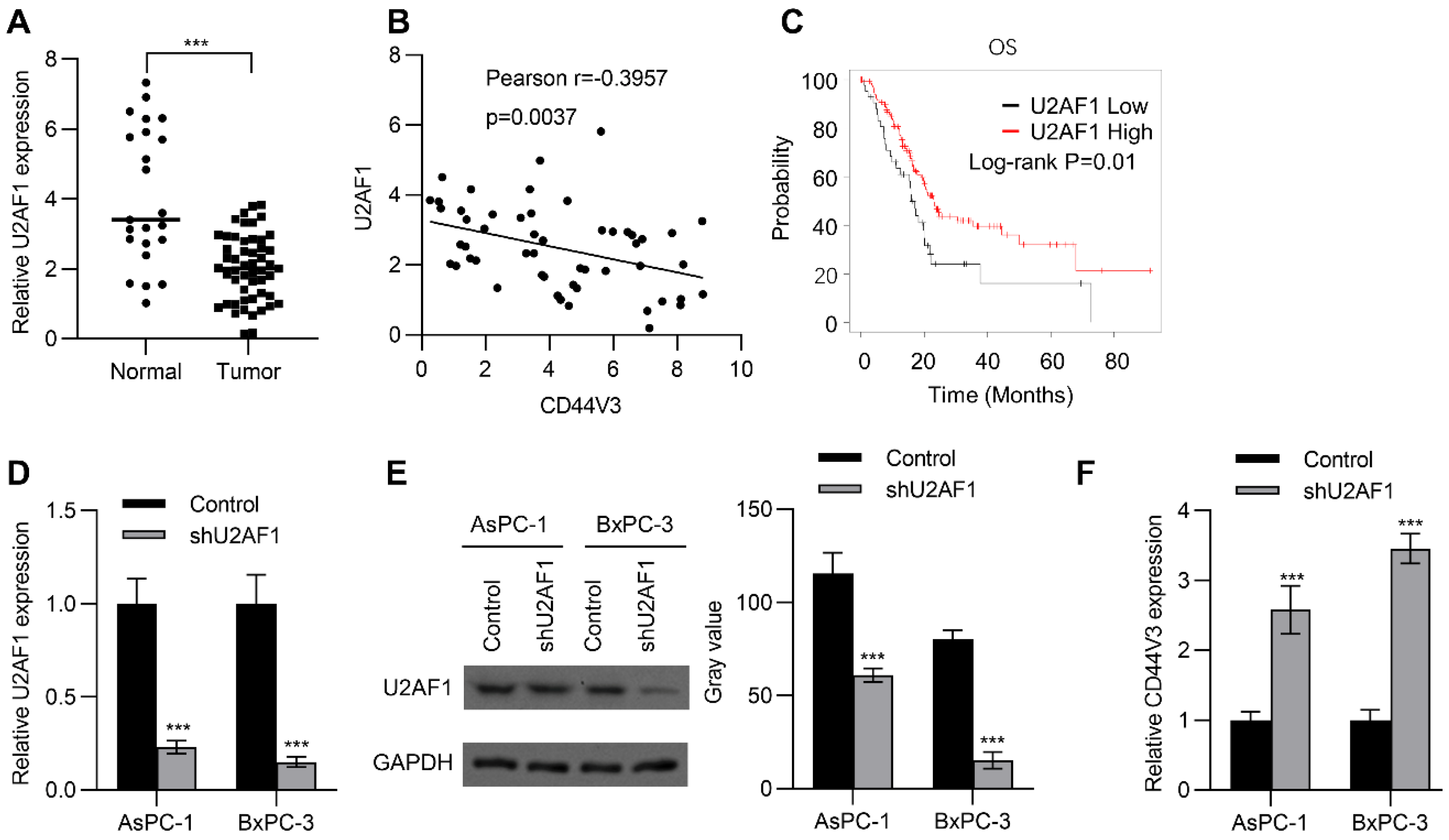
U2AF1 is an RNA splicing complex component of the spliceosome that regulates pre-mRNA splicing [27]. Recent studies have further confirmed that it can regulate alternative splicing of mRNA [28]. Therefore, we wondered whether alternative splicing of CD44V3 is regulated by U2AF1. To this end, we examined the expression of U2AF1 in adjacent normal tissues and pancreatic tumor tissues by RT-qPCR, to explore whether U2AF1 was involved in the regulation of CD44V3 expression. As shown in Figure 2A, U2AF1 levels were significantly reduced in tumors compared to those in normal tissues. Interestingly, U2AF1 expression level was negatively correlated with CD44V3 expression level in pancreatic tumor tissues (Figure 2B). Moreover, Kaplan–Meier analysis showed that higher U2AF1 levels predicted better OS for pancreatic cancer patients (Figure 2C). Since lower U2AF1 expression predicted poorer prognosis, we knocked down U2AF1 in BxPC-3 and AsPC-1 pancreatic cancer cells using shRNA, which significantly reduced both the mRNA and protein levels of U2AF1 (Figure 2D,E), and then measured the expression of CD44V3. As expected, U2AF1 knockdown could significantly increase the expression of CD44V3 in both AsPC-1 and BxPC-3 cells (Figure 2F). The above results indicated that U2AF1 regulated the splicing of CD44 and inhibited the expression of CD44V3.
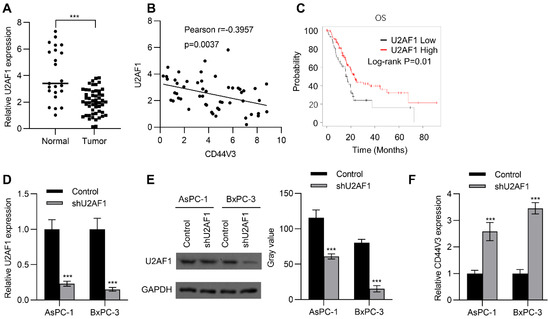
Figure 2.
U2AF1 regulates CD44V3 splicing. (A) The U2AF1 levels in normal and pancreatic tumor tissues were analyzed by qPCR. (B) The correlation between U2AF1 and CD44V3 in pancreatic cancer patients was determined by qPCR. (C) Kaplan–Meier plots of overall survival in pancreatic cancer patients stratified according to their U2AF1 levels. The Kaplan–Meier Plotter was used to analyze the data. (D,E) The U2AF1 levels in AsPC-1 or BxPC-3 cells transfected with U2AF1 shRNA were analyzed by qPCR (D) or Western blot (E). (F) The CD44V3 levels in AsPC-1 or BxPC-3 cells transfected with U2AF1 shRNA were analyzed by qPCR. The data are presented as mean ± S.D. *** p < 0.001.
2.3. Downregulation of CD44V3 Inhibits Pancreatic Cancer Cell Proliferation
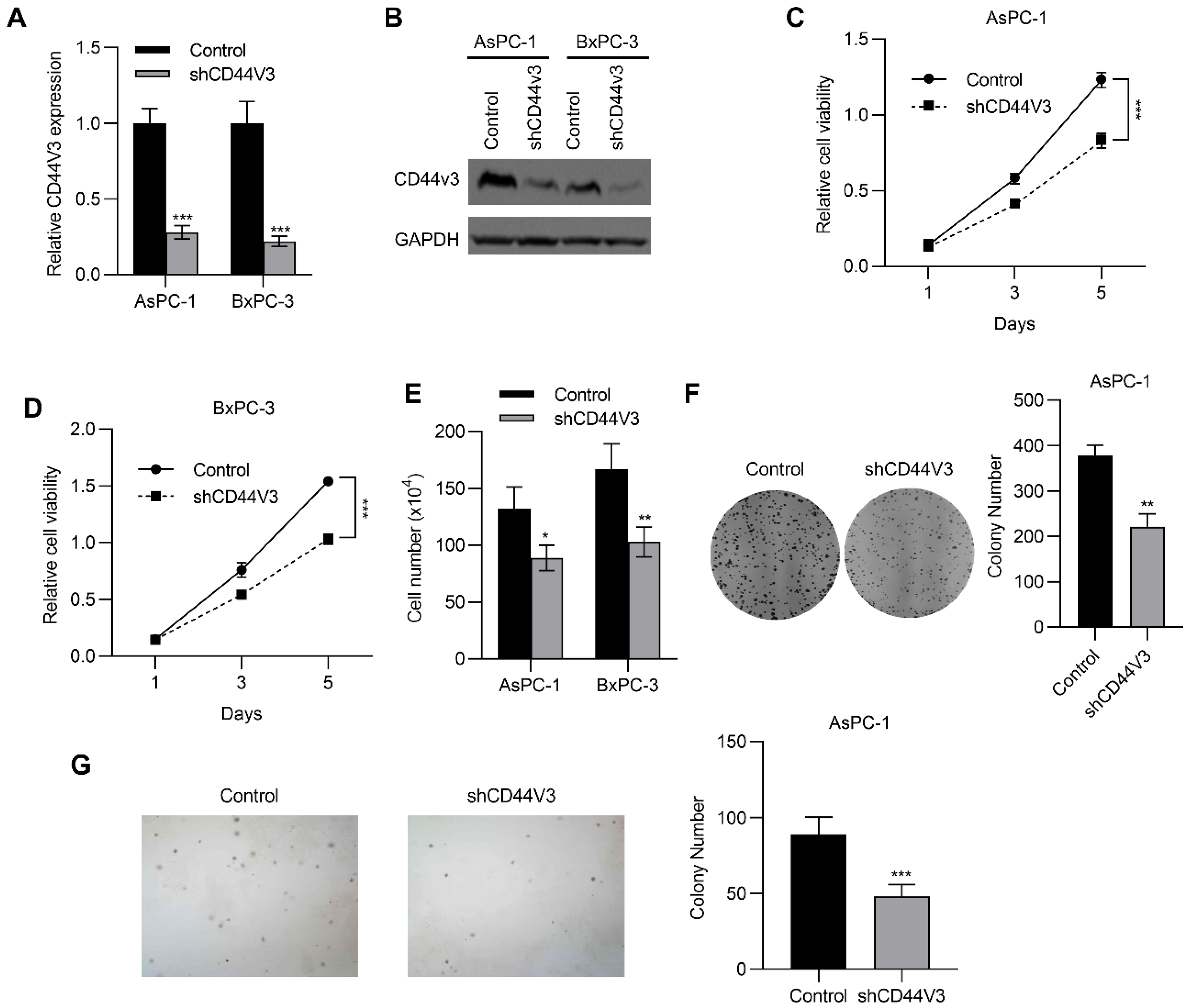
To investigate the regulatory role of CD44V3 in pancreatic cancer progression, we knocked down CD44V3 in BxPC-3 and AsPC-1 cells using shRNA and examined the changes in proliferative ability. As shown in Figure 3A,B, both mRNA and protein levels of CD44V3 were markedly reduced. Accordingly, the MTT assay showed that CD44V3 knockdown significantly suppressed proliferation of the two pancreatic cancer cell lines (Figure 3C,D). Cell count assay results indicated that CD44V3 knockdown markedly reduced the viability of both AsPC-1 and BxPC-3 cells (Figure 3E). We performed cell colony formation and soft agar assays to assess the cell viability and anchorage-independent cell growth ability with or without CD44V3. The results showed that knockdown of CD44V3 significantly reduced the colony formation of AsPC-1 cells, while interestingly, the average size of the clone was smaller than that in the control group (Figure 3F,G), indicating that CD44V3 deficiency inhibited the proliferation of pancreatic cancer cells.
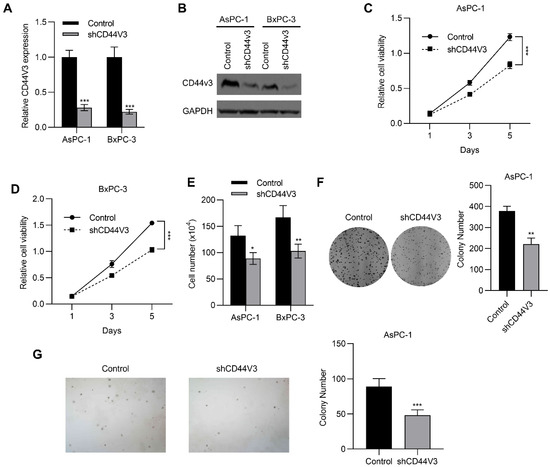
Figure 3.
Downregulation of CD44V3 inhibits pancreatic cancer cell proliferation. (A,B) The CD44V3 levels in AsPC-1 or BxPC-3 cells transfected with CD44V3 shRNA were analyzed by qPCR (A) or Western blot (B). (C,D) Cell viability of AsPC-1 (C) or BxPC-3 (D) cells transfected with CD44V3 shRNA was determined by MTT assay. (E) A cell count assay was used to determine the viability of AsPC-1 or BxPC-3 cells transfected with CD44V3 shRNA. (F) The foci formation assay was used to determine the viability of AsPC-1 cells transfected with CD44V3 shRNA. (G) The cell viability of AsPC-1 cells transfected with CD44V3 shRNA was determined by a soft agar assay. The data are presented as mean ± S.D. * p < 0.05; ** p < 0.01; *** p < 0.001.
2.4. Downregulation of CD44V3 Suppresses Pancreatic Cancer Cells Invasion and Stemness
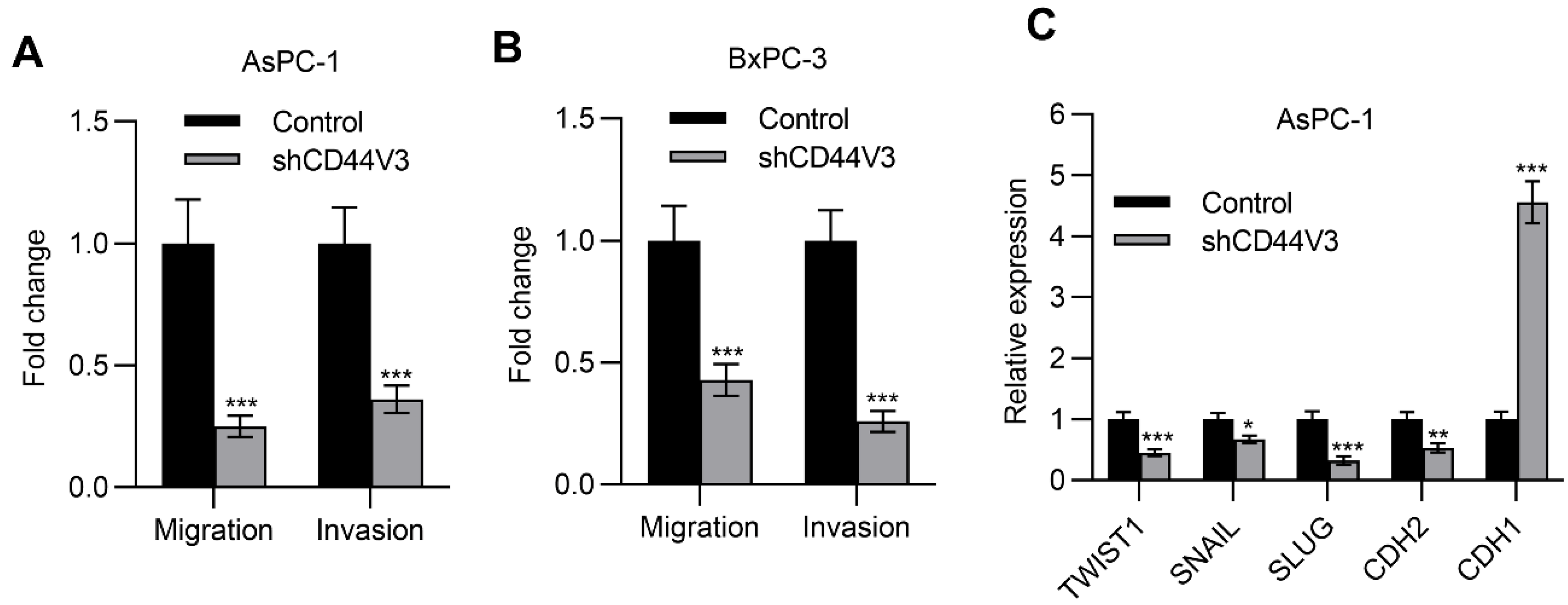
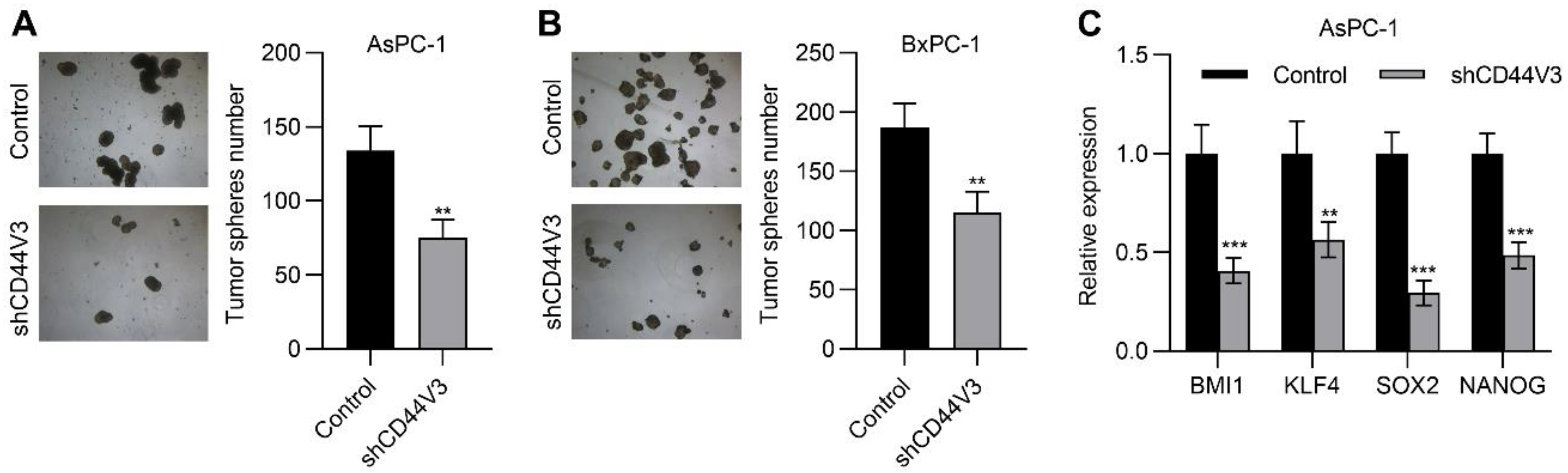
Next, we examined the pancreatic cancer cell invasion ability with or without CD44V3 by using transwell migration and invasion assays. The knockdown of CD44V3 greatly suppressed the migration and invasion capacity of AsPC-1 cells (Figure 4A). A similar inhibitory effect of cancer cell invasion was observed in BxPC-3 cells (Figure 4B). Furthermore, we measured a panel of EMT markers in control and CD44V3 knockdown AsPC-1 cells and found that mesenchymal markers (TWIST1, SNAIL, SLUG, and CDH2) were decreased in CD44V3 knockdown cells, whereas the epithelial marker CDH1 was significantly increased (Figure 4C). We further detected the cancer cell stemness ability of CD44V3 knockdown in both AsPC-1 and BxPC-3 cells through a tumor sphere formation assay. The results showed that knockdown of CD44V3 could significantly reduce the average size of AsPC-1 and BxPC-3 spheres, indicating that CD44V3 deficiency suppresses the balloon-forming ability of pancreatic cancer cells (Figure 5A,B). Stem cell markers (BMI1, KLF4, SOX2, and NANOG) in AsPC-1 cells were measured by RT-qPCR, all of which were significantly decreased in CD44V3 knockdown cells compared to the control group (Figure 5C). All this data indicated that CD44V3 was able to promote cellular migration, invasion, and stemness of pancreatic cancer cells.
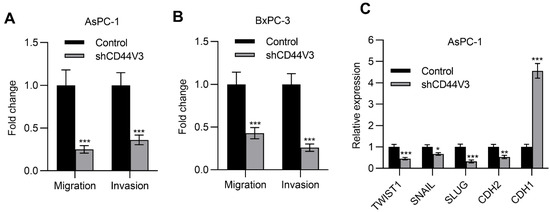
Figure 4.
Downregulation of CD44V3 inhibited pancreatic cancer cell invasion. (A) Transwell migration and invasion assay of AsPC-1 cell transfected with CD44V3 shRNA. (B) Transwell migration and invasion assay of BxPC-3 cell transfected with CD44V3 shRNA. (C) A panel of EMT markers in AsPC-1 cells transfected with CD44V3 shRNA was determined by qPCR. The data are presented as mean ± S.D. * p < 0.05; ** p < 0.01; *** p < 0.001.
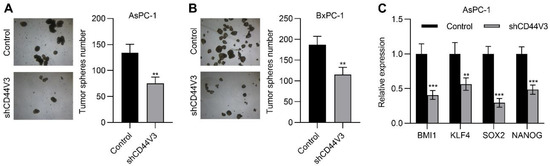
Figure 5.
Downregulation of CD44V3 inhibited pancreatic cancer cell stemness. (A) Tumor spheres formation assay of AsPC-1 cells transfected with CD44V3 shRNA. (B) Tumor sphere formation assay of BxPC-3 cells transfected with CD44V3 shRNA. (C) A panel of stem cell markers in AsPC-1 cell transfected with CD44V3 shRNA was determined by qPCR. The data are presented as mean ± S.D. ** p < 0.01; *** p < 0.001.
2.5. CD44V3 Increases the AKT Signaling Pathway
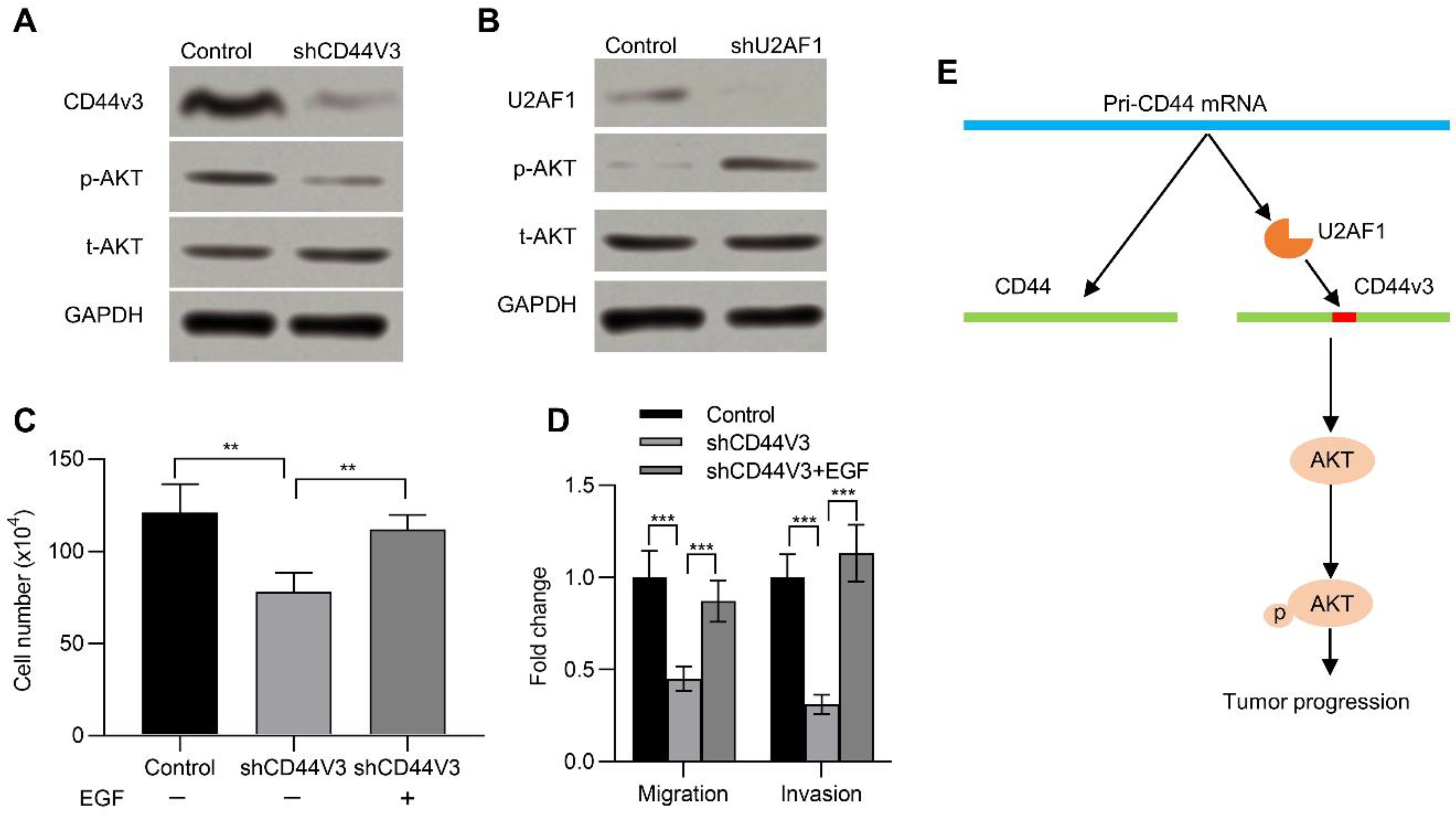
The AKT pathway is frequently activated in many kinds of human cancers and has a critical impact in neoplastic transformation [29,30]. Previous studies suggested that CD44 activated the AKT pathway [31]. We therefore wanted to determine if CD44v3 could also activate the AKT pathway. To this end, we examined its activation in pancreatic cancer cells with or without CD44V3. Knockdown of CD44V3 reduced the p-AKT level in AsPC-1 cells compared to that of control (Figure 6A). On the contrary, knockdown of U2AF1, which promoted the expression of CD44V3, could significantly increase the expression of p-AKT (Figure 6B). In addition, EGF, which activates the AKT signaling pathway and enhances tumor growth, invasion, and metastasis [32], recovered the CD44V3 knockdown-inhibited cell viability (Figure 6C). Moreover, EGF treatment could rescue the invasion and migration ability of pancreatic cancer cells, which was inhibited in CD44V3 knockdown pancreatic cancer cells (Figure 6D). Taken together, U2AF1 mediated the splicing of CD44V3. The upregulation of CD44V3 could activate the AKT signaling pathway, which eventually promoted pancreatic tumor progression (Figure 6E).
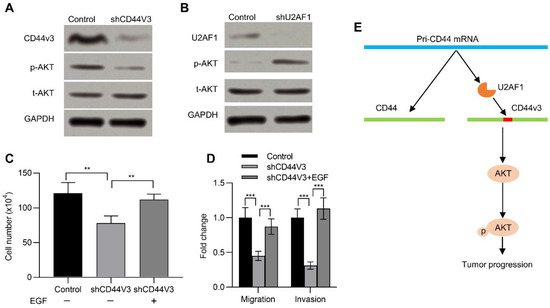
Figure 6.
CD44V3 increased the AKT signaling pathway. (A) The p-AKT and t-AKT levels in AsPC-1 cells transfected with CD44V3 shRNA were determined by Western blot. (B) The p-AKT and t-AKT levels in AsPC-1 cells transfected with U2AF1 shRNA were determined by Western blot. (C) Cell count assay of AsPC-1 cells transfected with CD44V3 shRNA and treated with or without recombinant EGF. (D) Transwell migration and invasion assay of the AsPC-1 cells transfected with CD44V3 shRNA and treated with or without recombinant EGF. (E) A graphical abstract of the role of CD44V3 in tumor progression. The data are presented as mean ± S.D. ** p < 0.01; *** p < 0.001.
3. Discussion
Cancer stem cells contribute to multiple tumor malignancies through their self-renewal and differentiation capacity, including heterogeneity, metastasis, recurrence, and therapeutic resistance. Therefore, cancer stem cells are considered important targets for cancer treatment. Cumulative evidence indicates CD44 is not only a cancer stem cell marker but also a critical regulator of cancer stemness [33,34,35]. Here, we studied the function of CD44 in pancreatic cancer progression and prognosis and found that the expression of one splicing variant, CD44V3, instead of CD44, was significantly increased in tumor tissues, and its level correlated with poor prognosis. Mechanistically, CD44V3 expression was downregulated by splicing factor U2AF1, an important component of the spliceosome, and the expression of U2AF1 was reduced in pancreatic cancer cells and tissues [27,28]. Elevated CD44V3 enhanced cell proliferation and invasion and promoted tumor migration and stemness of pancreatic cancer cells, which might be partially mediated through the AKT signaling pathway.
Both standard and variable CD44 isoforms are widely expressed in most human normal and carcinomatous tissues [19,20,36]. Alternative splicing of CD44 can generate more than 1000 isoforms theoretically, ranging from 85 to 250 kDa [20]. CD44 isoforms (CD44V) play important roles in cancer, including cancer cell stemness, metastasis, and tumor initiation. For example, in comparison with adjacent normal tissues, CD44V4-5 is highly expressed in breast cancer tissues [37]. In colorectal carcinoma, elevated CD44V6 promoted cancer progression to later stages through activating the Stat6 signaling pathway [38,39]. Knockdown of CD44V4-7 blocks tumor cell matrix crosstalk and reduces the metastatic potential of rat ASML cells [40]. In line with the above findings on CD44V, we demonstrated that CD44V3 expression level was markedly increased in pancreatic cancer tissues and was closely correlated with poor OS and RFS in 52 patients. Knockdown of CD44V3 could inhibit cell proliferation, invasion, and migration of pancreatic cancer cells. Furthermore, CD44V3 knockdown significantly suppressed the expression of multiple stemness markers, including BMI1, KLF4, SOX2, and NANOG [41], indicating that, in accordance with CD44V, CD44V3 possesses stemness-maintaining ability and then promotes pancreatic cancer migration and invasion.
The alternative splicing of CD44 is being revealed gradually [42]. Recent studies have indicated the relationship between autocrine growth factor signaling and alternative splicing regulated by epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2) [43]. Studies have also showed that ESRP1 and ESRP2 are involved in the splicing regulation of CD44V6-10 and V8-10 [43,44]. However, overexpression of ESRP1 cannot induce a differential CD44V splicing pattern in breast cancer cells [45], which suggests that ESRP1 is not the major regulator of CD44V splicing. In this study, we examined another splicing factor, U2AF1, in pancreatic cancer progression and found it was significantly downregulated in pancreatic tumor tissues. In contrast to CD44V3, U2AF1 expression level was correlated with better prognosis of pancreatic cancer patients. Moreover, knockdown of U2AF1 could significantly increase the expression level of CD44V3, which indicates that the splicing and expression of CD44V3 are regulated by U2AF1 in pancreatic cancer cells. In future studies, it would be interesting to look into methods to modulate the activation of the U2AF1 and/or CD44V3 axis for pancreatic cancer treatment.
4. Materials and Methods
4.1. Patients
Pancreatic cancer tissues and the paired adjacent non-tumor tissues were collected from 52 patients who received surgical resection of pancreatic tumors at the Hangzhou First People’s Hospital. Table S1 contains information on cancer type, histology subtype, and tumor stage. The excised tissue samples were frozen by liquid nitrogen and stored at −80 °C. For gene expression analysis, total RNA was extracted from tumor and adjacent non-tumor tissues. All the patients provided informed consent following a protocol approved by the ethics board of Hangzhou First People’s Hospital (#2019-61-01, 25 February 2019), and this study was conducted in accordance with the ethical standards formulated in Hangzhou First People’s Hospital.
4.2. Cell Culture
Two human pancreatic cancer cell lines, AsPC-1 (CRL-1682™) and BxPC-3 (CRL-1687™), were obtained from ATCC (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 μg/mL of streptomycin, 100 IU/mL of penicillin, and 2 mM L-glutamine. Cells were placed in the humidified incubator with 5% CO2 at 37 °C. Human epidermal growth factor (EGF) was purchased from Sigma-Aldrich (St. Louis, MO, USA). AsPC-1 cells with or without CD44V3 were treated by recombinant EGF (50 nmol/L) for 24 h, then transwell migration and invasion assays were performed.
4.3. Gene Knockdown in Pancreatic Cancer Cells
The expression of CD44V3 and U2AF1 in pancreatic cancer cells was inhibited through shRNA-mediated gene knockdown. Control, CD44V3, and U2AF1 shRNAs were ordered from GenePharma (Shanghai, China). The shRNA sequence for CD44V3: GATCCGAAGATAAAGACCATCCAATTCAAGAGATTGGATGGTCTTTATCTTCTTTTTTGGAAA. The shRNA sequence for U2AF1: CCGGGAAAGTGTTGTAGTTGATTGACTCGAGTCAATCAACTACAACACTTTCTTTTT. To knockdown the target genes, AsPC-1 and BxPC-3 cells were transfected with control shRNA, CD44V3 shRNA (shCD44V3), and U2AF1 shRNA (shU2AF1) constructs, respectively, by using the Lipofectamine 3000 reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. The knockdown of CD44V3 and U2AF1 was verified by RT-qPCR.
4.4. qRT-PCR
Total RNA of normal and pancreatic tumor tissues and treated pancreatic cancer cells was extracted by using the TRIzol™ Reagent (Invitrogen, Waltham, MA, USA), according to the manufacturer’s instructions. The first-strand cDNA was synthesized using the M-MLV Reverse Transcriptase (Beyotime, Shanghai, China). RT-qPCR was conducted using the CFX Opus Real-Time PCR System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The relative expression of target genes was normalized to GAPDH and calculated by using the 2-ΔΔCT method. The specific qPCR primers for CD44V3 are: forward-TTAGGGTGCTACCAGTAC; reverse-GTCTCTGGTGCTGGAGATAA. The qPCR primers for CD44 are: forward-CCAGTCATAGTATAACGCTTC; reverse-TCTTTATCTTCTTCCAAGCCT; U2AF1: forward-TTTCAAAATTGGAGCATGTCG; reverse- TGCATCTCCACATCGCTCA; and GAPDH: forward-TGACTTCAACAGCGACACC; reverse-TTGCTGTAGCCAAATTCGTT.
4.5. Western Blot
Control and CD44V3 or U2AF1 knockdown AsPC-1 cells were lysed using the lysis buffer (Beyotime) with a protease inhibitor cocktail (Promega, Madison, WI, USA). Target protein expression was determined by Western blot as previously described [31]. The primary antibodies were ordered from Cell Signaling (Danvers, MA, USA), and the detailed information was listed below: phospho-AKT (p-AKT, Ser473) (#9271, 1:2000 dilution), total-AKT (#4691, 1:2000 dilution), and GAPDH (#5174, 1:3000 dilution).
4.6. Cell Viability
The cell viability of AsPC-1 and BxPC-3 cells with or without CD44V3 was determined by cell count assay and MTT assay. In brief, control and CD44V3 knockdown cells were placed into a 6-well plate at a density of 1 × 105 cells per well, and the total cell numbers of each well were counted three times on day 3 using the Beckman Coulter Vi-Cell (Brea, CA, USA). For the MTT assay, control and CD44V3 knockdown cells were seeded in a 96-well plate at a density of 1 × 103 cells per well. Cell viability on days 1, 3, and 5 was measured using MTT cell proliferation and cytotoxicity assay kits (Beyotime, Shanghai, China).
4.7. Cell Colony Formation and Soft Agar Colony Formation Assays
Control and CD44V3 knockdown AsPC-1 cells were seeded into 35 mm plates (1 × 103 cells per well) and cultured with fresh media every 3 days. When they reached appropriate confluency, cells were stained with crystal violet (0.2%)/formaline (10%), and the signal of crystal violet was quantified at 700 nm using the GloMax® Discover Microplate Reader (Promega, Madison, WI, USA). The anchorage-independent growth ability of control and CD44V3 knockdown AsPC-1 cells was determined by a soft agar colony formation assay as previously described [46].
4.8. Migration and Invasion Assay
The invasion and migration of control and CD44V3 knockdown AsPC-1 cells were measured by using a 6.5 mm transwell chamber (8 μm pore, BD, San Jose, CA, USA). Briefly, serum-free medium was used to suspend cells (3 × 104 cells per well), which were subsequently cultured in the upper chamber. Complete culture medium was then added into the lower chamber. After 24 h of culture, the migrated/invaded cells (lower chamber) were fixed with 4% paraformaldehyde, and then stained with crystal violet (0.1 mg/mL) and counted using a Nikon microscope. Three visual fields were counted randomly for each well, and the average was presented.
4.9. Tumor Sphere Formation Assay
Control and CD44V3 knockdown AsPC-1 or BxPC-3 cells were suspended at a density of 5 × 104 cells/mL in complete culture medium and seeded into 6-well plates (2.5 mL per plate) coated with 1.2% polyhema. Fresh culture medium was changed every 3 days, and cells were passaged every week. As previously described, tumor spheres were measured and counted using Zeiss Axiovision software (Carl Zeiss, Jena, Germany) [47].
4.10. Statistical Analysis
The correlation between pancreatic cancer progression and gene expression of CD44 or U2AF1 was analyzed by using the Kaplan–Meier Plotter. Statistical analyses were performed by using the GraphPad Prism. Two- or one-way analysis of variance (ANOVA) and Student’s t-test were used to analyze the differences between groups. The data were presented as mean ± standard deviation (SD), * p < 0.05, ** p < 0.01, and *** p < 0.001 compared to the control group.
5. Conclusions
This study highlights CD44V3 as an important cancer-promoting factor in pancreatic cancer progression, supporting its role as a potential therapeutic target for pancreatic cancer intervention.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/ijms232012061/s1.
Author Contributions
Conceptualization, H.Z. and C.J.; validation, H.Z. and C.J.; investigation, C.J.; resources, C.J.; data curation, H.Z., W.Z., Y.W., J.L. and K.G.; writing—original draft preparation, H.Z., W.Z., Y.W., J.L., K.G. and C.J.; writing—review and editing, H.Z., W.Z., Y.W., J.L., K.G. and C.J.; supervision, C.J.; project administration, C.J.; funding acquisition, C.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (LY21H160016) and the Medicine and Health Science and Technology Plan Projects of Zhejiang Province (2021KY231).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the ethics board of Hangzhou First People’s Hospital (#2019-61-01, 25 February 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data could be obtained upon reasonable request to the corresponding author.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.P.; Brune, K.A.; Petersen, G.M.; Goggins, M.; Tersmette, A.C.; Offerhaus, G.J.; Griffin, C.; Cameron, J.L.; Yeo, C.J.; Kern, S.; et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004, 64, 2634–2638. [Google Scholar] [CrossRef] [PubMed]
- Ojajarvi, I.A.; Partanen, T.J.; Ahlbom, A.; Boffetta, P.; Hakulinen, T.; Jourenkova, N.; Kauppinen, T.P.; Kogevinas, M.; Porta, M.; Vainio, H.U.; et al. Occupational exposures and pancreatic cancer: A meta-analysis. Occup. Environ. Med. 2000, 57, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Amundadottir, L.; Kraft, P.; Stolzenberg-Solomon, R.Z.; Fuchs, C.S.; Petersen, G.M.; Arslan, A.A.; Bueno-de-Mesquita, H.B.; Gross, M.; Helzlsouer, K.; Jacobs, E.J.; et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009, 41, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef]
- Ilic, M.; Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Schizas, D.; Charalampakis, N.; Kole, C.; Economopoulou, P.; Koustas, E.; Gkotsis, E.; Ziogas, D.; Psyrri, A.; Karamouzis, M.V. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat. Rev. 2020, 86, 102016. [Google Scholar] [CrossRef]
- Torphy, R.J.; Fujiwara, Y.; Schulick, R.D. Pancreatic cancer treatment: Better, but a long way to go. Surg. Today 2020, 50, 1117–1125. [Google Scholar] [CrossRef]
- Gillen, S.; Schuster, T.; Meyer Zum Buschenfelde, C.; Friess, H.; Kleeff, J. Preoperative/neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010, 7, e1000267. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yin, S.; Brobbey, C.; Gan, W. Ubiquitination in cancer stem cell: Roles and targeted cancer therapy. STEMedicine 2020, 1, e37. [Google Scholar] [CrossRef]
- Jaggupilli, A.; Elkord, E. Significance of CD44 and CD24 as cancer stem cell markers: An enduring ambiguity. Clin. Dev. Immunol. 2012, 2012, 708036. [Google Scholar] [CrossRef] [PubMed]
- Pietras, A.; Katz, A.M.; Ekstrom, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef]
- Choi, E.S.; Kim, H.; Kim, H.P.; Choi, Y.; Goh, S.H. CD44v8-10 as a potential theranostic biomarker for targeting disseminated cancer cells in advanced gastric cancer. Sci. Rep. 2017, 7, 4930. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Zhang, H.; Brown, R.L.; Wei, Y.; Zhao, P.; Liu, S.; Liu, X.; Deng, Y.; Hu, X.; Zhang, J.; Gao, X.D.; et al. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019, 33, 166–179. [Google Scholar] [CrossRef]
- Grimme, H.U.; Termeer, C.C.; Bennett, K.L.; Weiss, J.M.; Schopf, E.; Aruffo, A.; Simon, J.C. Colocalization of basic fibroblast growth factor and CD44 isoforms containing the variably spliced exon v3 (CD44v3) in normal skin and in epidermal skin cancers. Br. J. Dermatol. 1999, 141, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Anand, V.; Khandelwal, M.; Appunni, S.; Gupta, N.; Seth, A.; Singh, P.; Mathur, S.; Sharma, A. CD44 splice variant (CD44v3) promotes progression of urothelial carcinoma of bladder through Akt/ERK/STAT3 pathways: Novel therapeutic approach. J. Cancer Res. Clin. Oncol. 2019, 145, 2649–2661. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.D.; Kim, H.S.; Lee, J.; Choi, J.K.; Han, E.; Jeong, J.E.; Cho, Y.S. ESRP1-Induced CD44 v3 Is Important for Controlling Pluripotency in Human Pluripotent Stem Cells. Stem Cells 2018, 36, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Yaffe, M.B.; Sharp, P.A. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006, 20, 1715–1720. [Google Scholar] [CrossRef]
- Weg-Remers, S.; Ponta, H.; Herrlich, P.; Konig, H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001, 20, 4194–4203. [Google Scholar] [CrossRef]
- Graubert, T.A.; Shen, D.; Ding, L.; Okeyo-Owuor, T.; Lunn, C.L.; Shao, J.; Krysiak, K.; Harris, C.C.; Koboldt, D.C.; Larson, D.E.; et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2011, 44, 53–57. [Google Scholar] [CrossRef]
- Kim, S.P.; Srivatsan, S.N.; Chavez, M.; Shirai, C.L.; White, B.S.; Ahmed, T.; Alberti, M.O.; Shao, J.; Nunley, R.; White, L.S.; et al. Mutant U2AF1-induced alternative splicing of H2afy (macroH2A1) regulates B-lymphopoiesis in mice. Cell Rep. 2021, 36, 109626. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Roy, H.K.; Olusola, B.F.; Clemens, D.L.; Karolski, W.J.; Ratashak, A.; Lynch, H.T.; Smyrk, T.C. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis 2002, 23, 201–205. [Google Scholar] [CrossRef]
- Lin, Y.H.; Yang-Yen, H.F. The osteopontin-CD44 survival signal involves activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. J. Biol. Chem. 2001, 276, 46024–46030. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Bjerkvig, R.; Tysnes, B.B.; Aboody, K.S.; Najbauer, J.; Terzis, A.J. Opinion: The origin of the cancer stem cell: Current controversies and new insights. Nat. Rev. Cancer 2005, 5, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mayea, Y.; Mir, C.; Masson, F.; Paciucci, R.; ME, L.L. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Semin. Cancer Biol. 2020, 60, 166–180. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells - what challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497–512. [Google Scholar] [CrossRef]
- Fox, S.B.; Fawcett, J.; Jackson, D.G.; Collins, I.; Gatter, K.C.; Harris, A.L.; Gearing, A.; Simmons, D.L. Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res. 1994, 54, 4539–4546. [Google Scholar]
- Best, A.; Dagliesh, C.; Ehrmann, I.; Kheirollahi-Kouhestani, M.; Tyson-Capper, A.; Elliott, D.J. Expression of Tra2 beta in Cancer Cells as a Potential Contributory Factor to Neoplasia and Metastasis. Int. J. Cell Biol. 2013, 2013, 843781. [Google Scholar] [CrossRef]
- Wielenga, V.J.; Heider, K.H.; Offerhaus, G.J.; Adolf, G.R.; van den Berg, F.M.; Ponta, H.; Herrlich, P.; Pals, S.T. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993, 53, 4754–4756. [Google Scholar]
- Li, B.H.; Xu, S.B.; Li, F.; Zou, X.G.; Saimaiti, A.; Simayi, D.; Wang, Y.H.; Zhang, Y.; Yuan, J.; Zhang, W.J. Stat6 activity-related Th2 cytokine profile and tumor growth advantage of human colorectal cancer cells in vitro and in vivo. Cell. Signal. 2012, 24, 718–725. [Google Scholar] [CrossRef]
- Klingbeil, P.; Marhaba, R.; Jung, T.; Kirmse, R.; Ludwig, T.; Zoller, M. CD44 variant isoforms promote metastasis formation by a tumor cell-matrix cross-talk that supports adhesion and apoptosis resistance. Mol. Cancer Res. 2009, 7, 168–179. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Y.; Zhang, X. Stemness-Related Markers in Cancer. Cancer Transl. Med. 2017, 3, 87–95. [Google Scholar] [PubMed]
- Prochazka, L.; Tesarik, R.; Turanek, J. Regulation of alternative splicing of CD44 in cancer. Cell. Signal. 2014, 26, 2234–2239. [Google Scholar] [CrossRef]
- Warzecha, C.C.; Sato, T.K.; Nabet, B.; Hogenesch, J.B.; Carstens, R.P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell 2009, 33, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, K.; Sakamoto, K.; Koinuma, D.; Semba, K.; Inoue, A.; Inoue, S.; Fujii, H.; Yamaguchi, A.; Miyazawa, K.; Miyazono, K.; et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene 2012, 31, 3190–3201. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, e51998. [Google Scholar] [CrossRef]
- Grimshaw, M.J.; Cooper, L.; Papazisis, K.; Coleman, J.A.; Bohnenkamp, H.R.; Chiapero-Stanke, L.; Taylor-Papadimitriou, J.; Burchell, J.M. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008, 10, R52. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).